Abstract
Regardless of the route of transmission, it is generally accepted that the human immunodeficiency virus type 1 (HIV-1) quasispecies transmitted from an infected individual to an uninfected individual is genetically homogeneous. This finding and the observation that HIV-1 genotypes in recipients are minor variants in the donors suggest strongly that selection for specific variants occurs. However, most analyses have been limited to the V3 region of env. In addition, the exact time at which most new infections occurred was not known, making it almost impossible to analyze virus populations present in donor-recipient pairs at the time of HIV-1 transmission. To circumvent this problem, three chimpanzees were inoculated with a genetically defined stock of cell-free HIV-1/JC499 by one of three routes: intravenously or via the cervical or penile mucosa. PCR products of the C2-to-V5 region of env were amplified from both proviral DNA and virion RNA in blood samples collected soon after infection and were screened by heteroduplex analysis (HDA). Those PCR products with distinct HDA banding patterns were cloned and sequenced. In all three animals, transmitted variants encoded one of two V3-loop populations identified in the inoculum, indicating relative homogeneity in this region. However, different virus populations, defined by combinations of specific V4 and V5 sequences, were found when variants in the animal inoculated intravenously (at least 13 V4-plus-V5 combinations) were compared with those in the two animals inoculated by the mucosal routes (limited to only four V4-plus-V5 combinations). The only V4-plus-V5 population in variants found in all three chimpanzees was the major population in the inoculum, which contained viruses with more than 30 different V4-plus-V5 combinations. That the majority of the V4-plus-V5 genotypes in variants transmitted to all three animals were minor populations in the inoculum indicated that selective transmission defined by the V4-plus-V5 regions had occurred but that distinct populations were transmitted by parenteral versus mucosal routes. These results indicate that the putative homogeneity of HIV-1 variants in a newly infected individual might be an artifact of the region of the env gene evaluated and that regions other than V3 might play a major role in selective transmission.
One hallmark of the human immunodeficiency virus type 1 (HIV-1) is its extreme genetic diversity. Not only are HIV strains that are isolated from each person distinct but also each individual is infected with a heterogeneous population of related viruses, termed a quasispecies, that continually evolve (18). Exceptions to the excessive diversity of HIV-1 have been described in persons newly infected with the virus. Although the HIV-1 quasispecies in a person transmitting the virus is genetically heterogeneous, early after infection many recipients, whether infected from male-to-male, male-to-female, or mother-to-child transmission, have a relatively homogeneous HIV-1 population (1, 24, 43, 54, 57–59). This finding has led to the speculation that either specific variants are selectively transmitted or the most prevalent member(s) of the quasispecies has the highest probability of establishing infection and being amplified (58). However, many examples of transmission of multiple variants have been reported (3, 8, 26, 38, 51). Regardless of whether the transmitted viruses are genetically homogeneous or heterogeneous, the nucleotide sequences of HIV-1 in recipients early after infection often are derived from minor species in the donor, indicating that some selective transmission of specific variants must occur (1, 16, 33, 43, 55, 58, 59).
In most transmission studies to date conclusions were based on sequences in the V3 loop, or to a lesser extent, on the region encompassed by V1 to V3 (V1-V3) or C2 to V5 (C2-V5) (23, 27, 38, 39, 55, 58, 59). In addition, in some, but not all, instances the relative degree of homogeneity in specific regions of gp120env has been reported not to extend to the gp41env, p17gag, and nef genes (57, 58). However, even though the extent of diversity in the latter genes was greater than that in the V3 region, in some studies the diversity of these other genes was still less than 1% (33, 57, 58). In contrast, the intrapatient diversity of V3 sequences defined as relatively homogeneous has been as high as 5% (55). Therefore, whether the transmitted viruses are defined as homogeneous or heterogeneous is dependent not only on which region of the HIV-1 genome is analyzed but also on the definition of homogeneous relative to percent diversity. However, that a specific region is homogeneous in the context of a diverse background does imply that there is selection for and/or selective amplification of viruses encoding particular amino acid sequences. An additional consideration is that unless multiple recipients of the same HIV-1 inoculum are evaluated at the same time early after infection, it is impossible to identify with certainty specific regions of the genome that might select for or facilitate infection after, for example, mucosal versus parenteral exposure.
The use of an animal model is one way to circumvent problems associated with identifying events that occur during transmission of an infectious agent or as disease progresses. Two major advantages of using an animal model are (i) that the exact time of infection is known so that blood samples can be obtained at specific times and (ii) that the genetic make-up of variants within the inoculum at the time of infection can be determined. The animal model most closely related to HIV-1 infection of humans is HIV-1 infection of chimpanzees (12, 34). Most chimpanzees infected with HIV-1 appear to be healthy (22, 25); however, some animals infected for more than 10 years or for shorter times with viruses derived from a chimpanzee that died of AIDS have developed signs of disease, as evidenced by loss of CD4+ lymphocytes and high viral burdens (5, 35, 37). To determine whether selective transmission of specific HIV-1 genotypes occurs and whether selection is a function of route of infection, we inoculated three chimpanzees either intravenously or by exposure of the cervicovaginal or penile mucosa to HIV-1/JC499. This strain of HIV-1 is a complex quasispecies that is genetically well characterized, is cytopathic for chimpanzee CD4+ lymphocytes, and causes loss of CD4+ lymphocytes after inoculation of chimpanzees (5, 52). Our results show that selection for specific variants occurs during mucosal, and to a lesser extent, parenteral transmission of HIV-1/JC499 and that this selection process appears to be mediated by combinations of specific V4 and V5 genetically defined populations of env.
MATERIALS AND METHODS
Virus stocks.
The HIV-1/JC499 virus stock was generated by coculturing phytohemagglutinin-stimulated healthy human peripheral blood mononuclear cells (PBMC) with PBMC that were obtained from chimpanzee C-499 approximately 22 months before its death due to AIDS (36). The culture supernatant was collected on day 15, filtered, aliquoted, and stored in liquid nitrogen vapor. This virus stock (HIV-1/JC499-P0) was titered in healthy human PBMC and then used to inoculate chimpanzees. Because of the limited amount of HIV-1/JC499-P0, an aliquot of HIV-1/JC499-P0 was used to infect phytohemagglutinin-stimulated healthy human PBMC, and the resulting passage-1 stock (HIV-1/JC499-P1) was used for some of the analyses described herein.
Animals.
One male and two female chimpanzees were housed in biosafety level 2 facilities at either the Laboratory for Experimental Medicine and Surgery in Primates, New York University, or the Coulston Foundation (Alamogordo, New Mexico) in accordance with the Animal Welfare Act, institutional guidelines, and standard practices for the humane care and use of chimpanzees in biomedical research. C-384 was inoculated intravenously with approximately 500 50% tissue culture infective doses (TCID50) of HIV-1/JC499-P0; C-166 was exposed without trauma via the cervical os to about 600 TCID50, as described previously (5, 16). The male chimpanzee, C-1323, was inoculated initially by the rectal route with 2,000 TCID50 of HIV-1/JC499-P0. When no evidence was obtained through 8 months of follow-up that HIV-1/JC499-P0 actually established infection (13), C-1323 was reinoculated via the penile route with the same dose of HIV-1/JC499-P0 using small-diameter flexible tubing attached to a syringe. Before virus inoculation or collection of blood samples, the animals were anesthetized with an intramuscular inoculation of ketamine.
PCR and quantification of proviral DNA and viral RNA.
For amplification and quantification of proviruses, genomic DNA was extracted from PBMC and single-cell suspensions of lymph node tissues using QIAamp DNA blood kits (Qiagen, Chatsworth, Calif.). To amplify virion RNA, EDTA-treated plasma was centrifuged at low speed to remove debris, and then 140 to 170 μl of clarified plasma was loaded on a column from a QIAamp viral RNA kit (Qiagen) and 50 μl of RNA was eluted with preheated (80°C) RNase-free water. The reverse transcription (RT) reaction was performed with 1 to 10 μl of RNA using a first-strand cDNA synthesis kit (Clontech Laboratories Inc., Palo Alto, Calif.) and was terminated by heating the reaction mixtures at 94°C for 5 min. Proviral DNA or cDNA was amplified as a 660-bp fragment in the C2-V5 region of env by PCR using primers C/H and D/F (14). Determination of proviral copy numbers was done using a nested PCR-based limiting dilution assay (PLDA), employing a two-pass strategy and the QUALITY statistical computer program (40, 53). HIV-1 virion RNA in EDTA-treated plasma was measured with the Amplicor HIV-1 Monitor test kit (Roche Diagnostics Corp., Indianapolis, Ind.) according to the manufacturer’s instructions.
Cloning and sequence analyses.
PCR products were recovered from agarose gels with a QIAquick gel extraction kit (Qiagen) and cloned into the PCR2.1 vector using a TA cloning kit (Invitrogen, San Diego, Calif.). Selected clones were sequenced using the D and B3A primers described previously (52), either on an ABI Prism 377 automated sequencer (Applied Biosystems, Inc., Foster City, Calif.) or using the Sequenase 2 protocol (U.S. Biochemicals, Cleveland, Ohio) and a Sequi-Gen sequencing cell (Bio-Rad Laboratories, Hercules, Calif.). There was an approximately 100-bp overlap between the sequences determined with the D and B3A primers. Consensus sequences were generated with the Clustal W algorithm and AssemblyLIGN (50% base designation threshold) programs in MacVector 6.5.3. Consensus amino acid sequence alignments were generated with the protein scoring matrix prefilter, BLOSUM30 scoring matrix, and PAM250 scoring matrix in MacVector 6.5.3, with minor modifications in the V4 and V5 Env regions. Pairwise nucleotide distances were estimated by using the Kimura two-parameter model with a transition/transversion ratio of 2. The neighbor-joining method was used to analyze sequence relationships and to construct phylogenetic trees, which were evaluated statistically by 100 bootstrap replicates (PHYLIP, version 3.572). Gaps in sequences were excluded from the analysis.
