Abstract
The tegument of herpesvirus virions is a distinctive structure whose assembly and function are not well understood. The herpes simplex virus type 1 VP22 tegument protein encoded by the UL49 gene is conserved among the alphaherpesviruses. Using cell biology and viral genetics, we provide an initial characterization of the pseudorabies virus (PRV) VP22 homologue. We identified three isoforms of VP22 present in PRV-infected cells that can be resolved by polyacrylamide gel electrophoresis. The predominant form is not phosphorylated and is present in virions, while the other two species are phosphorylated and excluded from virions. VP22 localized to the nucleus by 6 h postinfection, as determined by immunofluorescence and cell fractionation. VP22 immunofluorescence in the nucleus was both diffuse and in punctate structures. The punctate nuclear localization was the most pronounced form of staining and did not localize exclusively to sites of viral DNA replication. Unexpectedly, a VP22 null mutant had no obvious phenotypes during tissue culture infections and was similar to the wild type in all respects. Moreover, the VP22 null mutant was as virulent and neuroinvasive as the wild-type virus after infection of the rodent eye and spread to the brain using both anterograde and retrograde neuronal circuits.
Herpesvirus virions are characterized in part by the presence of tegument, the distinctive proteinaceous layer between the nucleocapsid and envelope. Despite the fact that tegument is an important structural component of virions, there exists rudimentary and often conflicting information on the site(s) of tegument assembly, and the organization, stoichiometry and function of tegument proteins. Recent reports of mRNAs in the virions of human cytomegalovirus (4) and herpes simplex virus (HSV) (45), a subset of which may be acquired along with tegument during final envelopment (22), underscore the complex biology inherent in the herpesvirus tegument. Tegument components are delivered to the cell immediately after fusion of the virus envelope with the plasma membrane and therefore can have an immediate effect on the cell. Some tegument proteins have been characterized as transcription activators, kinases, or RNases (42) affecting viral and host gene expression.
The alphaherpesviruses encode a conserved cluster of four tegument genes named UL46, UL47, UL48, and UL49 (1, 25, 34, 44, 54). These genes encode four tegument proteins, known in HSV-1 as VP11/12 (UL46), VP13/14 (UL47), VP16 (UL48), and VP22 (UL49) (34). Of these proteins, the transcription activator VP16 has been studied extensively, whereas considerably less is known about the function of the other three proteins in this cluster. Interestingly, VP16 interacts with each of the other three tegument proteins encoded in the cluster (7, 14, 24, 30, 55), but the structure and function of these complexes are not well characterized.
The HSV-1 VP22 protein has attracted recent attention because it has been shown to have the property of intercellular spread (11, 16, 38). VP22 also binds avidly to chromatin and is capable of binding to and stabilizing microtubules (17, 20). However, the biological relevance of such actions is not understood. In addition, the cellular localization of VP22 is somewhat controversial. In immunofluorescence assays with an antibody that recognizes phosphorylated and nonphosphorylated forms of HSV-1 VP22, the protein was found first in the cytoplasm early after infection and then in the nucleus late in infection (40). In contrast, a study using a monoclonal antibody that recognizes only the nonphosphorylated form of VP22 found that this form localizes to the nucleus early in infection and is predominantly cytoplasmic late in infection (35). An HSV-1 enhanced green fluorescent protein (EGFP)-VP22 fusion protein localized exclusively to the cytoplasm and was never found in the nucleus (19). Finally, the VP22 homologues expressed by bovine herpesvirus 1 (BHV-1) and Marek’s disease virus (MDV) were found predominantly in the nucleus during infection (12, 28). The reason for these discrepancies is unclear, and the importance of the subcellular localization and phosphorylation state of VP22 during infection remains to be established.
Despite considerable work describing the cellular localization of VP22, genetic studies of the UL49 gene encoding VP22 remain rudimentary. A VP22-null mutant was first constructed in BHV-1 and exhibited significantly reduced growth in tissue culture (28). Importantly, this VP22-null virus exhibited reduced virulence after intranasal infection of cattle (29). An HSV-1 VP22-null virus has not yet been reported, but an HSV-1 mutant expressing a truncated form of VP22 has been characterized (41). This report demonstrated that the carboxy-terminal domain of VP22 is necessary for virion incorporation and efficient cell-to-cell spread of the virus.
The purpose of this study was to provide an initial characterization of the pseudorabies virus (PRV) VP22 homologue with the following experimental goals: (i) investigate the subcellular localization of PRV VP22 during infection, (ii) identify the VP22 isoforms present in infected cells and purified virions, and (iii) construct a VP22-null mutant and revertant to test the role of VP22 in neuroinvasion and virulence after rodent eye infection.
(A portion of this work was completed by H.C.W. in fulfillment of a Princeton University undergraduate senior thesis.)
MATERIALS AND METHODS
Cells and virus strains.
The PK15 (pig kidney) cell line was used for propagation of all PRV strains. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), while viral infections were performed in DMEM supplemented with 2% FBS. For studies involving cell cycle arrest in G1 phase, CV-1 (African green monkey kidney) cells were incubated in DMEM with 1% FBS and 300 μM n-butyrate (Sigma) prior to and during the course of infection. For plaque formation during virus purification, PK15 cells were maintained in 1% Methocel in DMEM supplemented with 2% FBS.
Antiserum.
Rabbit polyclonal antiserum to VP22 and goat polyclonal antiserum to gC (282) have been described previously (3, 43). Rb133 is a rabbit polyvalent antiserum against acetone-inactivated PRV Becker and recognizes all of the major envelope glycoproteins (9). Rabbit polyclonal antiserum to EGFP was purchased from Clontech, and mouse monoclonal antibody to bromodeoxyuridine (BrdU) was purchased from Boehringer Mannheim.
Construction of plasmids.
A synthetic oligonucleotide linker was formed by annealing oligonucleotides 5′-CAGATCTAGGACCCTACCGC-3′ and 5′-GGTAGGGTCCTAGATCTGACGT-3′. This linker was cloned into the AatII and SacI sites of pGEM-5Zf to create pHW2, containing a novel BglII site. pGS138 contains the PRV Becker BamHI-1 fragment (2) cloned into pBeloBAC11. The UL49 gene with flanking sequence was subcloned from pGS138 by inserting a BamHI/SalI fragment into the BglII and SalI sites of pHW2, resulting in pTD12. Two oligonucleotides partially homologous to the EGFP gene with one containing an overhanging XhoI site (5′-AGTCTCGAGCAAGGGCGAGGAGCTG-3′) and the other an overhanging PstI site (5′-ACATCTGCAGTTGAGCTCAAGATCTGAGTCC-3′) were used for PCR.
