Abstract
The S2 gene of bluetongue virus, serotype 17, has been cloned, and the nonstructural protein NS2 has been expressed. Synthetic peptides matching regions within the amino acid sequence of NS2 were used to map three single-stranded RNA (ssRNA)-binding regions within the protein. A prokaryotic expression system was then used to generate a series of deletion mutants with the ssRNA-binding domains of NS2 removed, singly and in different combinations. These truncated proteins were expressed on a large scale and purified to near homogeneity. The affinity of each truncated protein towards ssRNA was then assayed by electrophoretic mobility shift assays. As a result, the three ssRNA-binding domains of BTV nonstructural protein NS2 have been conclusively localized, and removal of these three domains completely abrogates the ability of NS2 to bind to ssRNA.
Bluetongue virus (BTV) is a member of the Reoviridae family and is the prototype virus of the Orbivirus genus (9, 25). Transmitted by biting midges of the Culicoides genus, BTV is a pathogen of wild and domestic ruminants (1). The BTV genome consists of 10 double-stranded RNA (dsRNA) segments, which encode 11 viral proteins (19, 20). These genomic segments are enclosed by a double-shelled capsid, composed of seven structural proteins (24). Additionally, four nonstructural proteins have been identified inside BTV-infected cells (5). The role of each of the nonstructural proteins within the viral life cycle has not been conclusively determined, but they are presumed to be involved with the various steps of viral morphogenesis.
The 41-kDa nonstructural protein NS2 binds single-stranded RNA (ssRNA), is associated with the viral inclusion bodies, and is the only phosphorylated BTV protein (7). Similar ssRNA-binding proteins, such as the ϕNS of reovirus and the NS35 (NSP2) of rotavirus, appear to be a common feature of the members of the family Reoviridae (6, 11). They are suspected to be involved in the transport and condensation of the viral mRNAs, but their precise functions have not yet been established. Therefore, this study focused upon the ssRNA-binding domains of NS2 of BTV serotype 17 in order to more fully elucidate the roles of this protein within the replication of BTV.
MATERIALS AND METHODS
Preparation of viral dsRNA.
A seed stock for U.S. BTV serotype 17 (BTV-17) was obtained from the USDA Arthropod-Borne Animal Disease Research Laboratory, and then propagated and plaque purified as described by Kowalik and Li (12). Viral particles were purified on sucrose gradients (13), and then total dsRNA was extracted with sodium dodecyl sulfate (SDS)-KCl (16).
Cloning of BTV cDNAs.
A full-length cDNA for the BTV-17 S2 gene was generated from the purified BTV dsRNA by the Clamp-R method (13) of reverse transcription-PCR (RT-PCR). “Anchored” primer pairs [S2EXP(+) (5′-TCC-CCC-GGG-ATC-CAT-GGA-GCA-AAA-GCA-A-3′) and S2EXP(−) (5′-CGG-GAT-CCC-GGG-GTA-AGT-GTA-AAA-TCC-C-3′)], complementary to the termini of the S2 gene and containing a novel BamHI restriction site (26), were used to amplify the full-length S2 cDNA by PCR. A Techne PHC-3 Thermal Cycler was used for all PCRs.
The PCR product and the cloning vector pGEX2T (Pharmacia) were digested with BamHI (overnight digestion), and then the 5′ ends of the linearized vector were dephosphorylated by incubation with calf intestinal alkaline phosphatase. The BTV-17 S2 insert was ligated into the vector at a ratio of 8:1 by incubation with T4 DNA ligase. After ligation, 10 μl of the reaction mix was transformed to the XL-1 Blue strain of Escherichia coli (Stratagene) by the heat shock-calcium method adapted from Hanahan (3). Colonies were checked for the presence of plasmids containing the inserted gene by the procedure of Mangalathu and Bassett (17), and clones containing the inserted gene in the correct orientation were isolated.
Construction of BTV-17 S2 deletion mutants.
The S2Δ68 mutation was generated by digesting the full-length pGEX2T-B17S2 clone with StyI (restriction site at position 778 within the S2 sequence) and HindIII (site at position 944). Klenow fragment (5 U/μl; New England BioLabs) was then used to create blunt ends at each site by incubation with 33 μM deoxynucleoside triphosphates (dNTPs) for 15 min at 25°C. The ends were then ligated together with the 168-bp region removed.
The S2ΔN mutation was created by digestion of the full-length pGEX2T-B17S2 clone with EcoRI (restriction site at position 219 within the S2 sequence). The digested DNA was run on a 0.7% agarose gel along with DNA size markers, and a 904-bp band was cut out of the gel and purified with QuicKit GlasPac (National Scientific Supply). This DNA was then inserted into the EcoRI site of pGEX2T.
Primers S267(3) (5′-GAT-CTG-CAG-ATA-ATT-TGA-TCT-CA-3′) and S267(5) (5′-TAC-TGC-AGT-TAG-CGC-CAC-GGC-T-3′) were designed to flank the Δ67 region within the S2 sequence and introduced anchored PstI restriction sites. PCR amplification with the full-length S2 gene as a template was performed with the primers S2EXP(+) and S267(3), producing a 476-bp fragment, and S2EXP(−) and S267(5), producing a 605-bp fragment. The PCR products were digested with PstI, ligated together, and then digested with BamHI and electrophoresed on a 0.7% agarose gel. A band of 1,081 bp was cut out of the gel and ligated into pGEX2T at the BamHI site.
