Abstract
Molluscum contagiosum virus (MCV), a member of the human poxvirus family, encodes the MC159 protein that inhibits Fas-, tumor necrosis factor (TNF)-, and TNF-related apoptosis-inducing ligant (TRAIL)-induced apoptosis. We used site-directed mutagenesis to change charged or hydrophobic amino acid residues to alanines to identify regions of MC159 that are critical for protection from apoptosis and for protein-protein interactions. Surprisingly, while MC159 is thought to block apoptosis by binding to Fas-associated death domain (FADD) or caspase-8, several mutants that lost apoptosis blocking activity still bound to both FADD and caspase-8. Mutations in the predicted hydrophobic patch 1 and α2 regions of both death effector domains (DEDs) within MC159 resulted in loss of the ability to bind to FADD or caspase-8 and to block apoptosis. Amino acid substitutions in the RXDL motif located in the α6 region of either DED resulted in loss of protection from apoptosis induced by Fas, TNF, and TRAIL and abolished the ability of MC159 to block death effector filament formation. Thus, charged or hydrophobic amino acids in three regions of the MC159 DEDs (hydrophobic patch 1, α2, and α6) are critical for the protein’s ability to interact with cellular proteins and to block apoptosis.
Death receptors constitute a subgroup of the tumor necrosis factor (TNF) receptor superfamily, which are defined by the presence of a signaling domain with six α-helices in the cytoplasmic region that is termed the death domain (13). These receptors function to maintain homeostasis in the immune system by eliminating autoreactive cells, antigen-reactive T cells following an immune response, and virus-infected or malignant cells. In humans, six death receptors have been identified: Fas (also called CD95 and APO-1), TNF receptor 1 (TNFR1; also called TNFRSF1A and CD120a p55-R), DR3 (also called APO3, Wsl-1, TRAMP. and LARD), DR4 (also called TRAIL-R1 and Apo-2), DR5 (also called TRAIL-R2, KILLER, and TRICK2), and DR6 (17, 19).
Binding of a death receptor to its ligand initiates a change in the receptor complex, resulting in signaling through a series of protein-protein interactions that culminate in apoptosis (4, 22). Fas binding to its ligand (FasL) leads to the recruitment of the adapter molecule FADD (Fas-associated death domain) through interactions of the death domains in Fas and FADD. FADD contains another region known as the death effector domain (DED). The FADD DED also possesses six α-helices in a folded region similar to the death domains, but it forms distinct contacts only with other DED-containing proteins, whereas death domain-containing proteins interact chiefly with other death domain-containing adapter proteins.
Once FADD is recruited to Fas, its DED binds to the DEDs in the prodomain of caspase-8 or caspase-10 (1, 28). The complex containing Fas, FADD, and caspase-8 or caspase-10 is termed the death-inducing signaling complex (DISC). Recruitment of caspase-8 or caspase-10 into the DISC results in autocatalytic cleavage of the caspase into its active subunits and subsequent cleavage and activation of substrates, including other caspases, ultimately leading to apoptosis (15, 16). FADD also serves as an adapter molecule in other death receptor pathways (3, 5, 9, 10, 12, 24). Hence, death receptors may all work by a common mechanism involving recruitment of DED-containing caspases to form an active signaling complex.
Apoptosis affords the host a defense mechanism to eliminate virus-infected cells. This must have proven sufficiently effective, because viruses in turn evolved mechanisms to interfere with host apoptosis pathways. A family of proteins known as the viral FLICE-inhibitory proteins (v-FLIPs) inhibit the signaling pathways in death receptor-induced apoptosis. These include the molluscum contagiosum virus (MCV) MC159 protein, the equine herpesvirus 2 (EHV-2) E8 protein, and the bovine herpesvirus E2 protein (2, 25). v-FLIPs block apoptosis induced through the Fas, TNFR1, DR3, DR4, and DR5 pathways.
Apoptosis inhibition by the v-FLIPs stems from their two DEDs that can interact with the DEDs of FADD and caspase-8 (2, 11, 25, 27). The MCV MC159 protein is present in a complex with Fas when the receptor is activated (21). The EHV-2 E8 protein can be recruited to the DISC and may prevent activation of caspase-8 by blocking its recruitment to the DISC (25). v-FLIPs also inhibit formation of a cytoplasmic structure termed the death effector filament (18, 23). High-level expression of FADD or the prodomain of caspase-8 in cells results in their oligomerization as death effector filaments that recruit and activate caspases. Coexpression of v-FLIPs with FADD or the prodomain of caspase-8 blocks death effector filament formation and subsequent apoptosis. Despite these correlations, the precise inhibitory mechanism of v-FLIP, especially MC159, in the Fas signaling complex is unknown.
The MC159 v-FLIP protein contains a six-amino-acid N-terminal sequence followed by a 74-amino-acid DED, a 14-amino-acid linker region, an 83-amino-acid DED, and a 64-amino-acid carboxy-terminal tail. An RXDL motif, conserved among other DED-containing proteins, is present at the carboxy end of each DED. MC159 DEDs are homologous to other DED-containing proteins, including FADD. Alignment of the MC159 DED sequences onto the nuclear magnetic resonance (NMR)-determined structure of the FADD DED suggests that each MC159 DED consists of six α-helices and contains a highly conserved hydrophobic patch on its surface (6). The role that each of these protein motifs might play in apoptosis inhibition has not yet been defined.
Based on prior studies, v-FLIP is thought to prevent apoptosis by binding to FADD and caspase-8 (2, 21, 25, 26). In order to identify functionally important regions of MC159, we constructed a series of mutations in which charged amino acids were changed to alanines. Since charged amino acids are often on the surface of proteins, they are likely to participate in protein-protein interactions. Surprisingly, we found that the majority of the MC159 mutants that lost the ability to block apoptosis induced by Fas, TNF, and TNF-related apoptosis-inducing ligand (TRAIL) still bound FADD and caspase-8. The predicted hydrophobic patch 1 and α2 regions were important for FADD and caspase-8 binding. We also found that the conserved RXDL motif within the predicted α6 region is critical for protection from apoptosis and for inhibition of death effector filament formation.