The EGFP open reading frame was amplified from pEGFP-C1 (Clontech) by Pfu polymerase (Stratagene). The EGFP PCR product was digested with XhoI and PstI and cloned into complementary sites in pTD12 to create the VP22-null construct pTD14. The VP22-null construct was designed to fuse the fourth amino acid of VP22 to the second amino acid of the EGFP open reading frame so that there is only one start methionine. An in-frame stop codon is present in the EGFP DNA fragment prior to the PstI site of UL49.
Construction of VP22-null mutant virus and corresponding revertant.
PRV Becker DNA was isolated as previously described (48). The UL49 open reading frame was replaced with the EGFP open reading frame by homologous recombination after cotransfection of PRV Becker nucleocapsid DNA and pTD14 DNA on PK15 cells. After complete cytopathic effect was observed, the infected cells were harvested, lysed by freeze-thawing, diluted, and replated on PK15 cells to allow individual plaques to form. EGFP-positive plaques were picked and plaque purified three times. Potential recombinants were confirmed by Southern blot analysis.
The substitution of the UL49 gene with EGFP introduced novel BglII and BspEI sites and resulted in the loss of a StyI site. One such recombinant was proven to have the UL49 gene replaced by the EGFP open reading frame and was named PRV 175. This VP22-null mutant expresses EGFP under control of the viral VP22 promoter. A revertant of PRV 175 was constructed by marker rescue as follows. PRV 175 nucleocapsid DNA was cotransfected with pTD12 containing the wild-type UL49 gene. EGFP-negative plaques were picked and purified as before. One such viral isolate was demonstrated by Southern blot analysis to have the EGFP insertion replaced with the wild-type UL49 gene and was named PRV 175R.
Western blot analysis.
Monolayers of PK15 cells were infected with PRV Becker, PRV 175, or PRV 175R at a multiplicity of infection (MOI) of 10. For cell lysates, at 6 or 16 h postinfection, the cells were washed with phosphate-buffered saline (PBS) and harvested in lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 1% NP-40, 1% sodium deoxycholate, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]). Cell lysates were centrifuged at 2,000 × g for 3 min at 4°C, the supernatant was collected as the cytoplasmic fraction, and the nuclear pellet was washed in PBS and resuspended in an equal volume of PBS-0.2 mM PMSF.
Virus particles were purified from the pooled medium from 12 15-cm-diameter dishes at 16 h postinfection. The medium was clarified of cell debris by centrifugation at 1000 × g for 10 min at 4°C and pelleted through 30% sucrose in PBS at 100,000 × g for 60 min at 4°C. The viral pellet was resuspended in TNE (150 mM NaCl, 50 mM Tris [pH 7.4], 0.01 M EDTA), layered onto a step gradient of 6 ml of 20% tartrate (dipotassium salt)-2.5 ml of 50% tartrate in PBS, and centrifuged at 100,000 × g for 90 min at 4°C. The virus band was isolated with a needle syringe, diluted in PAE (PBS with 2 μg of aprotinin per ml and 1 mM EDTA, pH 8) and centrifuged at 75,0000 × g for 60 min at 4°C. The resulting pellet was resuspended in PAE and treated by sonication.
Purified virions were fractionated by treatment with 1% NP-40 or 1% NP-40 plus 1 M NaCl in PBS at 4°C for 30 min, layered on 30% sucrose in PBS, and centrifuged at 178,000 × g for 30 min (28). The supernatant was collected, and the virion pellet was resuspended in an equal volume of PAE. All protein extracts were combined with sample buffer, separated by sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel electrophoresis (PAGE), and transferred to nitrocellulose (Amersham-Pharmacia). The VP22 and gC proteins were visualized by incubation of the nitrocellulose with primary antibodies followed by enhanced chemiluminescence detection (Supersignal; Pierce).
Radioimmunoprecipitation assay.
Monolayers of PK15 cells were infected with PRV Becker or 175 at a multiplicity of infection of 10. Thirty minutes prior to the addition of radioactive label, the monolayers were washed once with PBS and overlaid with DMEM-2% FBS lacking cysteine and methionine or phosphate. At 5 h postinfection, the cells were labeled with 50 μCi of [35S] methionine-cysteine or 50 μCi of [33P]orthophosphate (Amersham-Pharmacia) per ml. For cell lysates, at 16 h postinfection, cells were washed in PBS and harvested in nuclease lysis buffer (300 mM NaCl, 50 mM Tris [pH 8.8], 100 mM MgCl2, 10 mM CaCl2, 0.5% Triton X-100, 2 μg of aprotinin per ml). Microccocal nuclease (USB) was added to a final concentration of 30 U/ml, and the lysates were incubated at room temperature for 30 min. At the end of this incubation, the lysates were supplemented with 0.2% SDS, 0.5% sodium deoxycholate, and 1 mM EDTA and incubated at 4°C for 15 min. The lysates were clarified by centrifugation at 14,000 × g for 5 min at 4°C.
Radiolabeled virions were prepared by incubating three 15-cm-diameter dishes of PK15 monolayers infected with PRV Becker with [35S]methionine-cysteine or [33P]orthophosphate as described above. At 16 h postinfection, the supernatant of three labeled 15-cm-diameter dishes was combined with an equal volume of nonlabeled infected cell supernatant, and virus particles were purified as described above. After the last centrifugation step, pelleted virions were resuspended in radioimmunoprecipitation assay (RIPA) buffer (nuclease lysis buffer plus 0.2% SDS, 0.5% sodium deoxycholate, 1 mM EDTA) and incubated at 4°C for 30 min. VP22 antiserum was coupled to protein A-Sepharose CL-4B (Amersham-Pharmacia) with dimethyl pimelimidate (Sigma) as previously described (23). Radiolabeled cell lysates were precleared with protein A-Sepharose by incubation at 4°C for 90 min. Coupled VP22 antiserum was then incubated with precleared radiolabeled cell lysates or virions overnight at 4°C. The antibody-coupled Sepharose with bound protein was washed twice with RIPA buffer and resuspended in sample buffer. Immunoprecipitated proteins were separated on an SDS-12.5% polyacrylamide gel (14 by 16 cm) by electrophoresis and detected by autoradiography.
Protein phosphatase treatment.