All other deletion mutants were generated by the techniques described above in different combinations. Inserts were checked by digestion with the appropriate enzymes, followed by electrophoresis on a 1% agarose gel along with DNA size markers. Orientations were confirmed by digestion with EcoRV (which has restriction sites at positions 238 and 478 within the S2 sequence and at position 4095 within pGEX2T). The clones pGEX2T-B17S2Δ67, pGEX2T-B17S2Δ68, and pGEX2T-B17S2Δ67/68 were confirmed by sequencing (Utah State University Biotechnology Center).
Upon isolation of each deletion mutant, plasmid DNA for each was purified from 100 ml of culture, aliquoted to microcentrifuge tubes, and stored at −20°C.
Expression of intact and truncated B17 NS2 proteins.
Each clone to be expressed was freshly transformed into the TOPP-Purple strain of E. coli (Stratagene) and grown into mid-log phase in 2-liter cultures. Isopropyl-β-d-thio-galactopyranoside (IPTG; Alexis Biochemicals) was then added to a final concentration of 2 mM to induce protein expression. After 4 h, the cells were harvested by centrifugation at 5,000 × g for 10 min with 1-liter bottles in a KA-9 rotor within a Sorvall RC-5B centrifuge. Cell pellets were transferred to 50-ml centrifuge tubes and stored at −80°C and then resuspended in 20 ml of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, and 1.4 mM KH2PO4 [pH 7.4]) and kept on ice. Lysozyme (500 mg) was added along with DNase I (50 mg), and the cells were lysed with a VirSonic-50 probe sonicator (Virtis). The cells were sonicated three times for 30 s each at 80% output power. The tubes were centrifuged at 4,000 × g for 10 min to pellet insoluble debris, and supernatants were stored at 4°C.
Protein analysis by SDS-PAGE and immunoblot assays.
Cell lysates were analyzed by discontinuous SDS-polyacrylamide gel electrophoresis (PAGE) (10% polyacrylamide) by the method of Laemmli (14). Gels were stained with Coomassie blue R250 (Sigma), or, alternately, proteins were transferred to nitrocellulose membranes from gels by a diffusional “sandwich” blot technique (4). After electrophoresis, SDS-PAGE gels were placed between two nitrocellulose membranes and compressed within a sandwich apparatus. This sandwich was then soaked for at least 24 h in a tank containing 25 mM Tris-HCl (pH 8.0), 192 mM glycine, and 20% methanol. The membranes were removed from the tank and blocked with 3% bovine serum albumin (BSA) in TBST buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) for 30 min. Each blot was then probed with antisera against bluetongue proteins, diluted in TBST at a ratio of 1:1,000 for polyclonal or oligoclonal antibodies or 1:5,000 for monoclonal antibodies. After incubation for 30 min on a rocking platform, the membranes were washed with TBST (three times for 5 min each), and then the appropriate secondary antibody (anti-rabbit or anti-mouse) conjugated to alkaline phosphatase (AP) was added at a 1:5,000 dilution. The membranes were incubated for another 30 min and then washed three times with TBST for 5 min each. Finally, the AP substrate solution was added for color development.
Purification of intact and truncated NS2 proteins.
Since the pGEX expression system was used, all proteins were produced as a fusion to glutathione S-transferase (GST) (21). An initial, partial purification was performed by anion-exchange chromatography with a POROS-20Q (quaternary amine) column on the BioCAD Perfusion Chromatography Workstation (PerSeptive Biosystems). Using isoelectric points predicted by ExPaSy for each deletion protein, the purification profile was run for each protein at pH 7.5 (to give each a net negative charge), at a flow rate of 20 ml/min, in 50 mM Tris-HCl buffer. With a double-step NaCl gradient elution, peaks were detected at A280 with a UV monitor, collected (1.5-ml fractions), and analyzed by SDS-PAGE. The characteristic peak fraction for each protein was then collected and pooled for further purification.
Each partially purified protein in the NS2 deletion series was dialyzed in PBS buffer to remove NaCl and then added to a column containing 10 ml of glutathione-conjugated Sepharose 4B (Amersham) in PBS, with the EconoSystem chromatography apparatus (Bio-Rad). With a flow rate of 1 ml/min, the resin was washed with at least 10 column volumes of PBS to remove unbound proteins, and then the GST fusion protein was eluted by the addition of 20 mM glutathione (reduced form) in 50 mM Tris-HCl (pH 8.0) plus 100 mM NaCl. Fractions were collected and analyzed at A280 in a spectrophotometer, and one peak was found to contain the desired GST fusion protein. The purification was repeated for each expressed protein in the deletion series, and then the concentration of each was determined by bicinchoninic acid assay (Pierce). Each protein was analyzed by SDS-PAGE, and the identity of each was confirmed by immunodetection.
In vitro transcription for synthesis of 32P-labeled BTV-17 S2 ssRNA transcripts.
The cDNA of BTV-17 S2 was cloned into the PstI site of the pGEM4Z vector (Promega). Recombinant plasmids with the inserted S2 gene in both orientations were obtained and then transformed into the XL-1 Blue strain of E. coli. Purified plasmids were linearized by either BamHI or HindIII digestion. BTV-17 S2 ssRNA transcripts were then transcribed and labeled by 32P in vitro with SP6 polymerase, as described by Hayama and Li (4, 18). Transcripts (both plus and minus sense) were treated with RNase-free DNase I, followed by phenol-CHCl3 extraction and isopropanol precipitation. A positive-sense BTV-11 S3 transcript was prepared in the same manner (by E. Hayama) and provided for use in dot blot analysis.