MATERIALS AND METHODS
Expression vectors.
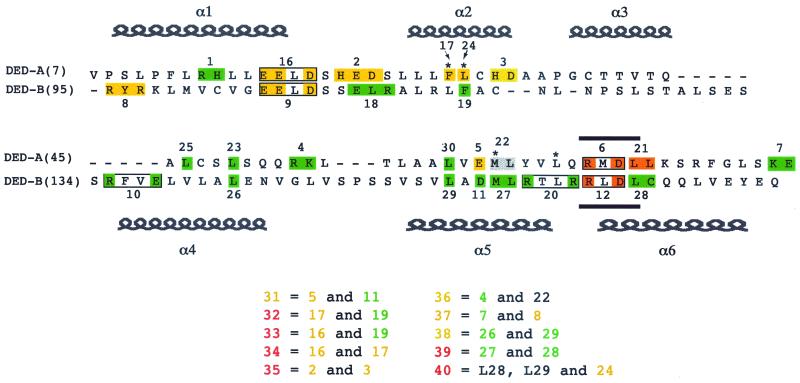
MCV159L was cloned into the pCI expression plasmid (Promega, Madison, Wis.) as described previously (2). Oligonucleotide site-directed mutagenesis was performed using the pCI-MC159 plasmid. Thirty-nine mutations that changed charged or hydrophobic amino acids to alanines were made in the MCV159L gene by the gene splicing by overlap extension (SOEing) method (8) and then cloned into the pCI expression vector (Fig. 1).
FIG. 1.
Structure of wild-type and mutant MC159 constructs. MC159 contains a short amino-terminal region followed by two DEDs and a carboxy-terminal domain. In the diagram, only the two MC159 DEDs are shown aligned with each other. The positions of the predicted α-helices in the DEDs, based on the structure of the FADD DED (6), are designated at the top of the diagram. The RXDL motif (denoted by bars) is located at amino acids 69 to 72 in DED-A and amino acids 166 to 169 in DED-B. The asterisks indicate the amino acids in hydrophobic patch 1. The numbered boxes in different colors indicate the locations of the mutations in mutants 1 to 30, in which charged or hydrophobic amino acids were changed to alanines (referred to as simple mutants). Three mutations located outside the DEDs are not shown: mutant 13 causes E207A, H208A, and E209A; mutant 14 contains E223A and R226A; and mutant 15 causes K5A and E6A. These three mutants showed no loss of protection from apoptosis. Mutants 31 to 40 are derived from combinations of two simple mutants and are referred to as complex mutants. The complex mutants and their composition are listed below the MC159 diagram. The colors of the mutants indicate their level of protection from apoptosis. Red indicates complete loss of protection, yellow indicates a partial loss of protection in either HeLa or Jurkat cells, and green indicates no loss of protection. Grey indicates a mutant that was not tested.
The MC159-hemagglutinin (HA) expression plasmid was constructed by using the oligonucleotides 159NHA (5′-CCGGAATTCAGCATGTACCCATACGACGTGCCAGACTACGCATCCGACTCCAAGGAGGTCCCTAGC3′) and 159XBA-C (5′-CCGTACTAGTCTAGACTAAGTCGTTTGCTCGGGGCTGTC-3′) to amplify MC159 from the pCI-MC159 expression plasmid. The MC159 PCR product was then cloned into the pCI expression vector at the EcoRI and XbaI restriction enzyme sites. The MC159-green fluorescent protein (GFP) expression plasmid was constructed by using the oligonucleotides pCI-T7 (5′-TAATACGACTCACTATAGG-3′) and 159C-SacII (5′-TCAGCCGCGGAGTCGTTTGCTCGGGGCTGTC-3′) to amplify MC159 from the pCI-MC159 expression plasmid. The MC159 PCR product was then cloned into the NheI and SacII restriction enzyme sites of the pEGFP-N1 expression vector (Clontech Laboratories Inc., Palo Alto, Calif.).
HeLa cell death assay.
HeLa cells were transfected with 400 ng of pCMVβ-gal (Stratagene, La Jolla, Calif.) and 800 ng of either pCI, pCI-MC159, or pCI-MC159 mutant expression vectors using Lipofectamine (Life Technologies, Gaithersburg, Md.). Cells were treated 28 h after transfection with either 500 ng of anti-human Fas monoclonal antibody (MAb) CH11 (Kamiya Biochemical, Seattle, Wash.) per ml and 5 μ g of cycloheximide per ml or 20 ng of recombinant (Escherichia coli) human TNF-α (Roche, Indianapolis, Ind.) and 15 μ g of cycloheximide per ml for 16 h. Control cells received cycloheximide treatment alone.
Cells were fixed in a solution containing 2% formaldehyde and 0.2% gluteraldehyde in phosphate-buffered saline (PBS) for 5 min at room temperature and stained for β-galactosidase expression with a solution containing 1 mg of 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal) per ml, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 2 mM MgCl2 in PBS for 6 to 8 h at 37°C. The total number of flat, blue-staining cells in five random fields of view was counted. Protection from Fas- and TNF-induced apoptosis was measured as the ratio of the number of flat, blue-staining cells in treated wells to that in control wells. The values were normalized so that the vector was equal to 0% protection. Percent cell death was calculated as 100% minus percent protection.
Jurkat cell death assay.