Lambda protein phosphatase has activity towards phosphorylated serine, threonine, and tyrosine residues and was used to dephosphorylate the different VP22 isoforms. VP22 from [35S]methionine-cysteine-labeled infected cell lysates was bound to antibody-coupled Sepharose as described above and washed twice with lambda protein phosphatase buffer (NEB). The antibody-coupled Sepharose with bound protein was resuspended in 100 μl of lambda protein phosphatase buffer-2 mM MnCl2 with 800 U of lambda protein phosphatase (NEB) and incubated at 30°C for 45 min. The Sepharose was washed twice with RIPA buffer, resuspended in sample buffer, and analyzed as described above.
Indirect immunofluorescence.
PK15 cells were grown on glass cover slips to about 40% confluence and infected with PRV Becker at a multiplicity of infection of 10. At 6 h postinfection, cells were rinsed with PBS, fixed with 2% paraformaldehyde for 10 min at room temperature, rinsed with PBS again, and permeabilized with PBS-3% bovine serum albumin-0.1% Triton X-100 at room temperature for 15 min. Cells were then incubated with VP22 antiserum for 30 min followed by incubation with Alexa-488 goat anti-rabbit immunoglobulin (Ig) secondary antibody (Molecular Probes) for 30 min.
For detection of viral DNA synthesis, CV-1 cells were incubated in DMEM-1% FBS-3 mM n-butyrate for 16 h prior to infection and maintained in DMEM-1% FBS-3 mM n-butyrate during infection (32, 46). At 6 h postinfection, BrdU was added to the medium for 15 or 60 min as specified by the manufacturer (Boehringer Mannheim, Labeling and Detection Kit I). The cells were then rinsed with PBS and fixed as before. In order to maintain VP22 staining while exposing the incorporated BrdU epitope, cells were permeabilized and incubated with VP22 antibody as before, washed, and fixed again with 2% paraformaldehyde for 15 min at room temperature. Following this, the cells were washed with PBS and treated with 4 N HCl for 5 min at room temperature. The cells were then incubated with BrdU antiserum as per the manufacturer’s instructions, followed by incubation with Alexa-488 goat anti-rabbit Ig and Alexa-546 goat anti-mouse Ig secondary antibodies (Molecular Probes). All cover slips were mounted on glass slides with Aqua Poly/Mount (Polysciences, Inc.), and images were collected with a Zeiss 510 confocal microscope.
Animal experiments, tissue processing, and immunohistochemistry.
Adult male Sprague-Dawley rats weighing 230 to 240 g were used. The rat ocular infection model has been described by Card et al. (8). Experimental protocols were approved by the Princeton University Animal Welfare Committee and were consistent with the regulations stipulated by the American Association for Accreditation of Laboratory Animal Care and those in the Animal Welfare Act (Public Law 99-198). During the course of the experiment, all animals were confined to a biosafety level 2 laboratory, and experiments were conducted with the specific safeguards described previously.
RESULTS
Construction of a VP22-null virus.
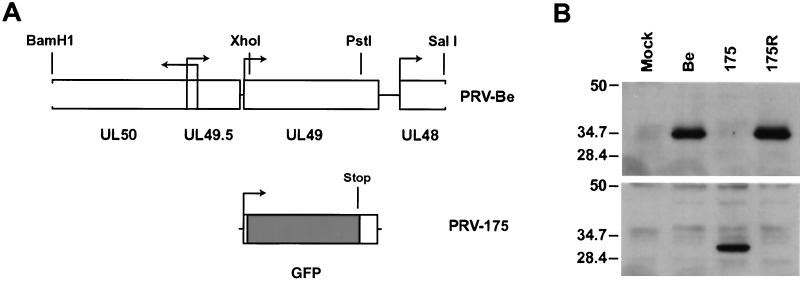
The gene organization and transcriptional activation surrounding the UL49 gene encoding VP22 is complex and not well studied in PRV. Moreover, the VP16 homologue encoded by UL48 is directly downstream of UL49 (34). Accordingly, care was taken to preserve the UL49 promoter and downstream transcription signals (Fig. 1A). The VP22-null mutant, PRV 175, was constructed by replacing a 0.63-kb fragment of the UL49 gene with the EGFP gene so that the EGFP open reading frame was fused to the first Met-Ser-Ser-Ser amino acids of VP22 (Fig. 1B). Therefore, EGFP transcription in PRV 175 is regulated by the UL49 promoter. A revertant of the VP22-null virus, called PRV 175R, was also constructed (Fig. 1B). Both PRV 175 and 175R were indistinguishable from the parental Becker strain in single-step growth after high-multiplicity infection of PK15 cells (data not shown).
FIG. 1.
PRV 175 is a VP22-null virus which expresses GFP. (A) Shown is a BamHI/SalI fragment of PRV Becker (Be) containing the UL49 gene. PRV 175 is identical to Becker except that the XhoI/PstI fragment of UL49 has been replaced with the EGFP gene. (B) Monolayers of PK15 cells were either mock infected or infected with PRV Becker (wild type), 175 (VP22-null), or 175R (revertant of PRV 175) at an MOI of 10 for 16 h prior to preparation of whole-cell lysates. The same infected cell lysates were separated on two polyacrylamide gels, and Western blot analysis was performed with either rabbit polyclonal antiserum to VP22 (top) or polyclonal antiserum to EGFP (bottom). The migration positions of molecular mass markers are shown on the left (in kilodaltons).
Cellular and viral compartmentalization of VP22.
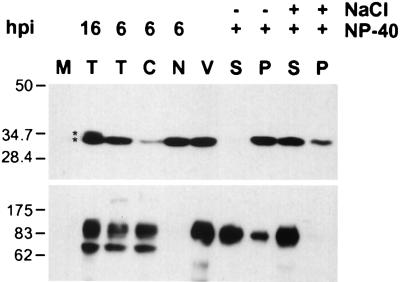
The subcellular localization of VP22 was investigated by cell fractionation. At 6 h postinfection, VP22 was present as a 34-kDa protein primarily in a nuclear fraction devoid of gC, while only a small amount of VP22 was found in the cytoplasmic fraction (Fig. 2). In agreement with previous results, the cytoplasmic fraction contained the 92-kDa mature form and the 74-kDa immature form of gC (Fig. 2) (43).
FIG. 2.
Compartmentalization of VP22 in infected cells and purified virions. Monolayers of PK15 cells were infected at an MOI of 10 for the times indicated (16 or 6 h) prior to preparation of cell lysates. Cells were either mock infected (M) or infected with PRV Becker from which total cell lysate (T), cytoplasmic (C), or nuclear (N) fractions were isolated. Purified extracellular virions (V) were treated with either 1% NP-40 or 1% NP-40 plus 1 M NaCl, from which soluble (S) and pellet (P) fractions were isolated by high-speed centrifugation. Western blot analysis was performed with either rabbit polyclonal antiserum to VP22 (top) or goat polyclonal antiserum to gC (bottom). The two forms present at 16 h postinfection (hpi) are indicated with asterisks. The migration positions of molecular mass markers are shown on the left (in kilodaltons).