Dot blot analysis for ssRNA-binding activity of NS2 synthetic peptides.
Eight oligopeptides were designed, according to the deduced amino acid sequence of the serotype 17 NS2 protein (26). The peptides were chosen at locations with clusters of positively charged amino acids and at regions with predicted surface accessibility by GCG software (Genetics Computer Group, Madison, Wis.). The peptides were synthesized in the multiple-antigen peptide (MAP) format at the Utah StateUniversity Biotechnology Center, with eight identical oligopeptides connected to a lysine core (22). After each synthetic peptide was dissolved in PBS at 1 μg/μl, each peptide was then dot blotted onto a nitrocellulose membrane in twofold serial dilutions from 5 μg. The membrane was blocked for 30 min at room temperature in Denhardt’s solution (0.02% Ficoll 400, 0.02% polyvinylpyrrolidone, 0.02% BSA). The membrane was incubated with 2 μg of 32P-labeled BTV-11 S3 transcript for 30 min and then washed three times with Denhardt’s solution. After the membrane was air dried, it was exposed to Kodak film for 24 h for autoradiography.
EMSAs for intact and truncated NS2 proteins.
Purified NS2 was analyzed for its ability to bind to both positive- and negative-sense BTV-17 S2 ssRNA transcripts by electrophoretic mobility shift assays (EMSAs) (2, 4). One microgram of protein was mixed with 0.4 μg of each 32P-labeled S2 transcript and then run on a 4% polyacrylamide gel. The gel was vacuum dried on filter paper and exposed to Kodak film for 24 h, and then the film was developed.
After each of the NS2 proteins in the deletion series was purified to near homogeneity, each was examined for its ability to bind ssRNA by EMSA. ssRNA size markers (New England BioLabs) were used at 1 μg per assay and mixed with each protein (0, 2, 4, 6, or 8 μg per assay) in 0.5× Tris-borate-EDTA (TBE). The tubes were incubated for 5 min at room temperature, and then loading buffer (0.1% bromphenol blue, 0.1% xylene cyanole, 15% Ficoll 400, in 0.5× TBE buffer) was added. Each assay was analyzed by electrophoresis on a 1% agarose gel stained with ethidium bromide (EtBr; 0.05 μg/ml). All buffers were prepared with diethyl pyrocarbonate-treated double-distilled H2O (ddH2O) to eliminate potential RNase activity.
RESULTS
Peptide mapping of ssRNA-binding domains.
Eight NS2 sequence-specific synthetic oligopeptides were assayed for their ability to bind to ssRNA. The peptides were applied to a membrane that was then probed with a 32P-labeled BTV-11 S3 positive-sense transcript, of which we had an abundant supply (courtesy of E. Hayama). Peptides N, 67, and 68 showed ssRNA-binding activity (Fig. 1), while the other five peptides did not demonstrate any affinity towards ssRNA. Identical results were found in EMSAs with yeast ssRNAs (data not shown). The sequences of each peptide and their corresponding locations within NS2 are shown in Fig. 2.
FIG. 1.
Dot blot analysis of ssRNA binding by synthetic NS2 peptides. Each synthetic peptide derived from the amino acid sequence of NS2 was applied to a nitrocellulose membrane in twofold dilutions from 5 μg. The membrane was probed with 2 μg of 32P-labeled BTV-11 S3 (positive sense) ssRNA and then exposed to film for 24 h.
FIG. 2.
Synthetic peptides derived from the amino acid sequence of NS2. Peptide sequences were chosen within the amino acid sequence of NS2 serotype 17 and then synthesized in the MAP format. All are shown in their positions relative to the full-length NS2 protein.
Construction of S2 deletions.
A series of deletions corresponding to the N, 67, and 68 domains were constructed within the BTV (serotype 17) S2 gene and inserted into the pGEX2T vector. Each deletion construct was designed to maintain the frame of the open reading frame (ORF) around the removed portion of the gene. All constructs were generated in the XL-1 Blue strain of E. coli. After selection for the presence of inserts by restriction digests and agarose gel electrophoresis, each clone was checked for the correct orientation of the insert.
Expression and purification of the NS2-GST fusion proteins.
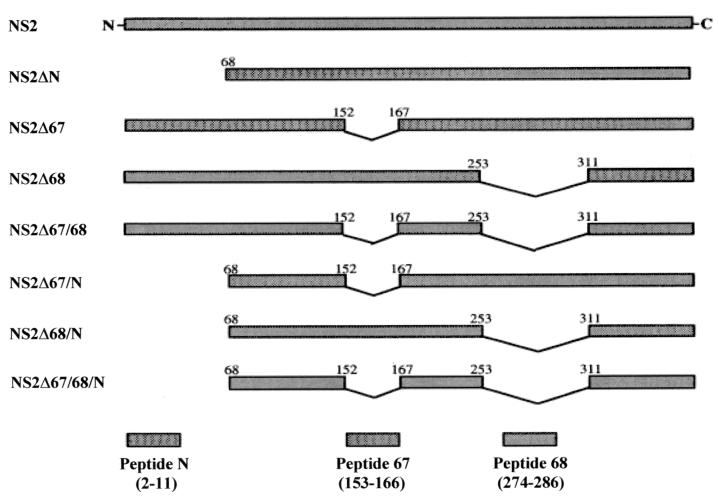
A schematic diagram is shown in Fig. 3 for all constructs within the NS2 deletion series. The sizes for each of the intact or truncated NS2 proteins are predicted in Table 1.