HDA and QHDA.
HDA was used not only to assist in evaluating the genetic populations in the HIV-1/JC499-P0 stock but also to confirm the results obtained by quantitative HDA (QHDA) for proviruses in chimpanzee PBMC and cell-free virion RNA in plasma. The probe, JC499 clone 6 (JC6), used in HDA and QHDA was generated from uncultured PBMC obtained from chimpanzee C-499 approximately 22 months before its death due to AIDS (52). The JC6 variant was a minor proviral population in uncultured PBMC and was not found in cultured PBMC. HDA was performed as described in an earlier study (52). For QHDA, 90 pmol of primer D was used in end-labeling reactions (37°C for 30 min) with 60 μCi of [γ-33P]ATP (ICN Biomedical, Inc., Costa Mesa, Calif.) and 4.5 U of T4 polynucleotide kinase (Amersham Life Science, Inc., Cleveland, Ohio) in a total volume of 15 μl. T4 polynucleotide kinase was then inactivated at 65°C for 10 min. A 33P-labeled single-stranded DNA (plus strand) was generated from the PCR amplification step with 2 μl of 5′-labeled primer D, 12 pmol of primer F, and 5 ng of the JC6 plasmid. The reaction was carried out in a PTC-100 programmable thermal cycler (MJ Research) and consisted of 22 cycles at 94°C for 45 s, 60°C for 45 s, and 72°C for 45 s, with a final extension at 72°C for 10 min. The [33P]ATP-labeled JC6 PCR products were diluted 1:8 in TE75 buffer (10 mM Tris-HCl [pH 7.5], 4 mM EDTA) and a 5-μl aliquot was mixed with 5 μl of each PCR-amplified sample to be tested. DNA molecules were denatured at 98°C for 3 min and then reannealed by cooling slowly to room temperature. The resultant hybrid molecules were electrophoresed in a 5% polyacrylamide gel containing 2.7 M urea in 1× TTE buffer (89 mM Tris base, 29 mM taurine, 0.54 mM EDTA) in a 21- by 40-cm Sequi-Gen sequencing cell. Gels were run overnight at 4 to 5 mA with constant current. Homo- and heteroduplexes were visualized by drying the gels onto 3-mm-thick paper (Whatman, Clifton, N.J.) and exposing them to X-ray film (Eastman Kodak Co., Rochester, N.Y.) overnight at −80°C with an LE intensifying screen (Eastman Kodak Co.). The densities and the proportion of each heteroduplex band in a lane were determined by imaging the film on an IS-1000 digital imaging system (Alpha Innotech Corp., San Leandro, Calif.) and using two different densitometry tools incorporated into the system.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in GenBank; the accession numbers are AF433426 to AF433635. Accession numbers for the HIV-1/JC499-P0 stock that were published previously are V56868, V56869, V56871, V56873 to V56875, V56878 to V56881, AF027776, and AF027779 to AF027784 (52).
RESULTS
Genetic characterization of HIV-1/JC499 quasispecies.
We previously reported an analysis of 52 diverse sequences of the HIV-1/JC499 C2-V5 env region using proviral DNA obtained from the original coculture of chimpanzee C-499’s PBMC with healthy human PBMC (52). This cell-free culture medium was the P0 virus stock used to inoculate the three chimpanzees. Due to the limited amount of HIV-1/JC499-P0, a first-passage stock, HIV-1/JC499-P1, was generated and included in the genetic analysis of the inoculum described here. Using clone JC6 as a probe and the same strategy used in our previous study (52), 273 clones of the C2-V5 env region in virion RNA isolated from the HIV-1/JC499-P1 stock were generated from the RT-PCR products, subjected to HDA, and grouped by HDA banding patterns. After sequencing from 1 to 23 clones in each group, 86 different C2-V5 env sequences were identified. Comparison of sequences from HIV-1/JC499-P1 and those obtained from the original coculture revealed that HIV-1/JC499-P1 contained a subset of variants in the HIV-1/JC499-P0 quasispecies, indicating that some genotypes were lost when HIV-1/JC499-P0 was amplified to generate HIV-1/JC499-P1. Therefore, neither viral sequences from HIV-1/JC499-P1 nor proviral sequences from C-499’s cultured PBMC alone represent all sequences in HIV-1/JC499-P0. To characterize the HIV-1/JC499-P0 inoculum more fully, all sequences obtained during the present and previous studies were combined. A total of 325 clones were prescreened by HDA, and of these, 104 clones (32.0%) were sequenced.
The C2-V5 Env fragment contains three hypervariable regions, V3, V4, and V5. Among the 325 clones, two basic V3 consensus amino acid sequences, designated A and B, were found (Table 1). These two consensus sequences contained 36 amino acids and had the same GYGR motif at the tip of the V3 loop and the same net charge. The major difference was four amino acids at the C terminus. In contrast, the amino acid sequences found in both V4 and V5 were more diverse. In V4, six distinct sequence types, designated 1, 2, 3, 5, 7, and 8 and consisting of 27 to 34 amino acids, were detected (Table 1). All of the V4 types had either three or four presumed N-glycosylation motifs (NXS/T). In V5 there were 11 major sequence types, designated a, b, c, d, f, i, j, k, l, n, and p, which consisted of 7 to 12 amino acids (Table 1). One N-glycosylation motif was located in all V5 sequence types, except for V5-a and V5-b, which contained no such motif.
TABLE 1.
Amino acid sequences of the V3, V4, and V5 populations detected in the HIV-1/JC499 inoculuma
| Population | Amino acid sequence |
|---|---|
| V3 (36 amino acids) | CTRPNYNETKRIRIHRGYGRSFVTVRKL?D?KQAHC |
| A | ............................G.R..... |
| B | ............................VNM.R... |
| V4 (27 to 34 amino acids) | CNSTKLFNSTWN--?T?ES---????E-N--ITLPC |
| 1 | .T..........--T.S..---D-TK.-.--..... |
| 2 | ............--D.T..---SGNG.-.--..... |
| 3 | .T..........--A.S..---DYPK.-.--..... |
| 5 | ............--G.V..---YNYG.-.--..... |
| 7 | ............VND.G.P----RYT.G.ST...Q. |
| 8 | ............VNS.WYATRESNYT.E.--..... |
| V5 (7 to 12 amino acids) | GP---????NET |
| a | -----DKTEH.. |
| b | .----HNTKK.. |
| c | ..---SGNK... |
| d | ..---DNNK... |
| f | .K---DNTN... |
| i | .K---ENTH... |
| j | .H---DKDK... |
| k | ..-SGNNIN... |
| l | ..-SGHNTN... |
| n | .H-DTSKNE... |
| p | ..SDKNNYH... |
Numbers and letters identifying specific V4 and V5 populations, respectively, are not sequential because clones found only in C-499’s PBMC, but not in cultured PBMC (P0) or HIV-1/JC499-P1, were omitted. Dots denote identity with the consensus sequences, dashes represent deletions or gaps to optimize alignments, and question marks are sites at which <50% of the sequences have the same amino acid.
To facilitate analysis, each genetic population in HIV-1/JC499-P0 was defined as the combination of one specific V4 sequence and one specific V5 sequence (e.g., 1+a, 1+b, 2+a) without regard to V3 and the sequences in the conserved regions between V3, V4, and V5. Using this classification method, 20 populations from HIV-1/JC499-P1 virion RNA and 16 proviral populations from C-499’s cultured PBMC were identified. Among these, 13 populations were observed only in the HIV-1/JC499-P1 stock, 9 populations were found only in cultured PBMC, and 7 populations were common to both sources, indicating that the HIV-1/JC499-P0 inoculum contained at least 29 populations (Table 2). However, only six V4-plus-V5 populations (1+d, 2+c, 2+l, 3+d, 5+a, and 7+c) were found in approximately 85% of the clones. Both of the V3 sequences were found in individual clones in these six V4-plus-V5 populations. Because of the differences in the number of amino acids in V4 and V5, overall C2-V5 fragment lengths varied from 654 to 681 residues. Genetic distances within each population consisting of two or more sequences were calculated. Even though these calculations included the entire C2-V5 region, the distance only ranged from 0.49 to 5.64%, indicating that our classification system for the HIV-1/JC499-P0 variants grouped closely related members of the quasispecies. Amino acid sequence alignment of the 29 distinct populations with the consensus sequence of all populations also confirmed that HIV-1/JC499-P0 could be characterized on the basis of different combinations of the V4 and V5 sequences (Fig. 1).
TABLE 2.