A 34-kDa form of VP22 was also detected in purified extracellular virions (Fig. 2). While cell lysates contained the mature and immature forms of gC, purified virions only contained the 92-kDa mature form, indicating that the virion preparation was free of immature viral proteins (Fig. 2). In order to determine what compartment of the virion VP22 was localized to, fractionation of purified virions was performed. It has previously been shown that the treatment of purified virions with NP-40 detergent solubilizes the envelope, while VP22 remains associated with the insoluble tegument and capsid structure (27, 28, 31, 49). Further treatment of virions with NP-40 and high concentrations of salt release significant amounts of tegument, including VP22, from the insoluble fraction.
To investigate whether VP22 in PRV virions exhibits similar biochemical behavior, purified virions were fractionated with 1% NP-40 or 1% NP-40 plus 1 M NaCl treatment. The viral envelope was solubilized with 1% NP-40, as evidenced by the presence of the majority of gC in the supernatant (Fig. 2). VP22 was not detectable in the 1% NP-40-solubilized envelope fraction. Treatment with 1% NP-40 plus 1 M NaCl resulted in the release of the majority of VP22 and complete solubilization of gC. A small amount of VP22 remained associated with the insoluble pellet fraction. Taken together, these data suggest that PRV VP22 was present in the tegument layer of purified virions.
Subcellular localization of VP22 by indirect immunofluorescence.
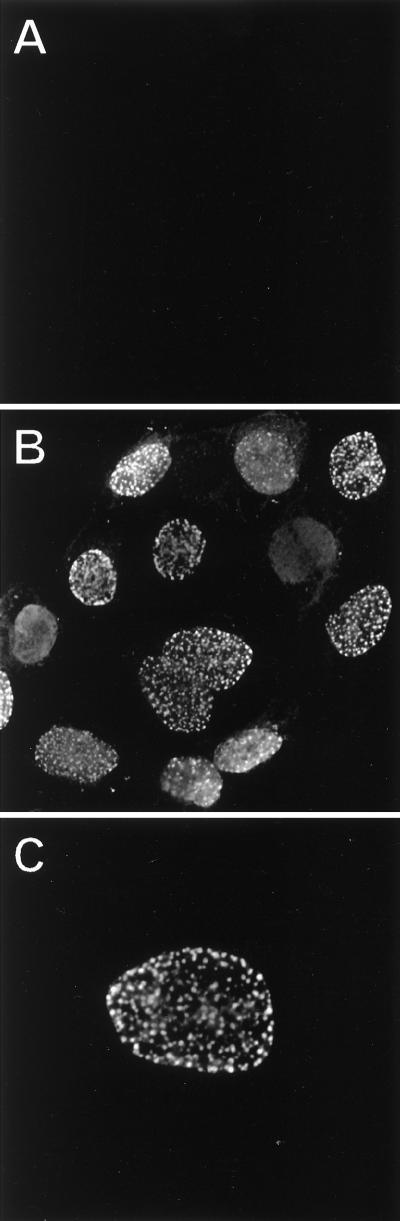
The nuclear localization of VP22 determined by cell fractionation was confirmed by indirect immunofluorescence. PK15 cells were infected at an MOI of 10 and fixed with 2% paraformaldehyde at 6 h postinfection. While mock-infected cells revealed no staining for VP22 (Fig. 3A), punctate nuclear localization of VP22 was seen in PRV Becker-infected cells (Figs. 3B and C). However, a diffuse nuclear stain was always evident and in some cases was also present in nuclei containing VP22 puncta. Some filamentous and punctate cytoplasmic staining was also visible, but to a much lesser extent than the signal from the nucleus (Fig. 3B).
FIG. 3.
Subcellular localization of VP22 by indirect immunofluorescence. PK15 cells grown on cover slips were mock infected (A) or infected at an MOI of 10 with PRV Becker (B and C) for 6 h prior to fixation. Cells were stained with rabbit polyclonal antiserum to VP22 and detected with an Alexa-488 secondary antibody. Magnifications: ×120 (A and B) and ×160 (C).
Tegument compartments in the nucleus have been to shown to localize adjacent to areas of viral DNA synthesis (52). We wished to determine whether the PRV VP22 puncta also localized near sites of viral DNA replication, similar to tegument compartments. To do this, viral replication centers were detected by incorporation of the nucleoside analog BrdU. In order to reduce or eliminate incorporation of BrdU into host chromatin, CV-1 cells were used, as they are susceptible to the effects of the cell cycle inhibitor n-butyrate (32). Treatment of cells with n-butyrate does not inhibit HSV-1 infection (46) and was found to be suitable for PRV infection as well (data not shown). CV-1 cells were incubated in reduced serum with 3 mM n-butyrate for 16 h prior to infection and maintained under the same conditions during infection. This treatment blocked cell entry into S-phase and reduced detectable cellular BrdU incorporation to less than 3% of the cells so that viral DNA replication centers could be visualized (data not shown).
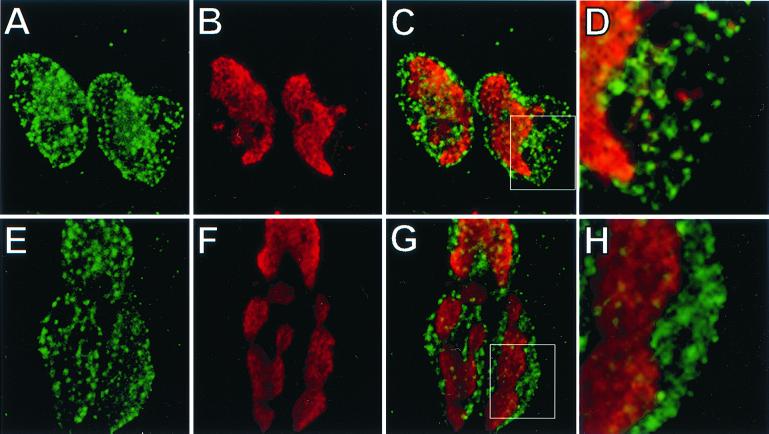
CV-1 cells were infected at an MOI of 10 with PRV Becker and labeled with BrdU at 6 h postinfection. The cells were then fixed with 2% paraformaldehyde and immunostained as described in Materials and Methods. Cells were labeled with BrdU for either 15 or 60 min to reveal different patterns of BrdU staining. Representative images reveal two types of staining with a 15-min BrdU label (Fig. 4B). Punctate BrdU staining is present near the periphery of the nucleus, while large regions of staining are present towards the center of the nucleus. The longer 60-min BrdU label resulted in the consolidation of the punctate staining into globular domains representative of viral replication compartments (Fig. 4F) (32, 51). Treatment of the CV-1 cells with n-butyrate did not have any apparent effect on the punctate nuclear localization of VP22 during infection (Fig. 4A and E). While there are examples where the VP22 puncta are adjacent to the BrdU puncta, there is little or no overlap of the two signals (Fig. 4D). VP22 is also clearly present in the periphery of the nucleus, where there is no BrdU signal with a 60-min label (Fig. 4H). The staining pattern of VP22 was localized throughout the nucleus and was not restricted to areas of globular BrdU staining (Fig. 4G).