FIG. 3.
Schematic diagram of all members of the NS2 deletion series. All constructs were expressed as carboxyl-terminal fusions to GST (not shown). The amino acids flanking each deletion site are indicated. The oligopeptides corresponding to each deleted region are shown below.
TABLE 1.
Predicted sizes of each NS2 deletion mutant and corresponding proteina
| Clone | Insert size (bp) | ORF | No. of amino acids | Protein size (kDa) | GST fusion protein size (kDa) |
|---|---|---|---|---|---|
| pGEX2T-B17S2 | 1,123 | 1062 | 354 | 41 | 67 |
| 2T-S2ΔN | 904 | 843 | 281 | 32 | 58 |
| 2T-S2Δ67 | 1,081 | 1020 | 340 | 39 | 65 |
| 2T-S2Δ68 | 955 | 894 | 298 | 34 | 60 |
| 2T-S2Δ67/68 | 913 | 874 | 291 | 33 | 59 |
| 2T-S2Δ67/N | 862 | 822 | 274 | 32 | 58 |
| 2T-S2Δ68/N | 736 | 696 | 232 | 27 | 53 |
| 2T-S2Δ67/68/N | 694 | 654 | 218 | 25 | 51 |
Each insert sequence for the S2 deletion mutants was entered into the SwissProt Tools at ExPaSy (Expert Protein Analysis System; www.expasy.ch). The software was then able to predict the number of amino acids and the size of each truncated protein from the ORF of each deletion mutant. The size of each GST fusion protein was also predicted (GST alone is 26 kDa).
Each truncated NS2 protein was expressed within and isolated from the XL-1 Blue strain of E. coli. However, we have found that the TOPP-Purple strain produced a higher yield of the protein for each transformed plasmid construct. Therefore, each plasmid was transformed to the TOPP-Purple strain and plated on Luria-Bertani agar. Colonies were picked from the plates and grown overnight for each round of expression on a 2-liter scale. When cells were in the mid-log phase of growth, expression of the fusion protein was induced by the addition of IPTG. A prior time course study established that optimal protein expression occurred at 4 h postinduction, so all induced cells were harvested 4 h after the addition of IPTG. The overexpressed fusion proteins accumulated within the bacterial cells, allowing isolation and concentration by centrifugation into a cell pellet.
Initial attempts to purify the NS2-GST fusion proteins by affinity chromatography, with glutathione-conjugated agarose beads (“G-beads”), were unsuccessful in isolating the proteins to homogeneity. An alternate purification strategy was therefore devised, utilizing a preliminary ion-exchange chromatography step. The BioCAD system provided a means for purification with a high flow rate, using POROS Perfusion beads containing an ion-exchange matrix. Since the isoelectric point for each fusion protein in the NS2 deletion series was predicted to be within a 5.4 to 5.6 range, a buffer in the pH 7.0 to 7.5 range, which would impart a net negative charge upon each protein, was chosen. Therefore, a POROS-Q (quaternary amine, an anion-exchanger) column was used with the BioCAD workstation for the purification of each protein. A double-step salt gradient was designed within an anion-exchange profile to optimize the resolution of the peaks eluted from the column, and a similar chromatogram was seen for each protein in the deletion series. Multiple purification runs were done for each protein, and fractions were collected over the elution gradient. The truncated proteins were not always found within the same elution fractions (due to the slight differences in predicted pI), but a characteristic “shape” was always observed for the elution profile, allowing collection of each truncated NS2 protein in one peak fraction. The eluted peaks were analyzed by SDS-PAGE, and those containing the desired proteins were pooled. A total of 60 ml (40 fractions) was collected for each protein, and the samples were stored at 4°C.
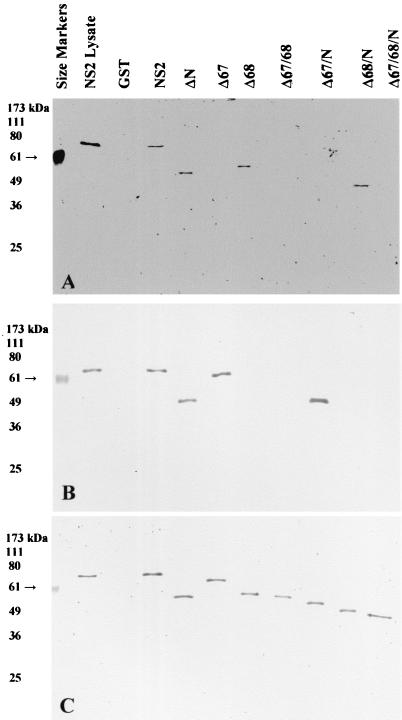
After this partial purification based on charge, affinity chromatography was again attempted. All samples were dialyzed in PBS to remove the salt from the anion-exchange elution gradient, and then 6 ml was applied to a column containing glutathione conjugated to Sepharose 4B rather than agarose. This resin allowed a higher flow rate for washing away unbound proteins and had been refined for a very specific affinity towards GST fusions by the manufacturer (Amersham). Using an EconoSystem chromatography apparatus, the column was washed by at least 10 column volumes of PBS. Bound proteins were then eluted by a “step” gradient of 20 mM glutathione in 50 mM Tris-HCl (pH 8) plus 100 mM NaCl. The extra washing, higher glutathione concentration, and presence of salt in the elution buffer resolved the eluted GST fusion proteins down to single bands per peak (as measured at A280), as shown by SDS-PAGE (Fig. 4). Peak fractions for each intact or truncated NS2 protein (and a GST control) were collected and analyzed by SDS-PAGE (Fig. 5) and were found to be of the correct size relative to size markers run in the same gel. Immunoblotting was performed after SDS-PAGE, and the blots were probed by oligoclonal antibodies generated by peptides 67 (Fig. 6A) and 68 (Fig. 6B) and by a series of monoclonal antibodies produced against NS2 (Fig. 6C). Immunodetection confirmed that the purified bands were members of the NS2 deletion series (Fig. 6C). The oligoclonal antibodies specific to regions 67 and 68 did not react with those truncated NS2 proteins in which the corresponding regions were deleted (Fig. 6A, lanes 6, 8, 9, and 11, and B, lanes 7, 8, 10, and 11, respectively).