Virus populations corresponding to unique V4-plus-V5 sequences in the HIV-1/JC499 inoculum
| V4-plus-V5 population | V3 populationa | Sourceb | % of HDA clonesc | % Genetic distance (no.)d |
|---|---|---|---|---|
| 1 + c | B | P1 | 0.92 | 1.35 (2) |
| 1 + d | B > A | C-PBMC + P1 | 6.41 | 0.50–5.05 (13) |
| 1 + l | B | C-PBMC + P1 | 0.62 | 1.68 (2) |
| 1 + n | B | P1 | 0.31 | |
| 2 + a | A | C-PBMC | 0.62 | |
| 2 + c | A > B | C-PBMC + P1 | 3.69 | 0.67–3.09 (6) |
| 2 + d | B | P1 | 0.31 | |
| 2 + f | A | C-PBMC | 0.31 | |
| 2 + i | A | C-PBMC | 0.31 | |
| 2 + k | B | C-PBMC | 0.31 | |
| 2 + l | A > B | C-PBMC + P1 | 3.69 | 1.33–4.30 (5) |
| 2 + n | A | C-PBMC + P1 | 0.62 | 4.96 (2) |
| 2 + p | A | C-PBMC | 0.31 | |
| 3 + a | B > A | P1 | 1.54 | 1.88–3.46 (3) |
| 3 + c | A = B | P1 | 1.54 | 3.26–4.69 (4) |
| 3 + d | A > B | C-PBMC + P1 | 8.00 | 0.67–4.67 (10) |
| 3 + j | B | P1 | 0.31 | |
| 3 + l | A = B | C-PBMC | 1.54 | 1.84 (2) |
| 3 + n | A | P1 | 0.62 | |
| 3 + p | A | P1 | 0.31 | |
| 5 + a | B > A | P1 | 4.92 | 0.68–5.64 (12) |
| 5 + c | B | P1 | 0.62 | 2.89 (2) |
| 7 + c | A > B | C-PBMC + P1 | 59.1 | 0.49–4.95 (25) |
| 7 + d | A | C-PBMC | 0.31 | |
| 7 + l | A = B | P1 | 1.23 | 2.98 (2) |
| 7 + p | A | C-PBMC | 0.31 | |
| 8 + a | B | C-PBMC | 0.31 | |
| 8 + b | A | P1 | 0.31 | |
| 8 + c | B | P1 | 0.62 | 3.34 (2) |
B > A, A > B, and A = B indicate that more B, more A, or equal numbers of the A and B V3 genotypes, respectively, were observed among sequenced clones in a particular V4-plus-V5 population.
Abbreviations: C-PBMC, C-499’s cultured PBMC only; P1, HIV-1/JC499-P1 stock only; C-PBMC + P1, both cultured PBMC and HIV-1/JC499-P1.
Percentage of HDA clones for each population among 325 clones generated from P0 and P1 stocks.
Genetic distances for populations containing at least two sequenced clones. Distances reflect diversity among all the ∼660-bp C2-V5 fragments of all clones with the same V4-plus-V5 combination. Numbers in parentheses are the numbers of clones sequenced.
FIG. 1.
Amino acid alignment of 29 genetic populations identified in the HIV-1/JC499-P0 stock. The consensus sequence at the top of the alignment was generated from the 104 sequences of all populations defined by specific V4-plus-V5 combinations. For seven populations for which there were multiple clones, consensus sequences were used in the alignment and are indicated by the number of clones shown in parentheses. Consensus sequences with open circles to the left were generated from sequences in both the HIV-1/JC499-P1 stock and cultured PBMC. Individual sequences, for which clone numbers are given, are shown for the remaining populations in the alignment. Solid circles to the left of population designations identify previously reported sequences (52). Dots denote identity with the consensus sequence, while dashes represent deletions or gaps introduced to optimize alignments. Question marks in the consensus sequence indicate sites at which fewer than 50% of the sequences share the same amino acid residue. Asterisks denote in-frame stop codons. The hypervariable V3, V4, and V5 regions of the HIV-1 envelope are underlined.
Semiquantitative detection of individual sequences by QHDA.
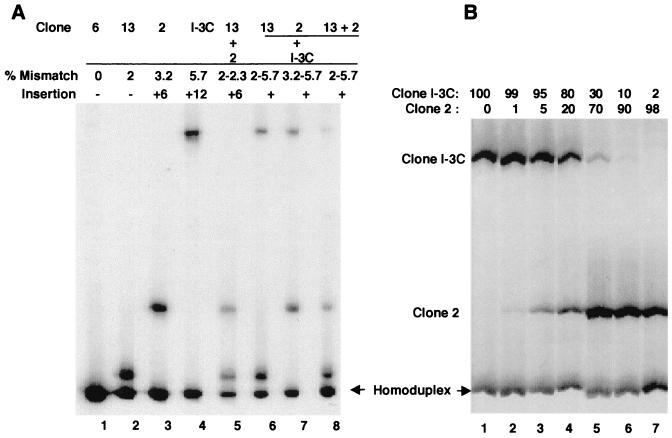
Before using QHDA to identify and quantify specific genotypes in the HIV-1/JC499-P0 inoculum that were transmitted to the three chimpanzees, the sensitivity of the technique was evaluated using known TA clones. A [33P]ATP-labeled single-stranded probe, generated by amplifying the JC6 plasmid with labeled and unlabeled primers D and F, respectively, was mixed with individual clones or mixtures of clones. HIV-1/JC499 clone 13 (JC13), which contains only 2% mismatched nucleotides compared to the JC6 probe, was detected easily by this assay (Fig. 2A, lane 2). In addition, insertions of 6 and 12 nucleotides (relative to the probe) in molecular clones JC2 and JCI-3C, respectively, noticeably reduced heteroduplex mobility (Fig. 2A, lanes 3 and 4). Similarly, when mixtures of two or three different clones were incubated with the probe, two (Fig. 2A, lanes 5, 6, and 7) or three (Fig. 2A, lane 8) heteroduplex bands, respectively, were detected. A significant correlation (r2 = 0.920, P = 0.0025; InStat 2.00, linear regression) between heteroduplex mobility and percentage of mismatched nucleotides was observed.
FIG. 2.
Detection of individual molecular clones by QHDA. (A) Heteroduplexes formed by probe JC6 with clones (JC13, JC2, and JCI-3C in lanes 2, 3, and 4, respectively) with known percentages of mismatch and different numbers of nucleotide insertions. Lanes 5 to 8 show heteroduplexes between JC6 and mixtures of 2 or 3 of the other clones. (B) Semiquantitative detection of two cloned sequences (clones JC2 and JCI-3C) in mixtures with defined proportions of each clone. The percentages of the two clones within each sample before QHDA-PCR are shown above lanes 1 through 7.
To assess the sensitivity of QHDA for detecting individual heteroduplexes within a sample, mixtures containing different proportions of clones JC2 and JCI-3C were subjected to PCR. Addition of [33P]ATP-labeled probe JC6 to the products of each PCR resulted in detection of both the JC2 and JCI-3C clones, even when these clones comprised only 1 or 10%, respectively, of the original mixture (Fig. 2B). This result indicated that QHDA was sensitive enough to detect specific sequences within a given sample, even when sequences were present in low amounts. It appeared that the sensitivity of detection decreased as the diversity between a clone and the probe increased. However, there was a significant correlation (r2 = 0.9084 to 0.9843; P = 0.0001 to 0.0009) between the actual percentages of the two clones in each mixture and the percentages that were determined by scanning and quantifying the bands based on their densities. Thus, specific nucleotide sequences within a given sample were detected semiquantitatively, validating the use of this method to determine the heterogeneity in a sample of PCR products.
Characteristics of chimpanzees.
As reported previously (5), inoculation of chimpanzees C-166 and C-384 with HIV-1/JC499 resulted in systemic infections characterized by high viral burdens, induction of high titers of serum antibodies, and gradual loss of CD4+ lymphocytes. In contrast, after penile exposure of C-1323 to HIV-1/JC499, virus was detected in PBMC only on two occasions (4 and 8 weeks after inoculation) and in lymph node cells obtained by biopsy at 8 weeks (Table 3). This limited detection of cell-associated virus in the periphery was consistent with detection of only 500 virion RNA copies/ml of plasma at 4 weeks, a number that was at least 104-fold lower than maximum amounts of cell-free virus in plasma from the other two chimpanzees (Table 3). Furthermore, during long-term follow-up of C-1323, serum antibodies to HIV-1 were detected by enzyme-linked immunosorbent assay (ELISA) on only six occasions during the first year after inoculation, never exceeded a titer of 200, and have remained undetectable since that time, indicating that HIV-1/JC499 underwent limited replication and dissemination to systemic sites.
TABLE 3.
Characteristics of and viral loads in HIV-1/JC499-infected chimpanzees
| Chimpanzee | Inoculation route | No. of wks after inoculation | Antibody titera | Proviral copies per μg of DNA (SE)b | RNA copies per ml of plasmac |
|---|---|---|---|---|---|
| C-384 | Intravenous | 1 | 200 | 3.11 (1.78) | 2.0 × 104 |
| 2 | 1,600 | 1,534 (326) | 2.1 × 107 | ||
| 4 | 102,400 | 1.1 × 106 | |||
| C-166 | Cervical | 1 | 800 | Neg | Neg |
| 2 | 800 | 10.77 (3.62) | 1.6 × 105 | ||
| 4 | 204,800 | 4.9 × 106 | |||
| 6 | 409,600 | 766 (466) | 5.0 × 106 | ||
| 12 | 204,800 | 818 (439) | |||
| 2,218 (1,634) | |||||
| C-1323 | Penile | 4 | 100 | 0.61 (0.25) | 5.0 × 102 |
| 6 | 100 | Neg | Neg | ||
| 8 | 1.52 (0.52) | Neg | |||
| 1.77 (0.66) | |||||
| 12 | 100 | Neg | Neg |
Antibody titers in serum are the reciprocals of maximum serum dilutions that gave a positive optical density reading, as determined by enzyme immunoassay.