Jurkat cells were electroporated with 2.5 μ g of pEGFP-N1 (Clonetech) and 10 μ g of either pCI, pCI-MC159, or pCI-MC159 mutant expression vectors and incubated for 24 h as described (7). Transfected cells were plated at 2 × 105 cells per well into a 96-well plate and treated with 100 ng of anti-human Fas MAb αAPO-1 (Kamiya Biochemical) per ml and 5% soluble protein A or 100 ng of TRAIL and 2 μ g of enhancer antibody (Alexis, San Diego, Calif.) per ml for 12 h. Control samples received either soluble protein A or enhancer antibody only. After the 12-h treatment, cells were stained with annexin-V-phycoerythrin (PE) (BD Pharmingen, San Diego, Calif.) for detection of apoptotic cells and analyzed by flow cytometry for green fluorescent protein and annexin-V-PE-positive cells on a FACScan cytometer (Becton Dickinson & Co., Franklin Lakes, N.Y.). Percent apoptosis was determined by the ratio of annexin-V-positive cells to GFP-positive cells.
Immunoprecipitations and immunoblotting.
293T cells were cotransfected with the indicated plasmids using Fugene-6. Cells were lysed 24 h after transfection for 30 min in lysis buffer (140 mM NaCl, 10 mM Tris [pH 7.2], 2 mM EDTA, 1% NP-40, and one complete protease inhibitor tablet [Roche] per 50 ml of buffer). The lysates were centrifuged at 14,000 rpm for 10 min at 4°C. An equal volume of 2× sodium dodecyl sulfate (SDS) loading solution (0.1 M Tris, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 100 mM dithiothreitol [DTT]) was added to an aliquot of each lysate, and the mixture was boiled.
For immunoprecipitations, 100 μ l of lysate was immunoprecipitated with 3 μ l of anti-MC159 polyclonal rabbit antiserum (21) followed by 40 μ l of protein A-Sepharose (50% mixture in lysis buffer) or with 1 μ g of murine anti-HA monoclonal antibody (Covance, Richmond, Calif.) and 30 μ l of goat anti-mouse immunoglobulin G (IgG) beads (Dynal, Lake Success, N.Y.) resuspended in 10 μ l of lysis buffer. Immune complexes were washed four times in lysis buffer, and the pellet was resuspended in 20 μ l of 2× SDS loading solution containing 100 mM DTT. Lysates and immunoprecipitates were electrophoresed on 4 to 20% Tris-glycine-SDS gels and transferred onto nitrocellulose. Blots were blocked with 5% nonfat dry milk for 20 min and incubated with 0.5 μ g of murine anti-FADD monoclonal antibody (Signal Transduction Laboratories, Lexington, KY), 0.4 μ g of murine anti-GFP monoclonal antibody (Roche), 5 to 7 μ g of murine anti-HA monoclonal antibody (Covance) per ml, or a 1:500 dilution of anti-MC159 polyclonal rabbit antiserum for 1 h. After being washed three times in PBS with 0.05% Tween 20, the blots were treated with 0.08 μ g of horseradish peroxidase-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa) or 0.1 μ g of horseradish peroxidase-conjugated anti-rabbit IgG antibody (Amersham Pharmacia Biotech, Piscataway, N.J.) per ml for 30 min, followed by three washes with PBS containing 0.05% Tween 20. Proteins were detected with SuperSignal chemiluminescent substrate according to the manufacturer’s instructions (Pierce Chemical Co., Rockford, Ill.).
Immunofluorescence assay.
HeLa cells on glass cover slips were transfected with 0.5 μ g of pFADD-GFP (23) and 1.5 μ g of either pCI-MC159 or pCI-MC159 mutant using Superfect (Qiagen, Chatsworth, Calif.). The medium was replaced 2 h after transfection with medium containing 50 μ M zVAD-fmk (Enzyme Systems Products, Livermore, Calif.), and 16 to 24 h after transfection, Hoechst dye (1 μ g/ml; Sigma) was added for 30 min at 37°C to label nuclei. The cells were fixed with 100% methanol at −20°C for 10 min, blocked in immunofluorescence assay buffer (PBS with 0.1% bovine serum albumin and 0.01% Tween 20) for 1 h, and incubated with a 1:250 dilution of anti-MC159 polyclonal rabbit antiserum for 30 min, followed by 6 μ g of Texas red-conjugated donkey anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories, Inc.) per ml. Cells were examined on an Axiophot microscope (Carl Zeis Inc., Thornwood, N.Y.) at 630× magnification, and images were acquired by a camera using the appropriate filters. The images were processed with Adobe Photoshop software.
RESULTS
A conserved motif (RXDL) near the carboxy portion of each DED is critical for protection against Fas- and TNF-induced apoptosis.
In order to determine the regions of the MC159 protein that are important for its activities, we used site-directed mutagenesis to change charged or hydrophobic amino acid residues to alanines. Twenty-nine independent MC159 mutants were constructed with a single (or a cluster of a few) amino acid change(s). These mutants are termed simple mutants. Ten additional mutants were derived by engineering two of the simple mutations into the same molecule (termed complex mutants) (Fig. 1). Cytoplasmic expression of each mutant was confirmed by transfection of plasmids into HeLa cells followed by immunofluorescence using anti-MC159 polyclonal rabbit antiserum (data not shown).
To determine which domains in MC159 are critical for its ability to block apoptosis, HeLa cells were transfected with a plasmid expressing β-galactosidase and either empty vector, a plasmid expressing wild-type MC159, or a plasmid expressing one of the 39 MC159 mutants. The cells were then treated with either anti-Fas antibody or TNF in combination with cycloheximide and stained for β-galactosidase expression. The percent death for each MC159 mutant was then determined relative to killing of cells transfected with empty vector (set at 100%), and mutants with values greater than 50% death were designated as not protective.