FIG. 4.
Localization of VP22 and sites of viral replication. CV-1 cells grown on cover slips were incubated in reduced serum and 3 mM n-butyrate for 16 h prior to infection. The cells were infected in reduced serum and 3 mM n-butyrate at an MOI of 10 with PRV Becker. At 6 h postinfection, BrdU was added to the medium for 15 min (A to D) or 60 min (E to H) prior to fixation. Cells were then stained with polyclonal antiserum to VP22, fixed again, treated with 4 N HCl, and stained with monoclonal antiserum to BrdU. Detection of BrdU was performed with an Alexa-546 secondary antibody and is shown in red (B and F), while VP22, shown in green (A and E), was detected with an Alexa-488 secondary antibody. Merged BrdU and VP22 staining is shown in C and G). A region of costaining (C and G, indicated by the inset) is shown under higher magnification (D and H). Magnifications: ×160 (A to C, E to G) and ×480 (D and H).
Posttranslational modification of VP22.
Initial detection of VP22 present in PRV Becker- and 175R-infected cell lysates suggested the presence of a larger form (Fig. 1B). To investigate this prospect further, infected cell lysates at 16 h postinfection were prepared, separated by 12.5% polyacrylamide gel electrophoresis, and subjected to Western blot analysis. Two migrating forms of VP22, 34 and 35 kDa in size, were present (Fig. 2B, asterisks). Only the smaller form was detectable at 6 h postinfection or in purified virions.
Since HSV-1 VP22 is a major virus-encoded phosphoprotein (13, 15, 18, 21), we investigated whether phosphorylation might account for the different forms of PRV VP22. PK15 cells were infected with either PRV Becker or 175 at an MOI of 10 and labeled with either [35S]methionine-cysteine or [33P]orthophosphate from 5 h to 16 h postinfection. Cell lysates and purified extracellular virions were prepared, immunoprecipitated with VP22 antiserum. and electrophoresed on an SDS-12.5% polyacrylamide gel.
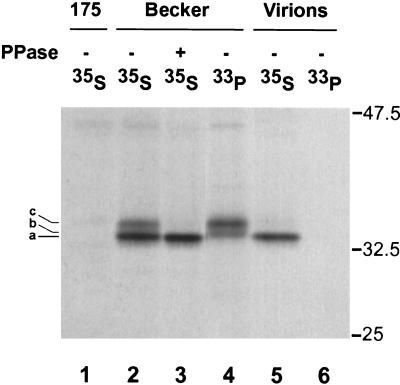
Incorporation of [33P]orthophosphate revealed the presence of two protein species, 34 kDa and 35 kDa in size (Fig. 5, lane 4). Labeling with [35S]methionine-cysteine revealed the presence of a major 33.5-kDa protein and a minor 35-kDa protein in PRV Becker-infected cell extracts, both of which were absent in PRV 175-infected cells (Fig. 5, lanes 1 and 2). Treatment of [35S]methionine-cysteine-labeled immunoprecipitated proteins with lambda protein phosphatase resulted in the loss of the 35-kDa protein band and no alteration of the 33.5-kDa band (Fig. 5, lane 3). Purified extracellular virions labeled with [35S]methionine-cysteine incorporated only the 33.5-kDa form of VP22, whereas the two [33P]orthophosphate-labeled forms were not detected in virions (Fig. 5, lanes 5 and 6). Thus, PRV VP22 exists as three distinct isoforms in infected cells (Fig. 5, a, b, and c). The major VP22 product, 33.5kDa in size (isoform a), is nonphosphorylated and incorporated into virions, while two larger species, 34 and 35 kDa in size (isoforms b and c), are phosphorylated and excluded from purified virions.
FIG. 5.
Radioimmunoprecipitation assay of VP22 in infected cells. Monolayers of PK15 cells were infected at an MOI of 10 with PRV Becker or 175 and labeled overnight in the presence of either [35S]methionine-cysteine or [33P]orthophosphate. Total cellular extracts (lanes 1 to 4) or purified wild-type extracellular virions (lanes 5 and 6) were prepared at 16 h postinfection and subjected to immunoprecipitation with rabbit polyclonal antiserum to VP22. Upon immunoprecipitation, one [35S]methionine-cysteine-labeled Becker sample was treated with lambda protein phosphatase (PPase) (lane 3). The immunoprecipitated products were separated by SDS-12.5% PAGE and visualized by autoradiography. The three observed electrophoretic forms of VP22 are indicated on the left (a to c). The migration positions of molecular mass markers are shown on the right (in kilodaltons).
Virulence and neuronal spread of a VP22-null virus.
The rat eye infection model tests both the virulence and neuronal spread of alphaherpesviruses across synaptically connected neurons in the peripheral and central nervous system (CNS) (8). The virulent PRV Becker strain used in our studies induces marked symptoms of sensory and autonomic nervous system infection after 48 h, followed by death at about 60 to 70 h postinfection (8). The virus invades the CNS and spreads to all retinorecipient areas of the brain. By contrast, the vaccine strain Bartha is much less virulent, as determined from a delayed onset of symptoms, a mean time to death of more than 120 h, and an inability to invade certain retinorecipient areas of the CNS (9). We have determined that PRV Becker-derived strains not expressing gE, gI, or Us9, all of which are deleted in the Bartha strain (33, 34), exhibit attenuated virulence and spread similar to Bartha in the rat eye infection model (6, 9, 50).