Proviral copy numbers are those in PBMC except for the second values for C-166 at 12 weeks and C-1323 at 8 weeks, which represent proviral copy numbers in lymph nodes. Neg, multiple PCRs using up to 4 μg of DNA/reaction were negative.
Neg, no detectable virion RNA in 1 ml of plasma using the Roche Amplicor HIV-1 Monitor kit.
PCR and RT-PCR amplification of proviral DNA and plasma RNA in infected chimpanzees.
Copy numbers of HIV-1/JC499 proviruses in PBMC were determined by PLDA. After the number of copies in 1 μg of genomic DNA in samples from C-166 and C-384 was ascertained, multiple independent PCRs were done using DNA that contained at least 100 proviruses in each reaction. Two exceptions were the PBMC DNA samples from C-166 (week 2) and C-384 (week 1) that had the fewest proviral copies by PLDA. For these two samples, a minimum of 40 proviruses was assumed to be present for amplification during the multiple PCRs. Then, one or more aliquots of the PCR products from each independent reaction were subjected to QHDA. Since PBMC and lymph node genomic DNA from C-1323 contained very low proviral copy numbers (Table 3), all of the PCR products resulting from the three positive samples were used for QHDA. Based on the PLDA quantification, these products had been amplified from a total of 2.7 to 8.54 proviral copies in multiple reactions.
The amount of cell-free virus in the various plasma samples from C-166 and C-384 varied as much as a thousandfold (Table 3). Because similar volumes (140 to 170 μl) of plasma were used to extract RNA for RT-PCR, the maximum number of viral RNA genomes that potentially could be amplified from any one sample ranged from 1,360 to 58,800. After amplification, the products from multiple independent reactions were analyzed by QHDA. Using the Roche HIV-1 Amplicor kit, 500 RNA copies per 1 ml of plasma were detected in samples from C-1323 at 4 weeks after inoculation (Table 3); however, none of the RT-PCRs performed on this sample were positive. From the combined analysis of the large numbers of proviral DNA and virion RNA genomes in multiple independent PCR and RT-PCR assays, it is likely that the sequences obtained from the repeated experiments were an accurate representation of those variants actually present in vivo.
Genetic composition of HIV-1/JC499 quasispecies in chimpanzees inoculated by mucosal routes.
Of the HIV-1/JC499 gp120env C2-V5 regions amplified from samples obtained from chimpanzees C-1323 and C-166, products of each PCR were divided into two equal parts, one of which was used in QHDA to analyze the overall genetic complexity. The other half was used for cloning and HDA to identify specific genotypes (6). Based on differences in QHDA banding patterns of all PCR products for each animal, TA clones were generated from each PCR whose bands exhibited a different pattern by QHDA, and then these clones were analyzed by standard HDA. The HDA results revealed that one heteroduplex band on RECOMBINATION AND EVOLUTIONthe QHDA gels represented a distinct group of bands on the HDA gels; that is, one QHDA band represented one population of variants with closely related sequences.
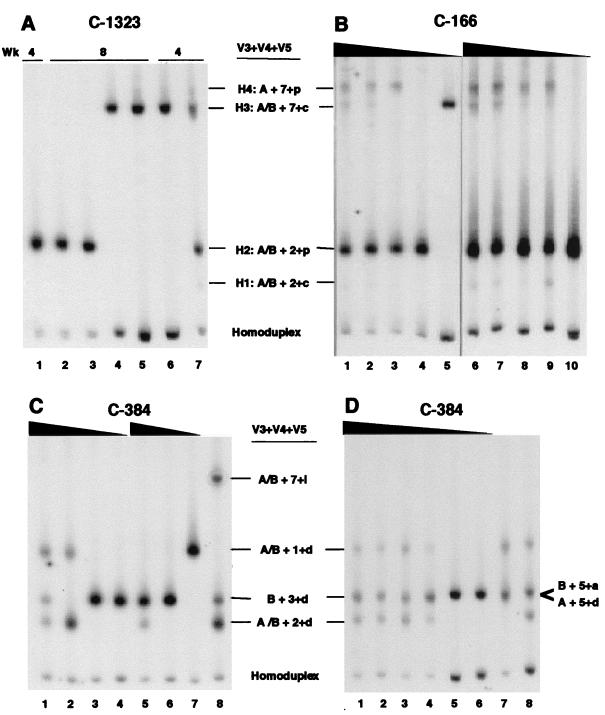
QHDA performed on PCR products generated using cellular DNA from C-1323 revealed that at 4 weeks after penile inoculation four heteroduplex bands (designated H1, H2, H3, and H4) with different mobilities and densities were present in PBMC (Fig. 3A, lanes 1, 6, and 7). However, at 8 weeks only one heteroduplex band (H2) was found in PBMC (Fig. 3A, lane 2) while two heteroduplex bands (H2 and H3) were detected in lymph nodes (Fig. 3A, lanes 3 [H2] and 4 and 5 [H3]). All of the TA clones subsequently evaluated by HDA corresponded to one of the four heteroduplex bands identified by QHDA. Of 141 clones generated from C-1323’s samples, 35 clones were sequenced, with the result that from 1 to 13 clones represented each of the four QHDA heteroduplex bands (H1 through H4). The four populations defined by specific V4 and V5 sequences that were transmitted to C-1323 were 2+c, 2+p, 7+c, and 7+p (Table 4), which by HDA constituted 3.69, 0.31, 59.1, and 0.31%, respectively, of the quasispecies in the HIV-1/JC499-P0 inoculum (see Table 2). These results showed that only a limited subset of the HIV-1/JC499 genetic populations were transmitted across the penile mucosa and that the transmitted viruses represented both major and minor populations in the HIV-1/JC499 inoculum.
FIG. 3.
Virus populations identified by QHDA in chimpanzees inoculated with HIV-1/JC499 by different routes. The JC6 probe was mixed separately with bulk PCR products amplified from proviral DNA in chimpanzee cells. (A) PBMC (lanes 1, 2, 6, and 7) and lymph node (lanes 3, 4, and 5) samples obtained from C-1323 at 4 and 8 weeks after penile inoculation. (B) PBMC obtained from C-166 at 6 weeks after cervicovaginal inoculation. Two independent limiting dilutions are shown (lanes 1 to 5 and 6 to 10). (C and D) PBMC obtained from C-384 at 1 (C) and 2 (D) weeks after intravenous infection. Samples in lanes below solid triangles are PCR products from independent limiting dilution series of genomic DNA instead of independent PCRs. The heteroduplex populations detected in panel C, lane 8, were products from a PCR in which 2 μg of DNA was used as template. In panel D, lanes 7 and 8, heteroduplexes from products of two independent PCRs, using 0.85 μg of DNA in each, are shown. The amount of DNA used for the five independent limiting dilution series shown in panels B, C, and D ranged from 1 μg (maximum) to 0.0016 μg (at the highest dilution). The specific V3, V4, and V5 region combinations (see Table 2) represented by the various heteroduplexes were determined by sequence analysis and are indicated. In panel D, in the heteroduplexes identified as B + 3+d, two additional populations (B + 5+a and A + 5+d) were identified after sequencing.
TABLE 4.
Identification and frequencies of HIV-1/JC499 populations detected in chimpanzee tissues early after infection
| Animal | No. of wks after inoculation | Sampleb | Frequency of HIV-1/JC499 populationa
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 + c | 2 + p | 7 + c | 7 + p | 1 + d | 1 + l | 2 + d | 2 + i | 2 + l | 3 + d | 5 + a | 5 + dc | 7 + ic | 7 + oc | 8 + a | 8 + dc | |||
| C-1323 | 4 | PBMC | 1/8 | 7/8 | 2/8 | 1/8 | ||||||||||||
| 8 | PBMC | 4/4 | ||||||||||||||||
| LN | 2/4 | 2/4 | ||||||||||||||||
| C-166 | 2 | PBMC | 1/21 | 21/21 | ||||||||||||||
| 4 | PL | 4/4 | ||||||||||||||||
| 6 | PBMC | 5/5 | 5/5 | 5/5 | 5/5 | |||||||||||||
| PL | 8/8 | 2/8 | ||||||||||||||||
| 12 | PBMC | 3/3 | 3/3 | 3/3 | 3/3 | |||||||||||||
| LN | 2/2 | 2/2 | 2/2 | 2/2 | ||||||||||||||
| C-384 | 1 | PBMC | 3/5 | 5/5 | 1/5 | 3/5 | 1/5 | |||||||||||
| 2 | PBMC | 6/6 | 6/6 | 6/6 | 2/6 | 2/6 | 1/6 | |||||||||||
| PL | 1/4 | 4/4 | 1/4 | 3/4 | 1/4 | 4/4 | 1/4 | 1/4 | 1/4 | 3/4 | 1/4 | |||||||
Number of independent PCRs in which a V4-plus-V5 population was detected, either as the only population or in combination with one or more other V4-plus-V5 combinations, out of the total number (denominator) of PCRs for a given sample.
LN, lymph node; PL, plasma.
Virus populations not found in the HIV-1/JC499-P0 stock.