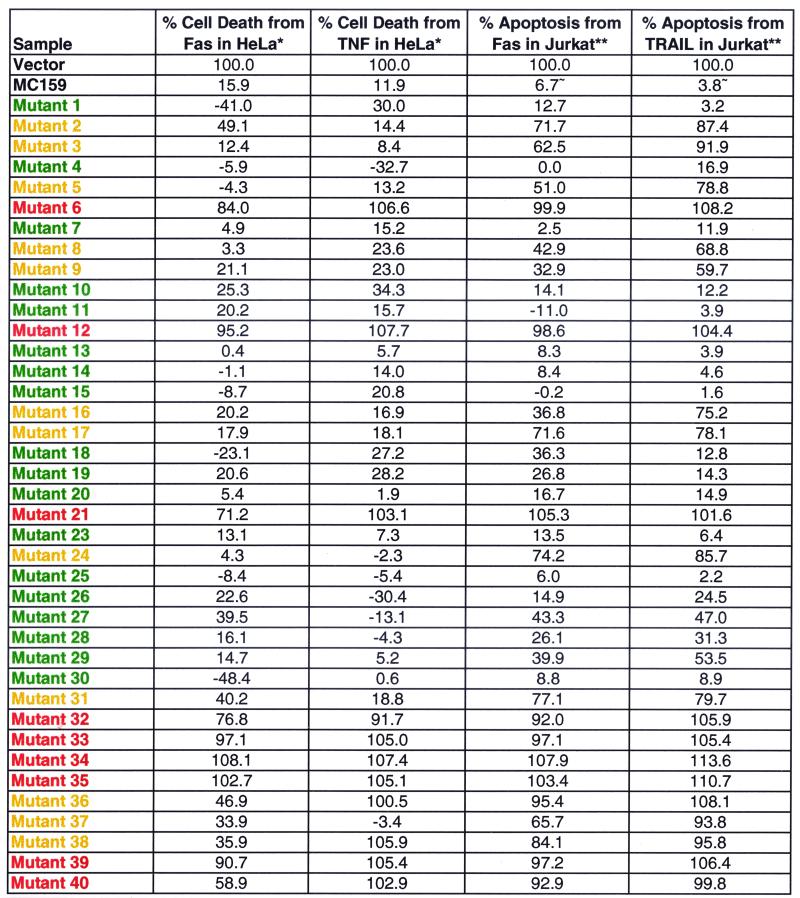
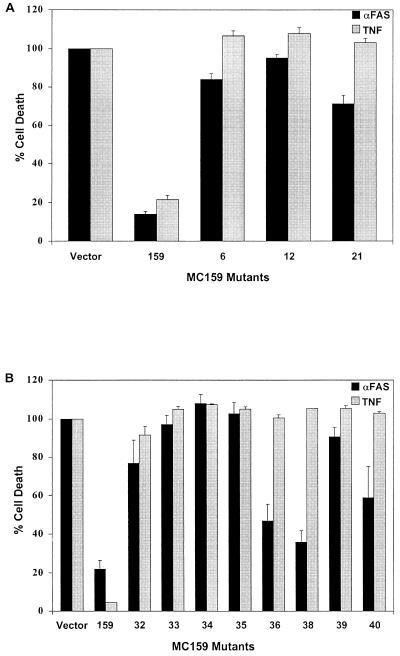
Thirty of 39 mutants remained fully or partially protective against Fas- and TNF-mediated apoptosis (Fig. 1; Tables 1 and 2). Mutants 6, 12, and 21 lost the ability to protect against Fas- and TNF-induced apoptosis in HeLa cells, allowing 84, 95, and 71% of cells to be killed by Fas and 107, 108, and 103% of cells to be killed by TNF, respectively, compared to cells transfected with the control plasmid alone (Fig. 2A; Table 1). All three mutants contain simple mutations located in the conserved RXDL region. The mutations in mutants 6 and 21 are located in the first DED of MC159, and that of mutant 12 is in the second DED (Fig. 1). These results indicate that the RXDL region of each DED is critical for protection against Fas and TNF apoptosis, whereas the vast majority of the other residues tested were tolerant of substitution. The only other mutants that lost the ability to protect against Fas- and TNF-induced apoptosis were all complex mutants. These complex mutants are all composed of pairs of simple mutations that are both located in the first DED, the second DED, or in one DED each (Fig. 1 and 2B).
TABLE 1.
MC159 alanine substitution mutantsa
Protection: red, complete loss; yellow, partial loss; green, no loss. Each value has been normalized to the vector, equal to 100% cell death or apoptosis. *, Value represents the average of all experiments performed for each mutant; **, value is from a representative experiment; ∼, average of the MC159 values for the representative experiments.
TABLE 2.
Structure and function comparisons for selected MC159 mutants
| Function | Mutant | DED | Positions of mutationsa |
|---|---|---|---|
| Loss of protection from Fas, TNF, and TRAIL | 6 | A | All are in the conserved RXDL motif of the α6 region |
| 12 | B | ||
| 21b | A | ||
| Loss of protection from TRAIL in Jurkat cells but partial protection from Fas in Jurkat cells | 8 | B | All are in the α1 region |
| 9 | B | ||
| 16 | A | ||
| Loss of protection from Fas in Jurkat cells but not in HeLa cells | 2 | A | Between α1 and α2 |
| 3 | A | α2 | |
| 5 | A | α5 | |
| 17 | A | α2 and hydrophobic patch 1 | |
| 24 | A | α2 and hydrophobic patch 1 | |
| 31 | A & B | α5 | |
| 37 | A & B | α1 and after α6 | |
| Reduced binding to FADD | 35 | A | Between α1 and α2; α2 |
| 40 | A | α2 region and hydrophobic patch 1 | |
| Loss of binding to caspase-8 | 32 | A & B | All are in α2 region and hydrophobic patch 1 |
| 40 | A | ||
| Inability to inhibit death effector filament formation | 6 | A | All are in the conserved RXDL motif of the α6 region |
| 12 | B | ||
| 21c | A |
Secondary structure is based on modeling the MC159 DEDs onto the NMR-determined structure of the FADD DED.