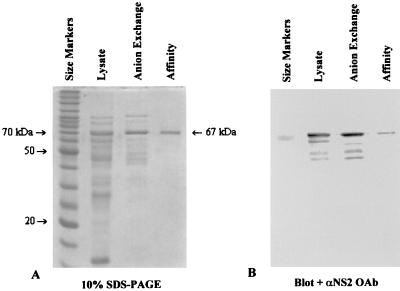
FIG. 4.
SDS-PAGE and immunoblot analysis of purified NS2. Samples (50 μl) were taken from cell lysate, from purified fractions after anion-exchange chromatography, or from fractions after affinity chromatography with glutathione-Sepharose 4B. Ten microliters of each sample was loaded per well of a 10% gel. The gels were stained with Coomassie blue (A) or blotted to nitrocellulose membranes (B) and probed with an oligoclonal antibody generated from NS2 peptide 67. The primary antibody was used at a 1:1,000 dilution, and the secondary antibody (conjugated to AP) was used at 1:5,000.
FIG. 5.
SDS-PAGE analysis of purified intact and truncated NS2 proteins. Samples (50 μl) were taken from each purified fraction, and 50 μl of loading buffer was added. The samples were boiled, and 10 μl was added to the wells of a 10% polyacrylamide discontinuous SDS-PAGE. All GST fusion proteins are shown relative to prestained BenchMark size markers (lane 1).
FIG. 6.
Immunoblot analysis of purified intact and truncated NS2 proteins. Nitrocellulose membrane blots, corresponding to the SDS-PAGE shown in Fig. 5, were probed with antibodies against NS2. Oligoclonal antibodies generated by peptide 67 (A) or peptide 68 (B) were used at a 1:1,000 dilution. A mixture of monoclonal antibodies (C) was used at a dilution of 1:5,000 for each (B87.29.7, B87.29.4, F13.20.2, and F12.17.2, all reacting against unknown epitopes within NS2). Appropriate secondary antibodies (anti-rabbit or anti-mouse), conjugated to AP, were used at a 1:5,000 dilution.
ssRNA-binding assays for the purified intact and truncated NS2 proteins.
Upon purification to near homogeneity, each NS2-GST fusion protein was dialyzed against PBS (three buffer changes over 2 days, 4°C) to remove salt from the elution step. The decision was made to assay the intact fusion proteins for affinity towards ssRNA, due to the difficulty in completely removing the GST portion of the protein by thrombin digestion. Prior experiments showed incomplete cleavage of NS2 from GST, even with extended incubation times and high concentrations of thrombin. GST has a negligible effect upon the behavior of a fusion protein (10). In the studies presented here, GST alone had no affinity towards ssRNA, and GST conjugated to NS2 was found to have no effect on the ability of NS2 to bind to ssRNA.
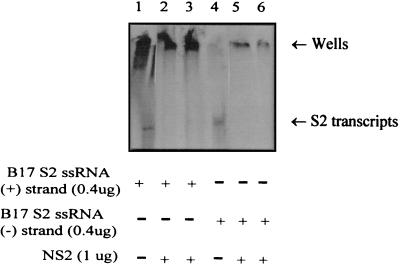
Earlier studies have shown nonspecific binding to poly(U)-Sepharose by NS2 isolated from cells infected by BTV (23). NS2 produced by recombinant baculovirus and purified from insect cells also exhibited nonspecific affinity towards ssRNA. Recombinant NS2 produced as a GST fusion protein showed the same ssRNA-binding activity towards both positive- and negative-sense viral ssRNA transcripts (Fig. 7).
FIG. 7.
Affinity of NS2 for positive- and negative-sense BTV-17 (B17) S2 ssRNA transcripts. EMSAs were used to show binding of 1 μg of GST/NS2 to 0.4 μg of 32P-labeled plus-sense BTV-17 S2 ssRNA (lanes 2 and 3) and to 0.4 μg of 32P-labeled negative-sense BTV-17 S2 ssRNA (lanes 5 and 6). The mobility of the radiolabeled ssRNA transcripts was retarded at the wells of the PAGE gel when the NS2 protein was present, while unbound ssRNAs are shown in lanes 1 and 4.
In order to establish the general ssRNA-binding activity of each intact or truncated NS2 protein, EMSAs were performed for each purified fusion protein. An ssRNA size ladder, a commercial product generated by in vitro transcription from seven different DNA templates (500 to 9,000 bp), was first used for the EMSAs. The denaturing sample buffer accompanying the size markers contained 7 M urea, which would potentially disrupt the interaction of the truncated proteins with the ssRNA, so it was not used. All assays were performed in 0.5× TBE buffer, and all reagents were prepared with diethyl pyrocarbonate-treated ddH2O. In accordance with the manufacturer’s recommendation, 1 μg (0.5 μg/μl) of ssRNA was used per assay, for optimal viewing with EtBr staining. After incubation with each protein, samples were analyzed by agarose gel electrophoresis. Affinity towards ssRNA resulted in the formation of a protein-RNA complex, which inhibited the ability of the RNA to move within the gel. Therefore, ssRNA binding by each protein was determined by the degree of retarded movement of the ssRNA in the gel as the protein concentration was increased.