The identical strategy was used to determine the virus populations transmitted by the cervical route to C-166. First, the products of individual nested PCRs or PCRs performed on each dilution of a limiting dilution series of PBMC, lymph node cells, or plasma obtained from 2 to 12 weeks after HIV-1/JC499 inoculation were subjected to QHDA. The QHDA banding patterns observed for samples from C-166 appeared very similar to those seen for C-1323 (Fig. 3B). After cloning the PCR products and performing HDA on 586 clones, 48 clones with heteroduplex bands that migrated similarly to the H1 to H4 bands indicative of clones from C-1323 were selected and sequenced. Despite the observed minor variations in mobility, only four genetic populations were identified among the 48 clones. At 2 weeks after inoculation only populations 2+p and 7+c were detected in PBMC, whereas all four populations were detected in PBMC at 6 and 12 weeks and in lymph nodes at 12 weeks (Table 4). In contrast, only two of the populations were represented in plasma virion RNA; 2+p was found at 4 weeks and both 2+p and 7+p were observed in plasma at 6 weeks. Again, these results showed that a limited number of variant populations defined by V4 and V5 in the HIV-1/JC499 stock were transmitted across the cervical mucosa and that these transmitted populations were the same as those transmitted to C-1323 by a different mucosal route.
The genetic diversity of specific populations within each individual sample (intrasample) and between all samples (intersample) from the chimpanzees was calculated. The intrasample genetic distances for populations 2+c and 7+c in C-1323 ranged from 1.18 to 3.09% and from 0.33 to 3.37%, respectively; these distances did not exceed those of the corresponding populations in the HIV-1/JC499 stock (see Table 2). The intersample range (0.33 to 5.47%) of distances for population 7+c in C-166 was also consistent with that for 7+c in the HIV-1/JC499 stock. Based on the HDA banding patterns and the genetic distances, the heterogeneity of populations 2+p in C-1323 and 2+p, 7+c, and 7+p in C-166 were relatively stable during the time that samples were evaluated. These results suggested that the same V4 (2 and 7) and V5 (c and p) sequences in various combinations, irrespective of the V3 or intervening constant region sequences, were more likely to be transmitted across mucosal surfaces, clearly demonstrating that selection of specific HIV-1/JC499 genotypes occurred.
Genetic composition of HIV-1/JC499 populations in a chimpanzee inoculated intravenously.
QHDA was performed with products amplified during multiple independent PCRs from proviral DNA in PBMC and viral RNA in plasma at 1 and 2 weeks after intravenous inoculation of C-384. This analysis primarily revealed that most of the PCR products from PBMC at 1 and 2 weeks and plasma at 2 weeks were comprised of one to three distinct heteroduplexes (Fig. 3C and D), later shown to represent genetic populations 1+d, 2+d, and 3+d. Clones were generated from PCR products with different QHDA heteroduplex banding patterns and subjected to HDA. In contrast to what was observed with samples from C-1323 and C-166, where HDA results were always consistent with the QHDA results, the number of populations estimated by the number of QHDA bands was sometimes less than the number subsequently identified by HDA banding patterns of individual clones. For example, three distinct V4-plus-V5 populations, 3+d, 5+a, and 5+d, were represented by only one heteroduplex band on QHDA gels (Fig. 3D). Two factors can explain this difference. First, the sensitivity of QHDA decreased as the diversity between the sample and probe increased, and second, QHDA detects only one DNA strand (minus strand) whereas HDA detects both DNA strands.
When clones from each HDA group generated from PBMC at 1 week after infection were sequenced and the sequences were aligned with the HIV-1/JC499 consensus sequence, five genetic populations defined by V4 and V5 (1+d, 2+d, 3+d, 2+i, and 7+i) were observed (Table 4). By 2 weeks after infection, the number of populations had increased to 6 (1+d, 2+d, 3+d, 5+a, 5+d, and 7+o) in PBMC and 11 (1+d, 1+l, 2+d, 2+l, 3+d, 5+a, 5+d, 7+c, 7+i, 8+a, and 8+d) in plasma. However, only five of the V4-plus-V5 combinations were common to both PBMC and plasma at 2 weeks and only three populations (1+d, 2+d, 3+d) were present in all samples. The ranges of genetic distance within populations 1+d, 2+d, and 3+d were 0.48 to 2.90%, 0.33 to 4.48%, and 0.5 to 2.20%, respectively. Thus, a combined total of 13 populations were detected in samples from C-384 at 1 and 2 weeks after intravenous infection; four of these (5+d, 7+i, 7+o, and 8+d) were not detected in the HIV-1/JC499 stock (Table 4).
The 7+c population, which was the most prevalent population in the inoculum, was the only one detected in all three chimpanzees, irrespective of the route of inoculation. Only 13 of the 33 populations (29 shown in Table 2 plus 4 populations not detected in the HIV-1/JC499 inoculum) defined by distinct V4-plus-V5 combinations appeared to establish infection after intravenous exposure. This result indicated that selective transmission of specific HIV-1/JC499 genotypes also occurred during intravenous infection. However, these genotypes were different from and less restricted than those selected during mucosal infection.
Frequency of HIV-1/JC499 genetic populations early after transmission.
QHDA analyses of products amplified during multiple PCRs with PBMC, lymph node, or plasma samples were used to ascertain the relative frequencies with which viruses encoding specific V4-plus-V5 populations were transmitted. Of the four V4-plus-V5 populations transmitted to C-1323 and C-166, in both animals 2+p and 7+c were amplified more frequently than 2+c or 7+p during the first few weeks after infection (Table 4). The relative abundance of each V4-plus-V5 combination in C-1323’s 4-week PBMC sample, in which all four populations were amplified, also was determined by measuring the densities of each band in a lane of a QHDA gel and expressing the result as a percentage of the total density. This method indicated that 46% of the populations were 2+p, 38% were 7+c, 11% were 7+p, and 5% were 2+c, showing that the first two populations were more prevalent, consistent with the QHDA results. That QHDA and HDA always gave equivalent results for samples from C-1323 demonstrated that the frequency of specific sequences within a given sample could be quantified reliably by QHDA.
For C-166 the dominant population detected in plasma was 2+p; it was the only one identified at 4 weeks and the predominant one at 6 weeks after inoculation, at which time 2+p and 7+p were found at frequencies of 68 and 32%, respectively. Four populations (2+c, 2+p, 7+c, and 7+p) were observed consistently in all PCRs performed with DNA from PBMC at 6 and 12 weeks and lymph node cells at 12 weeks. The average percentages for 2+c, 2+p, 7+c, and 7+p from five PCRs performed on the 6-week PBMC and lymph node samples were 2.0, 68.7, 8.9, and 20.4%, respectively. At 12 weeks, the average percentages for 2+c, 2+p, 7+c, and 7+p in PBMC and lymph node cells were 9.1, 47.1, 14.3, and 29.6%, respectively. Taken together, in C-166 the 2+p population was dominant, followed by 7+p, 7+c, and 2+c, in order of prevalence. Of interest, although the 2+p population was only 0.31% of the total populations identified in the HIV-1/JC499 stock, it was the predominant population in both C-1323 and C-166, suggesting that this genotype was transmitted preferentially across chimpanzee mucosal surfaces.
Instead of measuring the intensity of bands on QHDA gels, we quantified V4-plus-V5 populations identified in C-384’s samples by determining the percentages of HDA clones that represented each population among all the clones. This method was necessary because on the QHDA gels on which samples from C-384 were analyzed, one heteroduplex band sometimes was shown by HDA and sequencing to represent multiple V4-plus-V5 populations. A total of 263 clones were generated from the products of a total of 11 PCRs performed with DNA from PBMC obtained at 1 and 2 weeks and of 4 RT-PCRs on virion RNA in plasma obtained at 2 weeks. Of the 13 different V4-plus-V5 populations found in samples from C-384, 86.6% were represented by 3+d, 2+d, and 1+d, at frequencies of 33.8, 30.4, and 22.4%, respectively. However, in the HIV-1/JC499 inoculum, these three populations represented only 6.41, 0.31, and 8.0% of the quasispecies. The prevalence of all other populations in C-384 ranged from 0.4 to 3.4%. These data suggested that the V5 sequence identified as “d” might be an important determinant for preferential replication of HIV-1/JC499 during intravenous infection. Of additional interest, while genotypes closely related to 7+c comprised 59.1% of all populations in the HIV-1/JC499-P0 inoculum, these variants were only a minor population in plasma from C-384 at 2 weeks.
Relationships of the inoculum and transmitted HIV-1/JC499 populations.