Complex mutants 32, 33, 34, 35, 36, 38, 39, and 40 also have this phenotype. Mutants 36 and 38 were nonprotective against Fas in Jurkat cells but were partially protective against Fas in HeLa cells.
Mutants 2, 9, 16, 17, 33, 34, 35, 36, and 40 also show partial or complete loss of inhibition of FADD death effector filament formation.
FIG. 2.
Eleven of 39 MC159 mutants lost the ability to protect against Fas- or TNF-induced apoptosis in HeLa cells. HeLa cells were cotransfected with a plasmid expressing β-galactosidase and empty vector or the indicated plasmid and treated with TNF or anti-Fas antibody. Percent protection was determined by counting the number of blue cells surviving after treatment with anti-Fas or TNF compared to the number of cells with no treatment. (A) Nonprotective simple mutants. (B) Nonprotective complex mutants. Values were normalized so that vector was equal to 0% protection. Percent cell death was calculated as 100% minus percent protection. The values represent the averages of at least two experiments.
MC159 mutants show differences in protection from apoptosis with different cell lines and different stimuli.
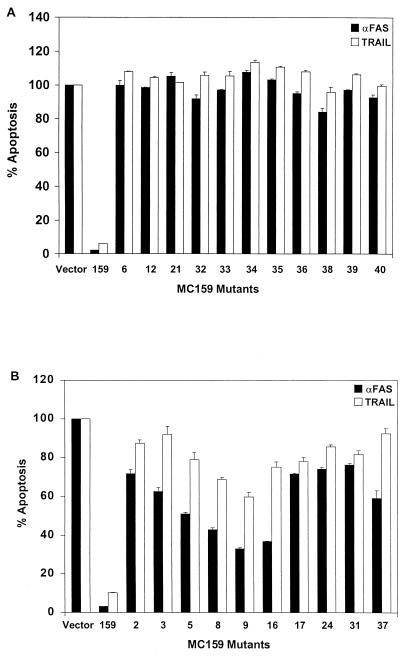
The correlation of functional deficits in specific regions of MC159 as defined in HeLa cells was also tested in T lymphocytes. Jurkat cells were cotransfected with the MC159 constructs and a plasmid expressing green fluorescent protein (pEGFP). The transfected cells were treated with either TRAIL or anti-Fas antibody, stained with annexin-V-PE, and analyzed by flow cytometry for GFP- and annexin-V-PE-positive cells. All the mutants that lost protection from apoptosis mediated by Fas and/or TNF in HeLa cells were also unable to protect against Fas- and TRAIL-induced apoptosis in Jurkat cells (Fig. 3A). Thus, the conserved RXDL motif that is altered in mutants 6, 12, and 21 is critical for protection from death receptor-mediated apoptosis in both HeLa and Jurkat cells.
FIG. 3.
Twenty-one MC159 mutants lost the ability to protect against Fas- and/or TRAIL-induced apoptosis in Jurkat cells. Jurkat cells were cotransfected with the indicated plasmid and a plasmid expressing GFP. The transfected cells were treated with anti-Fas antibody or TRAIL, stained with annexin-V-PE, and analyzed by flow cytometry for GFP- and annexin-V-positive cells. (A) MC159 mutants unable to protect against apoptosis in HeLa cells and Jurkat cells. (B) MC159 mutants able to protect against apoptosis in HeLa cells but not in Jurkat cells. Percent apoptosis was determined by the percentage of annexin-V-positive cells in the GFP-positive population. The values were normalized to empty vector, set at 100%. Each value represents one experiment done in duplicate, and the experiment was performed twice.
In addition, 12 other MC159 mutants that completely or partially protected from Fas-mediated apoptosis in HeLa cells (mutants 2, 3, 5, 8, 9, 16, 17, 24, 31, 36, 37, and 38) showed reduced protection against Fas- and/or TRAIL-induced apoptosis in Jurkat cells (Fig. 3; Table 2). Three of these mutants (mutants 8, 9, and 16) apparently lost the ability to protect against TRAIL but partially protected against Fas-induced apoptosis in Jurkat cells (Fig. 3B; Table 2). These three mutations are located in the predicted α1 region of MC159, suggesting that this region may be more important for protection against apoptosis mediated by TRAIL than by Fas.
Seven of the 12 mutants that protected against Fas-mediated apoptosis in HeLa cells but protected less well against Fas- and TRAIL-mediated killing of Jurkat cells (mutants 2, 3, 5, 16, 17, 24, and 36) have mutations located in the first DED of MC159 (Fig. 3; Table 1). In contrast, some mutations in the corresponding site of the second DED of MC159 retained the ability to protect against Fas- or TRAIL-induced apoptosis in Jurkat cells. Mutant 11 (D159A), mutated in the second DED of MC159, corresponds to mutant 5 (E62A), mutated in the first DED, and mutant 19 (F118A), mutated in the second DED, corresponds to mutant 24 (L31A), mutated in the first DED (Fig. 1). Mutants 5 and 24, mutated in the first DED, showed partial or complete loss of protection from Fas- and TRAIL-mediated apoptosis in Jurkat cells, while mutants 11 and 19, mutated in the second DED, retained protection in Jurkat cells (Table 1). Thus, protection against Fas- and TRAIL-mediated apoptosis in Jurkat cells appears to depend more on the first DED than on the second DED. Furthermore, these results indicate that the two DEDs of MC159 are not simply interchangeable.
RXDL motif located in the predicted α6 region is important for inhibition of FADD death effector filaments.