While the gE, gI, and Us9 membrane proteins clearly have an important role in virulence and neuronal spread, it is unclear what role tegument proteins may have in the rat eye infection model described above. Although it is currently unknown how viral capsid structures are able to interact with molecular motors to mediate transport of packaged viral genomes, one thought is that the tegument proteins can potentially serve to connect the capsid to cellular motors. We used the rat eye infection model to determine whether the VP22 tegument protein has any role in virus transport across neurons and the ability of PRV to cause disease in the CNS.
The mean time to death of PRV Becker- and 175-infected animals was 73.9 and 68.0 h, respectively (Table 1). The absence of VP22 had no effect on virulence: the appearance of various symptoms of disease, including ruffling of the fur, nasal discharge, hunched posture, labored breathing, uncoordinated movement, and scratching of the face surrounding the injected eye (50), were severe and indistinguishable from those caused by the virulent PRV Becker strain.
TABLE 1.
Virulence after intravitreal injections
| Virus | No. of animals | Time to death (h) | Symptomsa |
|---|---|---|---|
| PRV Becker | 3 | 72.6, 72.5, 76.6 | Severe |
| PRV 175 | 3 | 65.0, 68.1, 70.8 | Severe |
Symptom severity based on reference 50.
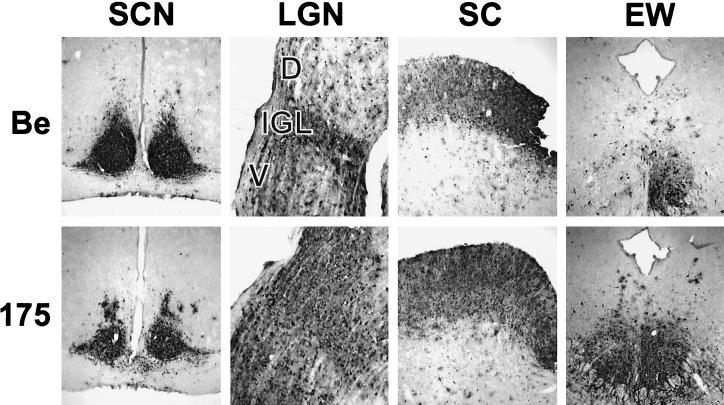
Neuroinvasiveness was scored by detecting viral antigen in areas of the brain known to be connected to the retina and eye musculature. Infected neuronal cell bodies were readily detected in fixed coronal brain slices with the polyclonal antibody Rb133 (9). As previously reported, PRV Becker infected the retina and spread to all retinorecipient neurons in the brain (anterograde spread). PRV 175 gave an identical pattern of anterograde invasion (Fig. 6). Spread to the brain via retrograde routes occurs when virus is taken up at axon terminals that innervate the eye musculature. For example, infection of the Edinger-Westphal (EW) nucleus occurs when PRV infects the ciliary body in the eye, and infection of the oculomotor nucleus occurs when virus infects the muscles that rotate the eye ball (8). Both PRV Becker and 175 gave extensive infections of the EW (Fig. 6). These experiments demonstrate that PRV VP22 plays no detectable role in promoting virulence or neuroinvasiveness in the rat eye infection model.
FIG. 6.
Anterograde and retrograde spread of PRV 175 in the rodent visual system. Approximately 5 × 105 PFU of PRV Becker (Be) or 175 was injected into the vitreous humor of Sprague-Dawley male rats. At the time of death, the brains were fixed and sliced into 35-μm-thick coronal sections with a freezing microtome. Viral antigen was detected with polyvalent antiserum Rb133. Representative sections containing suprachiasmatic nuclei (SCN), LGN including the dorsal (D) and ventral (V) aspects, as well as the intergeniculate leaflet (IGL), superior colliculus (SC), and the Edinger-Westphal nucleus (EW) are pictured. The infection of the ipsilateral EW nucleus by PRV Becker and of both nuclei by PRV 175 reflects the kinetics of multisynaptic spread from the pretectal nucleus across the posterior commissure to the contralateral EW nucleus. Intensity of infection of the contralateral nucleus varied from animal to animal.
DISCUSSION
Nuclear localization of PRV VP22.
The PRV VP22 homologue localizes to the nucleus at 6 h postinfection, as determined by cell fractionation and indirect immunofluorescence (Fig. 2 and 3). Nuclear localization of VP22 during infection appears to be conserved among the different alphaherpesvirus homologues (12, 28, 40). We also observed nuclear localization of PRV VP22 (Fig. 3), which is consistent with prior observations for nonphosphorylated HSV-1 VP22 (35, 40). However, two types of nuclear staining were observed, diffuse and punctate. BrdU analysis indicates that while some VP22 punctate staining is adjacent to punctate sites of viral DNA replication, as revealed by a 15-min BrdU label, there is little to no overlap of signal, and the colocalization is not exclusive (Fig. 4). Indeed, punctate VP22 staining in the nucleus partially colocalizes with the marginated host chromatin, where viral replication centers are not present (data not shown).
It is interesting that while some punctate BrdU staining is present at the periphery of the nucleus with a 15-min label, there is no BrdU signal near the nuclear periphery with a 60-min label (Fig. 4C and G). Over the course of the 60-min BrdU labeling, the peripheral BrdU staining appears to consolidate into the globular replication domains localized more centrally in the nucleus. While the significance of BrdU consolidation is unknown at this time, it is clear that the punctate nuclear VP22 localization is unaffected. One possibility is that the VP22 puncta may be in a distinct compartment of the nucleus that is not associated with viral replication. Moreover, the size of the puncta is consistent with VP22’s presence in a protein complex. We have not yet determined if other viral proteins colocalize with VP22 in the nucleus or if this punctate staining is associated with tegument compartments previously shown to contain VP16 (52).
Assembly of VP22 into the tegument.
Previous evidence that the PRV VP22 homologue was a tegument protein came primarily from immunogold electron microscopy (26) and by analogy to other VP22 homologues (12, 13, 28, 34). Based on our biochemical fractionation data, we confirm that PRV VP22 is a tegument protein (Fig. 2). VP22 was released from the tegument layer by high-salt treatment, suggesting that it was retained in the virion by noncovalent interactions.
Although the mechanism of assembling VP22 in the tegument is unknown, protein-protein interactions are likely to be important, as incorporation of HSV-1 VP22 into virions requires the VP22 carboxy-terminal region (41). The VP22 protein interactions affecting packaging in virions are likely to occur in the cytoplasm. For example, in HSV-1, the undermodified form of VP22 present in the cytoplasm is incorporated into virions (40). Furthermore, a (GFP)-VP22 fusion protein expressed during HSV-1 infection localizes exclusively to the cytoplasm and is efficiently incorporated into virions (19). Electron micrographs of PRV-infected epithelial cells show that extracellular and intracytoplasmic enveloped virions contain VP22, while immature, perinuclear virions are devoid of VP22 (26). In HSV-1-infected rat dorsal root ganglion neurons, virion particles in the cell body also contain VP22, whereas perinuclear virions do not (Anthony Cunningham, personal communication). Given these findings, the nuclear localization for PRV VP22 is noteworthy.