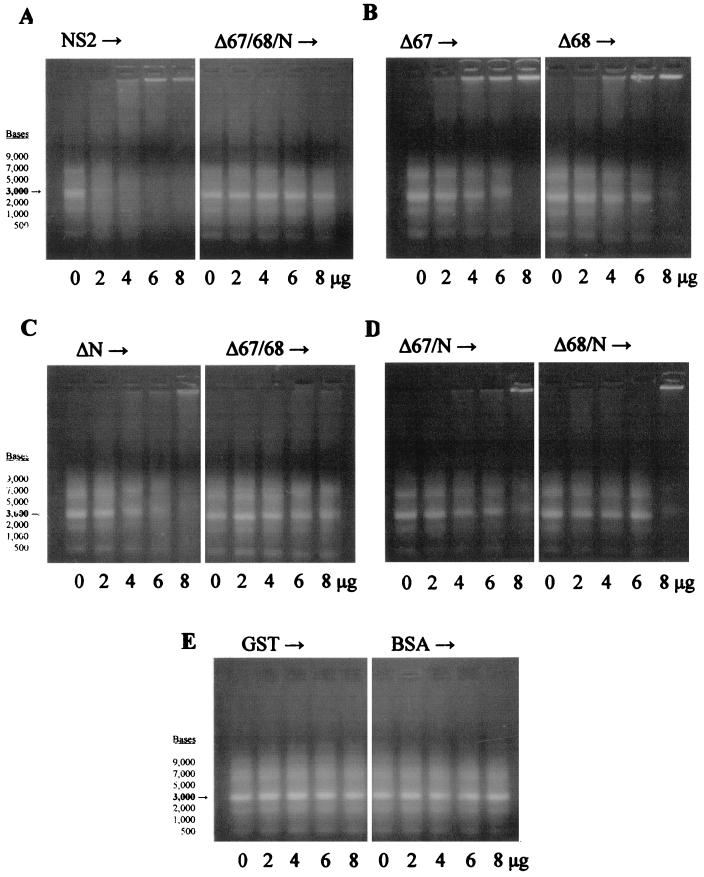
The results show complete retardation of 1 μg of the ssRNA size markers by the NS2 fusion protein at 6 μg, but no inhibition of mobility by the Δ67/68/N protein (Fig. 8A) over the same concentration range. With these three ssRNA-binding domains removed, NS2 completely lost its affinity towards ssRNA. This confirms the importance of these three domains towards binding ssRNA, but the affinity for ssRNA shown by each truncated protein may also be affected by the conformational changes resulting from the deletion of each domain.
FIG. 8.
EMSA for NS2 and truncated NS2 proteins with ssRNA size markers. The ssRNA markers (9,000, 7,000, 5,000, 3,000, 2,000, 1,000, and 500 bases) were used at 1 μg per assay, with each purified fusion protein added at 0, 2, 4, 6, or 8 μg per sample. Retardation of ssRNA mobility was demonstrated by the retention of movement relative to the loading wells (shown at the top of each panel). The 3,000-base ssRNA size marker is highlighted in each gel. Panel A shows the intact NS2 protein and the triple deletion Δ67/68/N, panel B shows the single deletions Δ67 and Δ68, panel C shows the single deletion ΔN and the double mutant Δ67/68, panel D shows the double deletions Δ67/N and Δ68/N, and panel E shows the control proteins GST and BSA.
Those proteins with the deletion of a single binding domain, either ΔN, Δ67, or Δ68, still demonstrated the ability to bind ssRNA, but showed less affinity than intact NS2 (Fig. 8B and C). At 8 μg, all three single-deletion proteins could completely retard the RNA. The Δ67 mutant appeared to have slightly greater affinity for the ssRNA size markers than did the ΔN or Δ68 mutants. This suggests a weaker contribution of the 67 region to ssRNA binding by NS2 relative to the other two domains.
The double-deletion proteins Δ67/68, Δ67/N, and Δ68/N demonstrated less binding to ssRNA than the single-deletion forms, and 8 μg did not completely inhibit ssRNA mobility (Fig. 8C and D). The truncated Δ67/68 protein showed lower affinity for the RNA markers than did the Δ67/N or Δ68/N proteins. This might indicate not only differences in the affinity of each region towards ssRNA, but a cooperative or sequential role for each domain in binding to ssRNA by the intact NS2 protein.
The GST (purified in the same manner as all the other fusion proteins) and BSA control proteins demonstrated no affinity towards the ssRNA markers (Fig. 8E).
Similar results were demonstrated in assays performed with yeast ssRNA (data not shown). It was necessary to use 10 μg of yeast ssRNA per assay for visualization with EtBr, and the yeast RNA (primarily tRNAs) was seen on agarose gels in the ∼100- to 200-bp range. Intact NS2 was able to bind the ssRNA in an apparent “double-stepwise” manner, with 80 μg of protein completely inhibiting mobility in the gel. This again seems to indicate cooperative or interactive binding between domains, with differential affinity towards ssRNA. Δ67/68/N showed no retardation of the yeast ssRNA in the gel over the concentration range assayed.