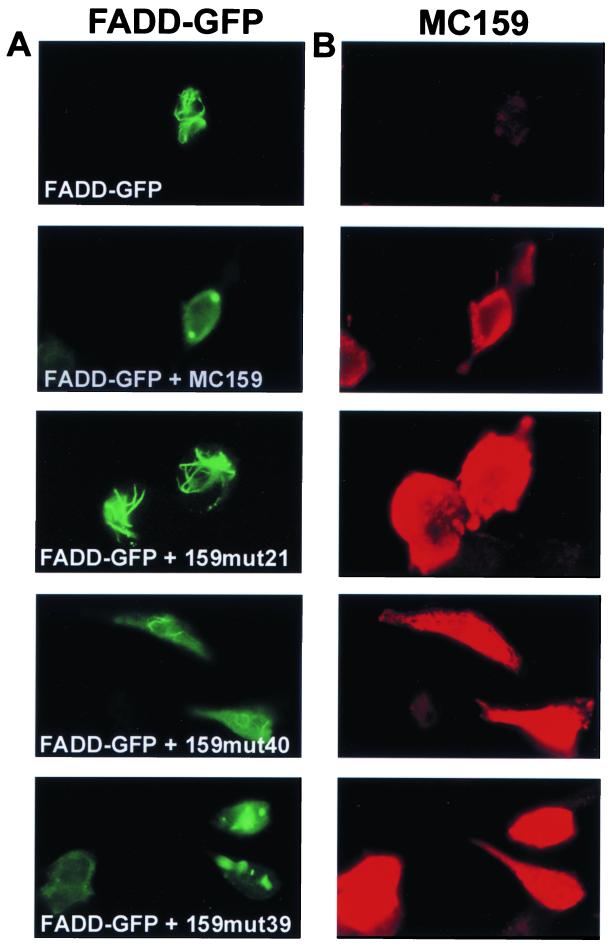
Death effector filaments are oligomerized DED-containing proteins that recruit caspases and cause apoptosis. Since MC159 blocks the ability of FADD to form death effector filaments (23), each of the above MC159 mutants was examined for its ability to block the formation of death effector filaments. HeLa cells were cotransfected with plasmids expressing each MC159 mutant and FADD-GFP and then stained with rabbit anti-MC159 antibody followed by Texas red-conjugated anti-rabbit IgG antibody. Twelve of 39 MC159 mutants showed complete (mutants 2, 16, 21, and 34) or partial (mutants 6, 9, 12, 17, 33, 35, 36, and 40) loss of the ability to block FADD death effector filament formation (Fig. 4; Table 2). In addition, these 12 mutants lost the ability to protect against death receptor-induced apoptosis in HeLa and/or Jurkat cells. The ability of mutant 39 to block death effector filament formation was comparable to that of wild-type MC159, while mutant 40 demonstrated partial death effector filament inhibition. Among these mutants, it was striking that mutants 6, 12, and 21, mutated in the α6 region and RXDL motif in either DED, lost some ability to block death effector filament formation (Table 2). Therefore, the α6 region of both DEDs contributes to inhibition of death effector filament formation.
FIG. 4.
Three patterns of death effector inhibition are produced by the MC159 mutants. HeLa cells were cotransfected with FADD-GFP and either wild-type MC159 or MC159 mutants. At 24 h after transfection, the cells were fixed and stained with anti-MC159 polyclonal rabbit antiserum followed by anti-rabbit IgG-Texas red antibody. The cells were visualized on a fluorescent microscope. (A) FADD-GFP expression in green. (B) MC159 expression in red. FADD-GFP expression alone shows the formation of death effector filaments, and the coexpression of FADD-GFP and MC159 shows complete inhibition of death effector filament formation. Mutant 21 shows no inhibition, mutant 40 shows partial inhibition, and mutant 39 shows complete inhibition of death effector filaments. The experiment was done twice, and a representative result is shown.
MC159 does not form oligomers.
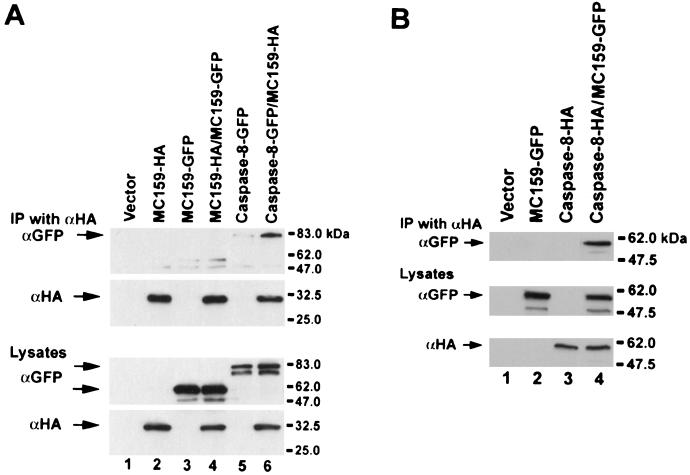
Since the DEDs of MC159 interact with the DEDs of caspase-8, which can form oligomers, we determined whether MC159 could also form self-oligomers. 293T cells were cotransfected with plasmids expressing MC159-HA and MC159-GFP, and cell lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-HA and anti-GFP antibodies. As a control, 293T cells were cotransfected with caspase-8-GFP and MC159-HA. While caspase-8-GFP coprecipitated with MC159-HA, MC159-GFP did not (Fig. 5A). As an additional control to show that the MC159-GFP fusion protein was functional, we found that the MC159-GFP coimmunoprecipitated with caspase-8 (Fig. 5B) and blocked apoptosis similar to wild-type MC159 (unpublished data). Therefore, MC159 does not self-oligomerize.
FIG. 5.