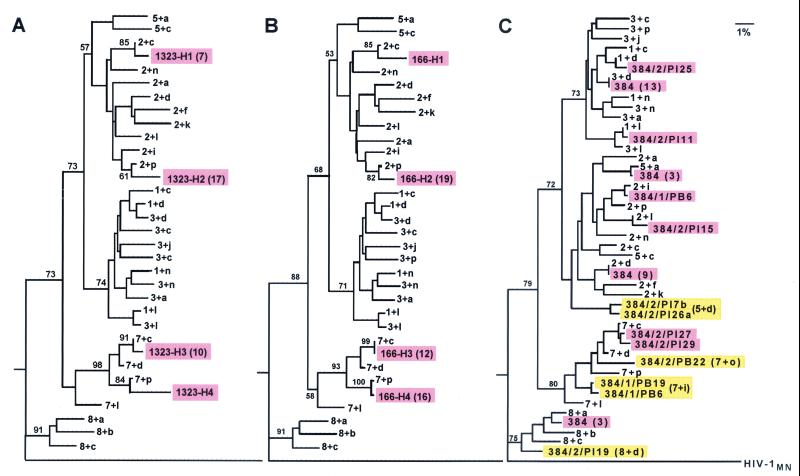
The relationships of sequences identified in each of the three chimpanzees to the parental sequences in the HIV-1/JC499 inoculum were evaluated by constructing phylogenetic trees with the 3′ half of the C2-V5 fragment. Because there were only two V3 consensus sequences and both were found in most V4-plus-V5 populations, inclusion of the V3 region obscured the branching of individual populations, which in general formed branches according to V4 sequences. Two sets of V4 sequences, V4-1 and V4-3, and to a lesser extent, V4-2 and V4-5, appeared more closely related. These populations branched together because the consensus sequences for the first and second sets differed by only two and four amino acids, respectively. That V5 appeared not to contribute significantly to branching order is probably because it is a shorter region and contains multiple gaps, which were excluded in generating the phylogenetic trees. The phylogenetic analysis confirmed that the same four V4-plus-V5 genetic populations (identified as H1, H2, H3, and H4 in Fig. 3) were transmitted by both of the mucosal routes (Fig. 4A and B). Furthermore, it is clear that the relationships of these four populations to those in the inoculum differed significantly from the V4-plus-V5 sequence variants transmitted intravenously, which were distributed throughout the tree (Fig. 4C).
FIG. 4.
Phylogenetic trees of V4-plus-V5 populations in the HIV-1/JC499 inoculum and transmitted by the penile (A), cervical (B), and intravenous (C) routes to chimpanzees C-1323, C-166, and C-384, respectively. Transmitted populations are identified by shaded boxes. Numbers in parentheses indicate the number of individual sequences within one population for which a consensus sequence was used to generate the trees; otherwise, individual sequences were used. In panel C, the first numbers shown after “384/” indicate the week after inoculation when the sample was obtained, the tissue (Pl, plasma; PB, PBMC) of origin, and the individual clone number. The clones identified by pink boxes were those in the inoculum, whereas the yellow boxes identify clones with V4-plus-V5 populations (in parentheses) not found in the inoculum.
DISCUSSION
The Env glycoprotein mediates binding of HIV-1 to a cell via the CD4 molecule and one of several chemokine receptors (7, 9); therefore, one would expect that if selective transmission occurred, it would be influenced primarily by Env. In addition to a relatively conserved CD4-binding region, the sequence and charge of the V3 loop is a major determinant of cell tropism, and in particular, of whether an HIV-1 variant can infect macrophages by interactions between CD4 and CCR5 (11, 45, 48, 56). Therefore, it seems reasonable that the Env V3 loop might be a major determinant in selective transmission. There is evidence, however, that the V1-V2 and V4-V5 regions also can influence coreceptor usage (21, 41). Thus, it is possible that selection for preferential infection and amplification of different HIV-1 variants could be mediated by interactions between one or more of the variable regions of Env and the relative numbers of cells in blood or mucosal tissues that express a given coreceptor.
Most studies to determine whether a specific subset of HIV-1 variants is transmitted either parenterally or across mucosal surfaces have evaluated proviruses in PBMC or virions in plasma samples collected weeks or months after exposure. Even when samples were collected at the time of acute disease and before seroconversion, several weeks might have elapsed since initial infection, which is especially true with respect to collection of samples from an infected person who transmitted the virus. Using HIV-1 infection of chimpanzees as a model system, we eliminated potential problems associated with time of sample collection. Our results demonstrated that selection for specific variants in a genetically defined HIV-1/JC499 stock occurred after exposure of the cervical or penile mucosa to cell-free virus. Furthermore, in a chimpanzee inoculated intravenously with the same virus stock, a larger, different set of variants successfully established systemic infection. The three major populations in peripheral blood from the animal inoculated intravenously were minor populations in the HIV-1/JC499 stock, suggesting that selection also occurred during parenteral transmission. However, selective pressures influencing mucosal versus parenteral transmission of specific variants are likely to differ.
Sequence analysis in the C2-V5 region of env of multiple clones from each chimpanzee revealed that the virus genomes from all animals, irrespective of route of inoculation, encoded one of only two distinct V3 regions. Of particular interest, comparison of the V4 and V5 regions in all clones showed that all viruses from the two chimpanzees infected by mucosal routes encoded only one of two V4 and two V5 regions, generating four V4-plus-V5 combinations. In the animal inoculated intravenously, 13 different V4-plus-V5 combinations were identified. The only V4-plus-V5 combination common to viruses in all three animals was 7+c, which was the most prevalent population (59.1% of 325 clones analyzed by HDA) in the inoculum. However, the 7+c population represented only 1.1% of the clones in the quasispecies identified in the animal inoculated intravenously and from 9 to 38%, depending on the animal and the time after infection, of the clones in the animals inoculated by a mucosal route. These results and the identification of more than 30 different V4-plus-V5 combinations in the HIV-1/JC499 inoculum strongly suggest that different, but specific, V4 and V5 combinations, and not the V3 loop, played a substantive role in selection during both mucosal and parenteral transmission. It is possible that this apparent selection as a function of particular V4 and V5 combinations might have been influenced by the presence of basically two V3 loop variant populations in the inoculum. That there appeared to be less-restricted selection during intravenous compared to mucosal infection is consistent with the observed diversity of transmitted viruses in both HIV-1 infection of humans and simian immunodeficiency virus (SIV) or simian/human immunodeficiency virus infection of macaques (19, 20, 28, 47, 49). In all these instances, the virus population identified early after infection by a parenteral route was more diverse than that observed after mucosal infection.
In general, our results are in agreement with those of many studies of HIV-1 infection in human transmission pairs. In the human studies, irrespective of whether transmission was between homosexual or heterosexual partners or from mother to child, the transmitted virus was a minor species among the donor’s diverse quasispecies (1, 15, 33, 55, 58, 59). Although we identified either 4 (mucosal) or 13 (intravenous) different V4-plus-V5 populations among viruses present early after inoculation, these populations were only a subset of the more than 30 V4-plus-V5 populations in the HIV-1/JC499 inoculum. However, the variant genotypes transmitted to all three chimpanzees included both major and minor species. Furthermore, in human studies it was concluded that most viruses that established systemic infections in any one recipient were relatively homogeneous in the V3 regions (24, 54, 57, 59). An apparent exception was found in cohorts of African women in whom heterogeneous virus populations were detected early after vaginal transmission (27, 39). However, in the study by Poss et al. (39), heterogeneity in V1-V3 was defined by pairwise differences of as little as 5.3%. In our study, the V3 region of the HIV-1 populations transmitted to all three chimpanzees exhibited minimal variation, whereas the closely related populations defined by specific V4-plus-V5 combinations, i.e., intrapopulation diversity, ranged from 0.49 to 5.64% (Table 2). Wolinsky et al. (55) also found up to 5% variation in V4 and V5 regions and up to 15% variation in the V3 region in viral sequences from infants born to infected mothers. However, despite this level of diversity, phylogenetic analysis and comparison of consensus sequences of each mother-infant pair led to the conclusion that the most prevalent genotype in each infant was derived from a single genotype in the mother. These examples illustrate the point that definitions of relative homogeneity can vary according to the region(s) of the HIV-1 genome being compared and one’s definition of homo- and heterogeneity.
An example of extreme homogeneity was reported in a study by Zhang et al. (57), in which the env V3-V4 region was evaluated, and essentially no variation (0% diversity) was found in either virion RNA or proviral DNA in eight individuals who were infected either by a parenteral or mucosal route. However, these authors did not use a technique such as HDA to assess the extent of diversity and to aid in selecting distinct clones for sequencing. They relied solely on amplification of viral sequences from end-point limiting dilution series, which does ensure that only one molecule is amplified in each PCR, but it also could select preferentially for the most prevalent variant(s). To obtain a more accurate indication of heterogeneity, we used a combination of three techniques, QHDA, HDA, and sequencing, to analyze the PCR and RT-PCR products generated from multiple limiting dilution assays. QHDA, employing a single-strand 33P-labeled probe, was used as an initial screen to assess the overall genetic complexity semiquantitatively. HDA, employing both strands of the probe, was used to identify cloned products with different heteroduplex patterns to ensure that sequences were obtained from at least one clone representing each distinct mobility pattern. A similar approach was used by Zhu et al. (59) to analyze variants transmitted to humans. Despite the use of three separate techniques to identify variant populations, there were four V4-plus-V5 populations (5+d, 8+d, 7+i, and 7+o) not found in the inoculum that were identified in C-384 within 2 weeks after intravenous inoculation. It is likely that these four populations were minor ones in the inoculum and were amplified in vivo.
It is generally accepted that the phenotype of transmitted HIV-1 variants is non-syncytium-inducing (NSI) and macrophage tropic (4, 58). However, there is substantial evidence to show that both NSI and syncytium-inducing (SI) strains can establish infection, irrespective of whether transmission is parenteral or mucosal or from mother to child (4, 10, 44). Therefore, it cannot be concluded that variants with V3 loop sequences characteristic of macrophage tropism always have a selective advantage. In the SIV or simian/human immunodeficiency virus macaque models, the efficiency of vaginal transmission was related to the inherent ability of a virus to replicate in vivo after intravenous inoculation rather than to virus phenotype (31). The HIV-1/JC499 inoculum is phenotypically SI and cytopathic for human and chimpanzee CD4+ lymphocytes. A molecular clone generated from PBMC obtained from C-166 2 weeks after infection preferentially uses CXCR4 but can also utilize CCR2, CCR3, and GPR15 for productive infection of cells in vitro (L. Yue and P. N. Fultz, unpublished data). All variants that established infection in the chimpanzees, whether by the parenteral or mucosal route, encoded one of the same two V3 regions, indicating that the V3 region did not discriminate between infection of lymphocytes in blood and infection of susceptible cells on or near the mucosal surface. C-166 and C-384 had comparably high proviral copy numbers in PBMC and virion RNA in plasma after cervicovaginal and intravenous inoculation, respectively. Yet, C-166 harbored a more restricted set of viruses, as defined by V4-plus-V5 combinations, than did C-384, which provides additional support for the conclusion that selection at a mucosal surface was not related to the V3 loop in Env.