The deletions of a single binding domain, ΔN, Δ67, and Δ68, resulted in a reduced affinity towards yeast ssRNA. The retardation of the yeast RNA within the gel also demonstrated a stepwise binding pattern. The truncated Δ67 protein had a slightly greater affinity towards the RNA than those of the other two single-deletion proteins.
Removal of two binding domains produced a further reduction in the affinity towards the yeast ssRNA. The Δ67/68, Δ67/N, and Δ68/N proteins could not completely retard the movement of the RNA even at 80 μg, but a double-step binding pattern was still observed in the gels. The Δ68/N mutant showed slightly less affinity towards the yeast ssRNA (i.e., not as much RNA was held in the gel wells at a high protein concentration), but much more RNA was bound in the first step, as compared to the patterns shown by the Δ67/68 and Δ67/N proteins. This again implies some degree of differential affinity and cooperative interaction between ssRNA-binding domains. In this case, removal of the 67 region has less of an effect on the overall affinity towards ssRNA, but removal of the other two domains affects the stepwise binding pattern shown towards the yeast ssRNA.
The control proteins GST and BSA again demonstrated no affinity towards the yeast ssRNA. The abilities of all of the intact or truncated NS2 proteins to bind the two types of ssRNAs tested are summarized in Table 2.
TABLE 2.
Summary of ssRNA binding by intact or truncated NS2 proteins
| Protein | Binding levela
|
|
|---|---|---|
| ssRNA markers | Yeast ssRNA | |
| NS2 | ++++ | ++++ |
| ΔN | +++ | ++ |
| Δ67 | +++ | +++ |
| Δ68 | ++ | ++ |
| Δ67/68 | + | + |
| Δ67/N | ++ | + |
| Δ68/N | ++ | ++ |
| Δ67/68/N | − | − |
| GST | − | − |
| BSA | − | − |
++++, Complete retention of ssRNA in wells at lower protein concentrations; +++, complete retention of ssRNA only at highest protein concentrations; ++, partial retention of ssRNA in wells at highest protein concentrations; +, some retardation of mobility observed in gel, ssRNA not held in wells; −, no binding to ssRNA.
DISCUSSION
Nonstructural proteins play key roles within the replicative cycle of viruses. While encoded by the viral genome and produced within infected cells, NS proteins are not part of the virions and do not move with the viral particles into subsequent host cells. At each round of infection, however, nonstructural proteins are vital for regulating different mechanisms of the host cell or for assisting in the assembly of progeny virions (15).
The nonstructural protein NS2 has been proposed to fulfill such a role within the BTV replication and assembly process. Prior investigations have established the affinity of NS2 towards ssRNA, its oligomerization, and its presence at the VIB matrix (7, 8). These findings suggest that BTV NS2 is interacting with the viral mRNAs at the sites of assembly of new viral core particles. The presence and function of NS2 at the VIBs had not been conclusively explained, however.
Our peptide mapping established three regions within the amino acid sequence of NS2 that could bind ssRNA in vitro, indicating that synthetic oligopeptides corresponding to these regions could “mimic” the nonspecific affinity for ssRNA shown by the native NS2 protein. Specifically, peptide N (EQKQRRFTKN, residue positions 2 to 11), 67 (DIREL-RQKIKNERE, positions 153 to 166), and 68 (EKVAKQIKLKDER, positions 274 to 286) were found to bind ssRNA, out of eight peptides chosen for their charged nature and predicted surface accessibility. The designation “N” was related to the N-terminal region of the NS2 protein, and the nomenclature for the peptides 67 and 68 was based on the order in which a series of peptides were synthesized for various regions, predicted as antigenic epitopes or as highly charged domains, within all 11 of the bluetongue proteins. The NS2 peptides were produced in the MAP format and were found to have affinity for both positive- and negative-strand BTV transcripts, yeast tRNAs, and poly(U)-Sepharose. The same three peptides demonstrated no affinity for dsRNA, ssDNA, or dsDNA, however. Peptides were also used to generate anti-BTV oligoclonal antibodies in rabbits, as well as for the investigation of the affinity of several BTV proteins towards different nucleic acid species.
In the present study, three ssRNA-binding domains have been localized within NS2. Removal of each domain reduced the affinity of NS2 towards ssRNA, and the deletion of all three domains completely eliminated the ability of NS2 to bind to ssRNA. Different degrees of affinity towards ssRNA were also observed among the different truncated proteins, indicating that each of these three domains has differential affinity. The deleted regions were of different sizes, and the degree of reduced affinity towards ssRNA did not seem to correspond to the size of each deletion. It seems unlikely that the effect is due simply to an alteration in the conformation of each truncated protein, but this possibility can’t be completely ruled out.
A stepwise binding pattern was observed for yeast ssRNA in EMSAs. As protein concentrations were increased, intermediate complexes could be seen before there was complete retardation of ssRNA mobility. This pattern was observed to different degrees for each truncated NS2 protein (except for Δ67/68/N, which did not bind to the yeast ssRNA). This might further indicate a differential affinity by each domain towards the ssRNA or a sequential binding to ssRNA by the three ssRNA-binding domains. The pattern may also be an artifact related to the use of yeast single-stranded tRNA for the assays, since it was not seen as readily with the ssRNA size markers. It also remains to be determined if NS2 binds to three mRNAs, one by each ssRNA-binding domain, or to one mRNA by all three domains.