MC159 does not oligomerize. 293T cells were cotransfected with the indicated plasmids, lysed, immunoprecipitated (IP) with anti-HA (αHA) monoclonal antibody, and immunoblotted with anti-HA and anti-GFP to determine binding. The top panels demonstrate binding, and the bottom panels show the expression levels of the proteins in the lysates. (A) MC159 does not form oligomers with itself. A small amount of MC159-GFP is detected in cells transfected with either MC159-GFP or MC159-GFP and MC159-HA after immunoprecipitation with anti-HA antibody. This is presumably due to incomplete washing of the immunoprecipitates. (B) MC159-GFP binds to caspase-8. The experiments were done twice with the same result.
Binding of MC159 to FADD and caspase-8 is not sufficient for protection against apoptosis.
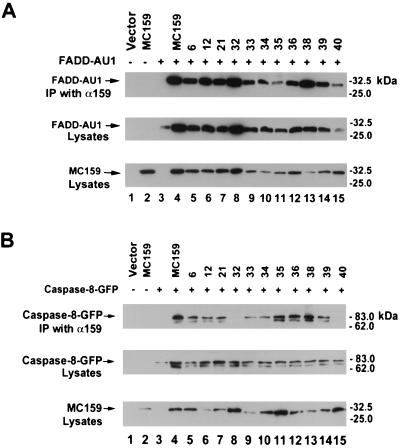
MC159 contains two DEDs that bind to the DEDs of FADD and caspase-8. To determine which portions of MC159 are required for interaction with each of these proteins, 293T cells were cotransfected with the MC159 mutants and either FADD-AU1 or caspase-8-GFP. Caspase-8-GFP contains the active-site mutation C360S to prevent spontaneous apoptosis. The cell lysates were immunoprecipitated with anti-MC159 polyclonal rabbit antiserum and immunoblotted with either anti-FADD antibody or anti-GFP antibody to determine the ability of the mutants to bind to FADD and caspase-8, respectively.
Surprisingly, nearly all the MC159 mutants that had lost the ability to protect against Fas-, TNF-, or TRAIL-mediated apoptosis were still able to bind to both FADD and caspase-8. Three mutants showed decreased binding to FADD and/or caspase-8 (Fig. 6). MC159 mutant 35 showed slightly reduced binding to FADD but still bound caspase-8. MC159 mutant 32 lost the ability to bind caspase-8 but still bound FADD. MC159 mutant 40 had slightly reduced binding to FADD and did not bind to caspase-8.
FIG. 6.
Binding of nonprotective MC159 mutants to FADD and caspase-8. 293T cells were cotransfected with either (A) FADD-AU1 or (B) caspase-8-GFP and the indicated plasmid. The cells were lysed, immunoprecipitated (IP) with anti-MC159 polyclonal rabbit antiserum, and immunoblotted with either anti-FADD MAb or anti-GFP MAb to determine binding. The top panel of each figure demonstrates binding to either FADD or caspase-8, the middle panel shows the level of FADD-AU1 or caspase-8 in the lysate, and the bottom panel shows the amount of MC159 in the lysate. The experiment was done four times with similar results, and a representative result is shown.
Comparison of the MC159 DED sequences with the structure of the FADD DED indicated that these three mutations involve amino acid substitutions in the predicted α2 region of the MC159 DEDs (Fig. 1; Table 2). The FADD DED contains a series of hydrophobic amino acids that contribute to hydrophobic patch 1 that is highly conserved among other DED-containing proteins and that was found to be important for FADD binding to caspase-8 (6). In addition to affecting the α2 region, the two MC159 mutations that resulted in the loss of binding to caspase-8 are also located in the predicted highly conserved hydrophobic patch 1 of the MC159 DED. These results indicate that MC159 binding to FADD or to caspase-8 is not sufficient to protect from apoptosis induced by the death receptors, since a number of mutants that retained binding to both FADD and caspase-8 had no antiapoptotic activity.
DISCUSSION
Using a series of mutants, we identified distinct regions of the MC159 protein that are required to block apoptosis, bind to caspase-8 and FADD, and inhibit formation of death effector filaments. The RXDL motif located in the α6 region, present near the carboxy portions of both DEDs, was critical for protection against apoptosis by Fas, TNF, and TRAIL. The RXDL motif is conserved among the DED-containing proteins. Both DEDs of most of the v-FLIPs (including MCV MC159, EHV-2 E8, and Kaposi’s sarcoma-associated herpesvirus K13), the two DEDs of the cellular FLIP, the FADD DED, and the first DED of caspase-8 have the motif RXDL (6, 27).
The RXDL motif was also important for inhibition of death effector filament formation. Mutants 6, 12, and 21, mutated in the RXDL motif, had partial or complete loss of the ability to inhibit death effector filament formation and failed to block apoptosis induced by all stimuli tested. Therefore, since a mutation in either DED resulted in a loss of function, the α6 region of both MC159 DEDs must remain intact to maintain inhibition of apoptosis and death effector filament formation. Surprisingly, while mutations in this motif were important for protection from apoptosis, these mutants retained the ability to bind to FADD and caspase-8. Thus, simply binding to FADD and caspase-8 is not sufficient to block Fas signaling. MC159 is thought to disrupt death effector filament formation by binding to the DED of FADD and blocking oligomerization of the protein. Interestingly, since the three mutations that caused slightly reduced binding to FADD or loss of binding to caspase-8 (mutants 32, 35, and 40) were in a different region than the RXDL mutations, our data also suggest that separate regions of MC159 DEDs are required for inhibition of death effector filament formation and for binding FADD and caspase-8.
Mutations in the first DED of MC159 (mutants 5 and 24) resulted in loss of protection against Fas- and TRAIL-mediated apoptosis in Jurkat cells. In contrast, two mutants with mutations at the corresponding site in the second DED of MC159 (mutants 11 and 19) were both able to protect against Fas- and TRAIL-induced apoptosis in Jurkat cells. The ability of similar mutations in the first but not the second DED of MC159 to impair protection from apoptosis indicates that the DEDs are not functionally equivalent.