While some cytoplasmic localization of PRV VP22 was observed, the steady-state localization at 6 h postinfection was predominantly nuclear (Fig. 2 and 3). If VP22 enters the tegument of egressing virions in the cytoplasm and cannot be detected in the nucleus by immunoelectron micrscopic analysis, why was VP22 localized to the nucleus in our studies? As VP22 localization by immunofluorescence can be affected by the use of different fixatives (5, 40) it may be that the fixation methods used for electron microscopy remove VP22 from the nucleus but not from virions. While certain fixation procedures may be well suited for the detection of virion-associated VP22, they may not be useful for the detection of nuclear localized VP22. Since VP22 has been suggested to associate with the nuclear matrix during infection (39), fixation methods which disrupt the nuclear matrix or other nuclear structures may also alter the localization of VP22.
Our fractionation experiments were performed on nonfixed cells and clearly showed that VP22 associated with the nuclear fraction (Fig. 2). While further work is necessary to understand both the structure and function of nuclear VP22, we propose that VP22 incorporated into virions originates from a cytoplasmic pool, while nuclear VP22 is nonstructural and has a role during infection that is yet to be defined.
Three isoforms of intracellular PRV VP22.
We identified three isoforms of PRV VP22 in infected cells. Importantly, only the nonphosphorylated 33.5-kDa form was present in purified extracellular virions (Fig. 5). The phosphorylation of tegument proteins upon entry into the cell is postulated to aid in the dissociation of the tegument structure (35, 36). VP22 in virions of both HSV-1 (15, 40) and PRV is underphosphorylated and may reflect a generally conserved mechanism of tegument packaging and dissociation upon entry among the alphaherpesviruses. The phosphorylated species of PRV VP22 migrated as distinct 34-kDa and 35-kDa species and were detected late but not early in infection (Fig. 2 and 5). As the majority of VP22 was localized to the nucleus by 6 h postinfection, we suggest that the nonphosphorylated species is localized to the nucleus at this time. The two phosphorylated proteins may accumulate slowly during infection. Furthermore, while less phosphorylated VP22 is present at 6 h postinfection, the ratio of phosphorylated to unphosphorylated VP22 appears different when Fig. 2 and 3 are compared. This difference may reflect the different techniques used to detect VP22. The significance of this difference is not clear. Furthermore, it is unknown if viral or cellular kinases phosphorylate VP22 and what role phosphorylation may have on the function or localization of PRV VP22.
VP22-null mutant virus has no observable phenotypes in vitro.
PRV 175 and its revertant PRV 175R were constructed to determine the role of VP22 protein during in vitro and in vivo infections. The strategy was to replace the UL49 coding sequence with the EGFP open reading frame so that EGFP expression is controlled by the endogenous VP22 promoter. The recombinant virus PRV 175 was readily isolated and propagated (Fig. 1). Moreover, the single-step growth profiles of PRV 175, 175R, and Becker were identical (data not shown). VP22 is also dispensable for the growth of BHV-1 in tissue culture, but the BHV-1 null mutant does not grow with wild-type kinetics (28). An HSV VP22-null mutant has not yet been reported. We do not know why the PRV VP22-null mutant grows like the wild type while the BHV-1 mutant does not. Perhaps PRV VP22 has a different role in infection, as has been noted for HSV and varicella-zoster virus (VZV) VP16. While VP16 is essential for a productive HSV-1 infection (37, 47, 53), the VZV VP16 homologue is not (10).
VP22-null mutant virus has no observable phenotypes in animal infections.
We tested the role of PRV VP22 in a rat eye model of infection. This assay reports on virulence (e.g., time to death and associated symptoms) as well as neuroinvasion (spread of virus from the periphery to the CNS). We specifically tested the hypothesis that VP22 may be important for capsid trafficking of virus across neuronal projections (17). PRV Becker and the VP22-null virus mutant were indistinguishable in virulence and neuroinvasion. Spread by neither the retrograde nor anterograde invasion route was compromised by a lack of VP22 expression.
Concluding remarks.
While this study has provided an initial characterization of the PRV VP22 homologue, we were unable to attribute any function to VP22 during infection. Others have reported that the HSV-1 VP22 protein has the property of intercellular spread (11, 16, 38). Accordingly, we looked for intercellular spread of PRV VP22 by indirect immunofluorescence and confocal microscopy. Intercellular spread was obvious when methanol was used as a fixative, but was not readily observed when 2% paraformaldehyde fixation was used (data not shown) (5, 40). Furthermore, intercellular spread of a PRV (EGFP)-VP22 chimeric protein was not observed in live cells during transient expression (data not shown) similar to HSV-1 (5). Thus, at this time, we have little evidence that PRV VP22 can spread from cell to cell, as reported for HSV-1 VP22. However, we cannot rule out the possibility that while HSV and BHV VP22 possess the property of intercellular spread, the PRV VP22 homologue lacks this property. If so, this difference may account for a lack of phenotype in our pathogenesis assay with a PRV VP22-null mutant. Alternatively, while the BHV-1 VP22-null virus was tested in the natural host, our PRV VP22-null mutant was not. A PRV VP22-null phenotype may only be evident in swine.
It is possible that the PRV genome encodes redundant functions that substitute for the loss of VP22. For example, the four proteins encoded by the tegument gene cluster are conserved among the alphaherpesviruses. They also exhibit similar subcellular localization and interact with each other. Indeed, our initial studies demonstrated that it is not possible to isolate a viable virus when all four tegument cassette genes are deleted from PRV (unpublished observations). Alternatively, the appropriate assay to test VP22 function during infection may not yet have been used. For example, the role of PRV VP22 in the establishment of latency, reactivation, or immunological responses during infection has not been investigated. The identification of cell- and virus-encoded proteins which interact with PRV VP22 may be particularly informative.
Acknowledgments
We gratefully acknowledge B. Klupp and T. Mettenleiter for providing the VP22 antiserum and PRV sequence data without which this work would not have been possible. Thanks to Greg Smith for providing a PRV subclone containing UL49 and Christoph Hengartner for assistance with the radioimmunoprecipitations. We also acknowledge J. Goodhouse for his expert technical assistance in confocal microscopy.