All intact and truncated NS2 proteins used for this project were produced by a prokaryotic expression system. Since BTV infects mammalian cells, experiments are in progress to examine how and where intact and truncated NS2 molecules are expressed inside of eukaryotic cells. We will also investigate the effect of heterologously expressed NS2 upon viral replication in cells that are infected by BTV. The presence or absence of the ssRNA-binding domains of NS2 will allow further elucidation of the role of this nonstructural protein within the BTV viral life cycle.
Acknowledgments
We thank I-Jen Huang for construction of the initial pGEX2T-B17S2 clone, and Emiko Hayama for production of the 32P-labeled BTV-11 S3 transcripts.
This work was supported in part by Utah Agricultural Experiment Station Project 537 and USDA grant 90-37266-5567.
Footnotes
This article has been approved as AES paper 7331.
REFERENCES
- 1.Barratt-Boyes, S. S., and N. J. MacLachlan. 1995. Pathogenesis of bluetongue virus infection of cattle. J. Am. Vet. Med. Assoc. 206:1322–1329. [PubMed] [Google Scholar]
- 2.Chodosh, L. A., R. W. Carthew, and P. A. Sharp. 1986. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol. Cell. Biol. 6:4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. [DOI] [PubMed] [Google Scholar]
- 4.Hayama, E., and J. K.-K. Li. 1994. Mapping and characterization of antigenic epitopes and the nucleic acid-binding domains of the VP6 protein of bluetongue viruses. J. Virol. 68:3604–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huismans, H. 1979. Protein synthesis in bluetongue virus-infected cells. Virology 92:385–396. [DOI] [PubMed] [Google Scholar]
- 6.Huismans, H., and W. K. Joklik. 1976. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology 70:411–424. [DOI] [PubMed] [Google Scholar]
- 7.Huismans, H., A. A. Van Dijk, and A. R. Bauskin. 1987. In vitro phosphorylation and purification of a nonstructural protein of bluetongue virus with affinity for single-stranded RNA. J. Virol. 61:3589–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyatt, A. D., and B. T. Eaton. 1988. Ultrastructural distribution of the major capsid proteins within bluetongue virus and infected cells. J. Gen. Virol. 69:805–815. [DOI] [PubMed] [Google Scholar]
- 9.Joklik, W. K. 1983. The members of the family Reoviridae, p.1–6. In W. K. Joklik (ed.), The Reoviridae. Plenum Press, New York, N.Y.
- 10.Kaelin, W. G., T. Chittenden, and D. M. Livingston. 1991. The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell 65:1073–1082. [DOI] [PubMed] [Google Scholar]
- 11.Kattoura, M. D., L. L. Clapp, and J. T. Patton. 1992. The rotavirus nonstructural protein, NS35, possesses RNA-binding activity in vitro and in vivo. Virology 191:698–708. [DOI] [PubMed] [Google Scholar]
- 12.Kowalik, T. F., and J. K.-K. Li. 1987. The genetic relatedness of United States prototype bluetongue viruses by RNA/RNA hybridization. Virology 158:276–284. [DOI] [PubMed] [Google Scholar]
- 13.Kowalik, T. F., Y.-Y. Yang, and J. K.-K. Li. 1990. Molecular cloning and comparative sequence analysis of bluetongue virus S1 gene by selective synthesis of specific full-length DNA copies of dsRNA genes. Virology 177:820–823. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 15.Levy, J. A., H. Fraenkel-Conrat, and R. A. Owens. 1994. Consequences of virus infection to the cell, p.271–316. In H. Fraenkel-Conrat and and R. A. Owens (ed.), Virology, 3rd ed. Prentice-Hall, Englewood Cliffs, N.J.
- 16.Li, J. K.-K., T. Johnson, Y.-Y. Yang, and V. Shore. 1989. Selective separation of virus proteins and dsRNA by SDS-KCl precipitation. J. Virol. Methods 26:3–16. [DOI] [PubMed] [Google Scholar]
- 17.Mangalathu, S. R., and C. L. Bassett. 1994. Flexibility in the boiling method of extracting plasmid DNA directly from cell culture. BioTechniques 16:376–380. [PubMed] [Google Scholar]
- 18.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertens, P. P. C., F. Brown, and, P. V. Sangar. 1984. Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virology 135:207–217. [DOI] [PubMed] [Google Scholar]
- 20.Roy, P. 1992. Bluetongue virus proteins. J. Gen. Virol. 73:3051–3064. [DOI] [PubMed] [Google Scholar]
- 21.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31–40. [DOI] [PubMed] [Google Scholar]
- 22.Tam, J. P. 1985. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 85:5409–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uitenweerde, J. M., J. Theron, M. A. Stoltz, and H. Huismans. 1995. The multimeric nonstructural NS2 proteins of bluetongue virus, African horsesickness virus, and epizootic hemorrhagic disease virus differ in their single-stranded RNA-binding ability. Virology 209:624–632. [DOI] [PubMed] [Google Scholar]
- 24.Verwoerd, D. W., H. J. Els, E.-M. De Villiers, and H. Huismans. 1972. Structure of the bluetongue virus capsid. J. Virol. 10:783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verwoerd, D. W., H. Huismans, and B. J. Erasmus. 1979. Orbiviruses, p.283–345. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 14. Plenum Press, New York, N.Y.
- 26.Yang, Y.-Y., J.-F. Chiou, G.-Y. Hwang, I.-J. Huang, and J. K.-K. Li. 1992. Evolutionary analyses of five US bluetongue viruses using the cognate S2 genes. Virus Res. 25:241–249. [DOI] [PubMed] [Google Scholar]