That chimpanzee C-1323, the animal inoculated via the penile mucosa, exhibited only transient viremia and failed to develop detectable serum antibodies to HIV-1 is not unusual after mucosal infection. We previously described low-level HIV-1 infection without seroconversion in chimpanzees exposed to the virus by the cervicovaginal route (16). Furthermore, similar results for macaques inoculated either vaginally or rectally with various SIV strains have been reported (30–32, 50). In some cases these transiently viremic, seronegative animals developed detectable viremia and virus-specific antibody responses several months or even years after the original inoculation (29, 50). During the period in which virus was not isolated from PBMC, some chimpanzees and macaques had lymphocytes that proliferated in response to viral antigens (16, 30). These findings are consistent with reports of HIV-specific T-cell responses in seronegative high-risk individuals, including female prostitutes, injection drug users, and partners of HIV-seropositive individuals (2, 17, 29, 42). The same HIV-1/JC499 genetic variants were transmitted to the two chimpanzees, C-166 and C-1323, inoculated by different mucosal routes, yet the animals had very different courses of infection, exemplified by chronic high-level versus transient low-level viremia. Thus, host factors appeared to play an important role in determining the type of infection that was established. Whether these factors were genetic, physiologic, or related to differences in the external or internal mucosal environments cannot be determined.
In summary, inoculation of three chimpanzees by one of two different mucosal routes or intravenously with a complex inoculum of cell-free HIV-1/JC499 resulted in selective transmission of genetically diverse viral populations, which were defined by different combinations of specific V4 and V5 sequences. The same two populations of V3 sequences were found in all variants in all three chimpanzees, but the V4-plus-V5 combinations in the parenterally and mucosally inoculated chimpanzees were mutually exclusive, with one exception: the most prevalent V4-plus-V5 population in the inoculum was present in all three animals. This finding, in conjunction with the observation that the same restricted V4-plus-V5 populations were transmitted by two different mucosal routes, suggests that both stochastic and selective factors played a role. That the various V4 and V5 populations exhibited up to ≈5% diversity but still could be grouped reliably into discrete populations reflects the extreme heterogeneity of the inoculum. Our results in the HIV-1 chimpanzee model and those in the SIV macaque model as well as some human studies are essentially the same. The observed inconsistencies in some human studies might be explained by two factors in addition to the different regions of the HIV-1 genome analyzed and one’s definition of homo- and heterogeneity, as discussed above. First, the nonhuman primate models used cell-free inocula, but transmission between humans is mediated by both cell-free and cell-associated virus (59). Second, the ongoing immune response to HIV-1 in the transmitter might influence which variants are transmitted (46). It is clear, however, that the general assumption that only minor, homogeneous populations of HIV-1 are transmitted is not valid and each case should be evaluated independently using diverse techniques.
Acknowledgments
We thank Jiangming Tang for advice in establishing the QHDA technique; Feng Gao, Yang Wang, David Nickle, and James Mullins for discussions about sequence analyses; Casey Morrow for comments on the manuscript; Pam May and Jackie Stallworth for isolating PBMC and generating single-cell suspensions of lymph nodes; and Zuhua Cao for part of the sequencing work.
This study was supported by grant AI28147 from the National Institute of Allergy and Infectious Diseases, NIH.
REFERENCES
- 1.Ahmad, N., B. M. Baroudy, R. C. Baker, and C. Chappey. 1995. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J. Virol. 69:1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beretta, A., S. H. Weiss, G. Rappocciolo, R. Mayur, C. DeSantis, J. Quirinale, A. Cosma, P. Robbioni, G. M. Shearer, J. A. Berzofsky, M. L. Villa, A. G. Siccardi, and M. Clerici. 1996. Human immunodeficiency virus type 1 (HIV-1)-seronegative injection drug users at risk for HIV exposure have antibodies to HLA class I antigens and T cells specific for HIV envelope. J. Infect. Dis. 173:472–476. [DOI] [PubMed] [Google Scholar]
- 3.Briant, L., C. M. Wade, J. Puel, A. J. L. Brown, and M. Guyader. 1995. Analysis of envelope sequence variants suggests multiple mechanisms of mother-to-child transmission of human immunodeficiency virus type 1. J. Virol. 69:3778–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelissen, M., G. Mulder-Kampinga, J. Veenstra, F. Zorgdrager, C. Kuiken, S. Hartman, J. Dekker, L. van der Hock, C. Sol, R. Coutinho, and J. Goudsmit. 1995. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J. Virol. 69:1810–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, I. C., M. Girard, and P. N. Fultz. 1998. Loss of CD4+ T cells in human immunodeficiency virus type-1-infected chimpanzees is associated with increased lymphocyte apoptosis. J. Virol. 72:4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257–1261. [DOI] [PubMed] [Google Scholar]
- 7.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. [DOI] [PubMed] [Google Scholar]
- 8.Dickover, R. E., E. M. Garratty, S. Plaeger, and Y. J. Bryson. 2001. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J. Virol. 75:2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein-coupled receptor. Science 272:872–877. [DOI] [PubMed] [Google Scholar]
- 10.Fiore, J. R., A. Bjorndal, K. A. Peipke, M. D. Stefano, G. Angarano, G. Pastore, H. Gaines, E. M. Fenyo, and J. Albert. 1994. The biological phenotype of HIV-1 is usually retained during and after sexual transmission. Virology 204:297–303. [DOI] [PubMed] [Google Scholar]
- 11.Fouchier, R. A. M., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fultz, P. N., H. M. McClure, R. B. Swenson, C. R. McGrath, A. Brodie, J. P. Getchell, F. C. Jensen, D. C. Anderson, J. R. Broderson, and D. P. Francis. 1986. Persistent infection of chimpanzees with human T-lymphotropic virus type III/lymphadenopathy-associated virus: a potential model for acquired immunodeficiency syndrome. J. Virol. 58:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fultz, P. N., Q. Wei, and L. Yue. 1999. Rectal transmission of human immunodeficiency virus type 1 to chimpanzees. J. Infect. Dis. 179(Suppl. 3):S418–S421. [DOI] [PubMed] [Google Scholar]
- 14.Fultz, P. N., L. Yue, Q. Wei, and M. Girard. 1997. Human immunodeficiency virus type 1 intersubtype (B/E) recombination in a superinfected chimpanzee. J. Virol. 71:7990–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuta, J., T. Bergstrom, G. Norkrans, and P. Horal. 1994. HIV type 1 V3 sequence diversity in contact-traced Swedish couples at the time of sexual transmission. AIDS Res. Hum. Retrovir. 10:1187–1189. [DOI] [PubMed] [Google Scholar]
- 16.Girard, M., J. Mahoney, Q. Wei, E. van der Ryst, E. Muchmore, F. Barre-Sinoussi, and P. N. Fultz. 1998. Genital infection of female chimpanzees with human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 14:1357–1367. [DOI] [PubMed] [Google Scholar]
- 17.Goh, W. C., J. Markee, R. E. Akridge, M. Meldorf, L. Musey, T. Karchmer, M. Krone, A. Callier, L. Corey, M. Emerman, and M. J. McElrath. 1999. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J. Infect. Dis. 179:548–557. [DOI] [PubMed] [Google Scholar]
- 18.Goodenow, M., T. Huet, W. Saurin, S. Kwok, J. Sninsky, and S. Wain-Hobson. 1989. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotide substitutions. J. Acquir. Immune Defic. Syndr. 2:344–352. [PubMed] [Google Scholar]
- 19.Goudsmit, J., V. V. Lukashov, E. J. C. Van Ameijden, F. Zorgdrager, R. Van Den Burg, and M. Cornelissen. 1998. Impact of sexual versus parenteral transmission events on the evolution of the gag and env genes of HIV type 1. AIDS Res. Hum. Retrovir. 14:1483–1486. [DOI] [PubMed] [Google Scholar]
- 20.Greenier, J. L., C. J. Miller, D. Lu, P. J. Dailey, F. X. Lu, K. J. Kunstman, S. M. Wolinsky, and M. L. Marthas. 2001. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J. Virol. 75:3753–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Q.-X., A. P. Barry, Z.-X. Wang, S. M. Connolly, S. C. Peiper, and M. L. Greenberg. 2000. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J. Virol. 74:11858–11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, B. K., G. A. Stone, M. S. Godec, D. M. Asher, D. C. Gajdusek, and C. J. Gibbs. 1993. Long-term observations of human immunodeficiency virus-infected chimpanzees. AIDS Res. Hum. Retrovir. 9:375–378. [DOI] [PubMed] [Google Scholar]
- 23.Kampinga, G. A., A. Simonon, P. Van de Perre, E. Karita, P. Msellati, and J. Goudsmit. 1997. Primary infections with HIV-1 of women and their offspring in Rwanda: findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology 227:63–76. [DOI] [PubMed] [Google Scholar]