T.D.R. is supported by NIH training grant GM07388. This work was supported by NINDS grant 2RO133506 to L.W.E.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Porat, T., and A. S. Kaplan. 1985. Molecular biology of pseudorabies virus, p.105–173. In B. Roizman (ed.), The herpesviruses. Plenum Publishing, New York, N.Y.
- 3.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373–2376. [DOI] [PubMed] [Google Scholar]
- 5.Brewis, N., A. Phelan, J. Webb, J. Drew, G. Elliott, and P. O’Hare. 2000. Evaluation of VP22 spread in tissue culture. J. Virol. 74:1051–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau, A. D., J. P. Card, and L. W. Enquist. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1–19. [DOI] [PubMed] [Google Scholar]
- 8.Card, J. P., M. E. Whealy, A. K. Robbins, R. Y. Moore, and L. W. Enquist. 1991. Two alphaherpesvirus strains are transported differentially in the rodent visual system. Neuron 6:957–969. [DOI] [PubMed] [Google Scholar]
- 9.Card, J. P., M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1992. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J. Virol. 66:3032–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, J. I., and K. Seidel. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 68:7850–7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dilber, M. S., A. Phelan, A. Aints, A. J. Mohamed, G. Elliott, C. I. Smith, and P. O’Hare. 1999. Intercellular delivery of thymidine kinase prodrug activating enzyme by the herpes simplex virus protein VP22. Gene Ther. 6:12–21. [DOI] [PubMed] [Google Scholar]
- 12.Dorange, F., S. El Mehdaoui, C. Pichon, P. Coursaget, and J. F. Vautherot. 2000. Marek’s disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 81:2219–2230. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, G. D., and D. M. Meredith. 1992. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J. Gen Virol. 73:723–726. [DOI] [PubMed] [Google Scholar]
- 14.Elliott, G., G. Mouzakitis, and P. O’Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, G., D. O’Reilly, and P. O’Hare. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140–145. [DOI] [PubMed] [Google Scholar]
- 16.Elliott, G., and P. O’Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223–233. [DOI] [PubMed] [Google Scholar]
- 17.Elliott, G., and P. O’Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott, G., D. O’Reilly, and P. O’Hare. 1999. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J. Virol. 73:6203–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott, G., and P. O’Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott, G., and P. O’Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson, W., and B. Roizman. 1974. Proteins specified by herpes simplex virus. Staining and radiolabeling properties of B capsid and virion proteins in polyacrylamide gels. J. Virol. 13:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greijer, A. E., C. A. Dekkers, and J. M. Middeldorp. 2000. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J. Virol. 74:9078–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow, E., and D. Lane. 1988. Immunoaffinity purification, p.526–527. In Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Kato, K., T. Daikoku, F. Goshima, H. Kume, K. Yamaki, and Y. Nishiyama. 2000. Synthesis, subcellular localization and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch. Virol. 145:2149–2162. [DOI] [PubMed] [Google Scholar]
- 25.Kingham, B. F., V. Zelnik, J. Kopacek, V. Majerciak, E. Ney, and C. J. Schmidt. 2001. The genome of herpesvirus of turkeys: comparative analysis with Marek’s disease viruses. J. Gen. Virol. 82:1123–1135. [DOI] [PubMed] [Google Scholar]
- 26.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie, J., F. J. Rixon, and J. McLauchlan. 1996. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology 220:60–68. [DOI] [PubMed] [Google Scholar]
- 28.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, X., B. Chow, and L. A. Babiuk. 1997. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine 15:1057–1064. [DOI] [PubMed] [Google Scholar]
- 30.McKnight, J. L., P. E. Pellett, F. J. Jenkins, and B. Roizman. 1987. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate alpha-trans-inducing factor-dependent activation of alpha genes. J. Virol. 61:992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269–276. [DOI] [PubMed] [Google Scholar]
- 32.McNamee, E. E., T. J. Taylor, and D. M. Knipe. 2000. A dominant-negative herpesvirus protein inhibits intranuclear targeting of viral proteins: effects on DNA replication and late gene expression. J. Virol. 74:10122–10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C., N. Lukacs, and H. J. Rziha. 1985. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J. Virol. 56:307–311.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C. 2000. Aujeszky’s disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99–115. [DOI] [PubMed] [Google Scholar]
- 35.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517–2528. [DOI] [PubMed] [Google Scholar]
- 36.Morrison, E. E., Y. F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phelan, A., G. Elliott, and P. O’Hare. 1998. Intercellular delivery of functional p53 by the herpesvirus protein VP22. Nat. Biotechnol. 16:440–443. [DOI] [PubMed] [Google Scholar]
- 39.Pinard, M. F., R. Simard, and V. Bibor-Hardy. 1987. DNA-binding proteins of herpes simplex virus type 1-infected BHK cell nuclear matrices. J. Gen. Virol. 68:727–735. [DOI] [PubMed] [Google Scholar]
- 40.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious Herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roizman, B., and E. Sears. 1996. Herpes simplex viruses and their replication, p.1043–1107. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven, Philadelphia, Pa.
- 43.Ryan, J. P., M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1987. Analysis of pseudorabies virus glycoprotein gIII localization and modification by using novel infectious viral mutants carrying unique EcoRI sites. J. Virol. 61:2962–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwyzer, M., D. Styger, B. Vogt, D. E. Lowery, C. Simard, S. LaBoissiere, V. Misra, C. Vlcek, and V. Paces. 1996. Gene contents in a 31-kb segment at the left genome end of bovine herpesvirus-1. Vet. Microbiol. 53:67–77. [DOI] [PubMed] [Google Scholar]
- 45.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol 75:8105–8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shadan, F. F., L. M. Cowsert, and L. P. Villarreal. 1994. n-Butyrate, a cell cycle blocker, inhibits the replication of polyomaviruses and papillomaviruses but not that of adenoviruses and herpesviruses. J. Virol. 68:4785–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smiley, J. R., and J. Duncan. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J. Virol. 71:6191–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tirabassi, R. S., and L. W. Enquist. 2000. Role of the pseudorabies virus gI cytoplasmic domain in neuroinvasion, virulence, and posttranslational N-linked glycosylation. J. Virol. 74:3505–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113–125. [DOI] [PubMed] [Google Scholar]
- 52.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 70:4623–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O’Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanagida, N., S. Yoshida, K. Nazerian, and L. F. Lee. 1993. Nucleotide and predicted amino acid sequences of Marek’s disease virus homologues of herpes simplex virus major tegument proteins. J. Gen. Virol. 74:1837–1845. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]