- 24.Kliks, S., C. H. Contag, H. Corliss, G. Learn, A. Rodrigo, D. Wara, J. I. Mullins, and J. A. Levy. 2000. Genetic analysis of viral variants selected in transmission of human immunodeficiency viruses to newborns. AIDS Res. Hum. Retrovir. 16:1223–1233. [DOI] [PubMed] [Google Scholar]
- 25.Koopman, G., A. G. M. Haaksma, J. T. Velden, C. E. Hack, and J. L. Heeney. 1999. The relative resistance of HIV type 1-infected chimpanzees to AIDS correlates with the maintenance of follicular architecture and the absence of infiltration by CD8+ cytotoxic T lymphocytes. AIDS Res. Hum. Retrovir. 15:365–373. [DOI] [PubMed] [Google Scholar]
- 26.Lamers, S. L., J. W. Sleasman, J. X. She, K. A. Barrie, S. M. Pomeroy, D. J. Barrett, and M. M. Goodenow. 1994. Persistence of multiple maternal genotypes of human immunodeficiency virus type 1 in infants infected by vertical transmission. J. Clin. Investig. 93:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long, E. M., H. L. Martin, J. K. Kreiss, S. M. J. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71–75. [DOI] [PubMed] [Google Scholar]
- 28.Lu, Y., P. Brosio, M. Lafaile, J. Li, R. G. Collman, J. Sodroski, and C. J. Miller. 1996. Vaginal transmission of chimeric simian/human immunodeficiency viruses in rhesus macaques. J. Virol. 70:3045–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazzoli, S., D. Trabattoni, S. LoCaputo, S. Piconi, C. Ble, F. Meacci, S. Ruzzante, A. Salvi, F. Semplici, R. Longhi, M. L. Fusi, N. Tofani, M. Biasin, M. L. Villa, F. Mazzotta, and M. Clerici. 1997. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat. Med. 3:1250–1257. [DOI] [PubMed] [Google Scholar]
- 30.McChesney, M. B., J. R. Collins, D. Ju, X. Lu, J. Torten, R. L. Ashley, M. W. Cloyd, and C. J. Miller. 1998. Occult systemic infection and persistent simian immunodeficiency virus (SIV)-specific CD4+-T-cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J. Virol. 72:10029–10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, C. J., M. Marthas, J. Greenier, D. Lu, P. J. Dailey, and Y. Lu. 1998. In vivo replication capacity rather than in vitro macrophage tropism predicts efficiency of vaginal transmission of simian immunodeficiency virus or simian/human immunodeficiency virus in rhesus macaques. J. Virol. 72:3248–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, C. J., M. Marthas, J. Torten, N. J. Alexander, J. P. Moore, G. F. Doncel, and A. G. Hendrickx. 1994. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J. Virol. 68:6391–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulder-Kampinga, G. A., A. Simonon, C. L. Kuiken, J. Dekker, H. J. Scherpbier, P. van de Perre, K. Boer, and J. Goudsmit. 1995. Similarity in env and gag genes between genomic RNAs of human immunodeficiency virus type 1 (HIV-1) from mother and infant is unrelated to time of HIV-1 RNA positivity in the child. J. Virol. 69:2285–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nara, P. L., W. G. Robey, L. O. Arthur, D. M. Asher, A. V. Wolff, C. J. Gibbs, Jr., D. C. Gajdusek, and P. J. Fischinger. 1987. Persistent infection of chimpanzees with human immunodeficiency virus: serological responses and properties of reisolated viruses. J. Virol. 61:3173–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novembre, F. J., J. de Rosayro, S. Nidtha, S. P. O’Neil, T. R. Gibson, T. Evans-Strickfaden, C. E. Hart, and H. M. McClure. 2001. Rapid CD4+ T-cell loss induced by human immunodeficiency virus type 1NC in uninfected and previously infected chimpanzees. J. Virol. 75:1533–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novembre, F. J., M. Saucier, D. C. Anderson, S. A. Klumpp, S. P. O’Neil, C. R. Brown, C. E. Hart, P. C. Guethner, R. B. Swenson, and H. M. McClure. 1997. Development of AIDS in a chimpanzee infected with human immunodeficiency virus type 1. J. Virol. 71:4086–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Neil, S. P., F. J. Novembre, A. B. Hill, C. Suwyn, C. E. Hart, T. Evans-Strickfaden, D. C. Anderson, J. de Rosayro, J. G. Herndon, M. Saucier, and H. M. McClure. 2000. Progressive infection in a subset of HIV-1-positive chimpanzees. J. Infect. Dis. 182:1051–1062. [DOI] [PubMed] [Google Scholar]
- 38.Pasquier, C., C. Cayrou, A. Blancher, C. Tourne-Petheil, A. Berrebi, J. Tricoire, J. Puel, and J. Izopet. 1998. Molecular evidence for mother-to-child transmission of multiple variants by analysis of RNA and DNA sequences of human immunodeficiency virus type 1. J. Virol. 72:8493–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poss, M., H. L. Martin, J. K. Kreiss, L. Granville, B. Chohan, P. Nyange, K. Mandaliya, and J. Overbaugh. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J. Virol. 69:8118–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodrigo, A. G., P. C. Goracke, K. Rowhanian, and J. I. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retrovir. 13:737–742. [DOI] [PubMed] [Google Scholar]
- 41.Ross, T. M., and B. R. Cullen. 1998. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc. Natl. Acad. Sci. USA 95:7682–7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowland-Jones, S., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, M. Takiguchi, T. Schultz, A. McMichael, and H. Whittle. 1995. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59–64. [DOI] [PubMed] [Google Scholar]
- 43.Salvatori, F., S. Masiero, C. Giaquinto, C. M. Wade, A. J. L. Brown, L. Chieco-Bianchi, and A. De Rossi. 1997. Evolution of human immunodeficiency virus type 1 in perinatally infected infants with rapid and slow progression to disease. J. Virol. 71:4694–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scarlatti, G., V. Hodara, P. Rossi, L. Muggiasca, A. Bucceri, J. Albert, and E. M. Fenyo. 1993. Transmission of human immunodeficiency virus type 1 (HIV-1) from mother to child correlates with viral phenotype. Virology 197:624–629. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu, N., Y. Haraguchi, Y. Takeuchi, Y. Soda, K. Kanbe, and H. Hoshino. 1999. Changes in and discrepancies between cell tropisms and coreceptor uses of human immunodeficiency virus type 1 induced by single point mutations at the V3 tip of the env protein. Virology 259:324–333. [DOI] [PubMed] [Google Scholar]
- 46.Shpaer, E. G., E. L. Delwart, C. L. Kuiken, J. Goudsmit, M. H. Bachmann, and J. I. Mullins. 1994. Conserved V3 loop sequences and transmission of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1679–1684. [DOI] [PubMed] [Google Scholar]
- 47.Sodora, D. L., F. Lee, P. J. Dailey, and P. A. Marx. 1998. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res. Hum. Retrovir. 14:171–181. [DOI] [PubMed] [Google Scholar]
- 48.Speck, R. F., K. Wehrly, E. J. Platt, R. E. Atchison, I. F. Charo, D. Kabat, B. Chesebro, and M. A. Goldsmith. 1997. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J. Virol. 71:7136–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trichel, A. M., E. D. Roberts, L. A. Wilson, L. N. Martin, R. M. Ruprecht, and M. Murphey-Corb. 1997. SIV/DeltaB670 transmission across oral, colonic, and vaginal mucosae in the macaque. J. Med. Primatol. 26:3–10. [DOI] [PubMed] [Google Scholar]
- 50.Trivedi, P., K. K. Meyer, D. N. Streblow, B. L. Preuninger, K. T. Schultz, and C. D. Pauza. 1994. Selective amplification of simian immunodeficiency virus genotypes after intrarectal inoculation of rhesus monkeys. J. Virol. 68:7649–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wade, C. M., D. Lobidel, and A. J. Leigh-Brown. 1998. Analysis of human immunodeficiency type 1 env and gag sequence variants derived from a mother and two vertically infected children provides evidence for the transmission of multiple sequence variants. J. Gen. Virol. 79:1055–1068. [DOI] [PubMed] [Google Scholar]
- 52.Wei, Q., and P. N. Fultz. 1998. Extensive diversification of human immunodeficiency virus type 1 subtype B strains during dual infection of a chimpanzee that progressed to AIDS. J. Virol. 72:3005–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei, Q., A. Javadian, N. Lausen, and P. N. Fultz. 2000. Distribution and quantification of human immunodeficiency virus type 1, strain JC499, proviral DNA in tissues from an infected chimpanzee. Virology 276:59–69. [DOI] [PubMed] [Google Scholar]
- 54.Wolfs, T. F. W., G. Zwart, M. Bakker, and J. Goudsmit. 1992. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189:103–110. [DOI] [PubMed] [Google Scholar]
- 55.Wolinsky, S. M., C. M. Wike, B. T. M. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137. [DOI] [PubMed] [Google Scholar]
- 56.Xiao, L., S. M. Owen, I. Goldman, A. A. Lal, J. J. deJong, J. Goudsmit, and R. B. Lal. 1998. CCR5 coreceptor usage of non-syncytium-inducing primary HIV-1 is independent of phylogenetically distinct global HIV-1 isolates: delineation of consensus motif in the V3 domain that predicts CCR-5 usage. Virology 240:83–92. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Leigh-Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science 261:1179–1181. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]