Recent studies (12, 24) indicate that the binding of TRAIL to its death receptors recruits FADD and caspase-8 to the DISC, resulting in activation of apoptosis. Earlier studies had suggested that an adapter molecule other than FADD might interact with TRAIL (29, 30). Our data suggest that FADD may be involved in the TRAIL pathway, since most of the MC159 mutants that blocked or lost the ability to block Fas- and TNF-induced apoptosis had a similar phenotype with TRAIL. However, three mutants (mutants 8, 9, and 16) were more protective against Fas- than TRAIL-mediated apoptosis in Jurkat cells. These findings suggest that that there may be additional differences between the Fas and TRAIL pathways in Jurkat cells.
Certain amino acid residues in the portion of hydrophobic patch 1 that is located in the α2 region of the MC159 DEDs are critical for the ability of the protein to bind to FADD and caspase-8. Mutant 32, which contains the mutations F30A and F118A, lost the ability to bind to caspase-8 and to block Fas-, TNF-, and TRAIL-induced apoptosis. The F30A mutation in MC159 corresponds to mutations at F25 of FADD that result in loss of FADD apoptotic activity and markedly reduce binding to caspase-8 (6). Thus, mutations at similar sites in MC159 and FADD interfere with both apoptotic activity and binding to caspase-8. In addition, both amino acid substitutions in mutant 32 correspond to amino acids in hydrophobic patch 1 in the FADD DED (6).
The MC159 mutations F30A and F118A (mutants 17 and 19, respectively) are both part of hydrophobic patch 1 but in different DEDs. Each of these mutations alone is able to bind to caspase-8 (data not shown), but when both mutations are present together, caspase-8 binding is lost. Thus, hydrophobic patch 1 in either DED of MC159 is important for binding to caspase-8. In addition, mutant 40, which has amino acid substitutions in approximately half of the α2 region in the first DED of MC159, also lost the ability to bind caspase-8. Thus, binding of MC159 to caspase-8 requires that most of the α2 region of the first DED remain intact.
The ability to form oligomers is a common theme for molecules containing death domains and DEDs. Oligomerization of caspase-8 in complex with oligomerized FADD in the DISC is critical for its activation by autoproteolysis (14, 16). High-level expression of proapoptotic cellular DED-containing proteins such as FADD and the DEDs of caspase-8 results in oligomerization, with formation of death effector filaments (18, 23). In contrast, our data indicate that MC159 does not form self-oligomers, consistent with prior observations that high-level expression of MC159 in cells did not result in formation of cytoplasmic filaments (23). Unlike the cellular proteins containing DEDs or death domains that mediate apoptosis, MC159 inhibits apoptosis. The lack of self-oligomerization of MC159 may favor heterodimer formation, with binding of MC159 to FADD, caspase-8, or an unknown protein, rather than homodimerization, and thereby inhibit the Fas-induced signaling complex by causing the formation of an altered, nonfunctional signaling complex.
MCV also encodes a homolog of MC159, termed MC160, that has two DEDs, which have 45 and 33% amino acid identity with the corresponding DEDs of MC159 (20). MC160 has a phenotype similar to some of the MC159 mutants in that MC160 does not block apoptosis induced by Fas and TNF but does bind to both FADD and caspase-8 (21). Thus, like the MC159 mutants, the ability of MC160 to bind to FADD or caspase-8 is insufficient to block apoptosis induced by the death receptors.
Our data suggest that a new model is needed to explain the ability of MC159 to block death receptor-induced apoptosis that involves more than simple competitive binding of MC159 with FADD and caspase-8. The majority of nonprotective mutants were still able to bind to both FADD and caspase-8. For example, mutant 39 lost the ability to block Fas-, TNF-, and TRAIL-induced apoptosis but maintained the ability to bind FADD and caspase-8 and to block death effector filament formation. This suggests that the antiapoptotic ability of MC159 requires novel functions in addition to its ability to block oligomerization of apoptotic signaling proteins containing DEDs.
Acknowledgments
We are grateful to J. Shisler for the gift of anti-MC159 polyclonal rabbit antiserum, S. Fesik for the coordinates of the FADD NMR structure, and E. Prikhodko for helpful discussions.
REFERENCES
- 1.Ashkenazi, A., and V. M. Dixit. 1998. Death receptors: signaling and modulation. Science 281:1305–1308. [DOI] [PubMed] [Google Scholar]
- 2.Bertin, J., R. C. Armstrong, S. Ottilie, D. A. Martin, Y. Wang, S. Banks, G. H. Wang, T. G. Senkevich, E. S. Alnemri, B. Moss, M. J. Lenardo, K. J. Tomaselli, and J. I. Cohen. 1997. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc. Natl. Acad. Sci. USA 94:1172–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodmer, J. L., N. Holler, S. Reynard, P. Vinciguerra, P. Schneider, P. Juo, J. Blenis, and J. Tschopp. 2000. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat. Cell Biol. 2:241–243. [DOI] [PubMed] [Google Scholar]
- 4.Chan, F. K., H. J. Chun, L. Zheng, R. M. Siegel, K. L. Bui, and M. J. Lenardo. 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288:2351–2354. [DOI] [PubMed] [Google Scholar]
- 5.Chinnaiyan, A. M., K. O’Rourke, G. L. Yu, R. H. Lyons, M. Garg, D. R. Duan, L. Xing, R. Gentz, J. Ni, and V. M. Dixit. 1996. Signal transduction by DR3, a death domain-containing receptor related to TNFR-1 and CD95. Science 274:990–992. [DOI] [PubMed] [Google Scholar]
- 6.Eberstadt, M., B. Huang, Z. Chen, R. P. Meadows, S. C. Ng, L. Zheng, M. J. Lenardo, and S. W. Fesik. 1998. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature 392:941–945. [DOI] [PubMed] [Google Scholar]
- 7.Fisher, G. H., F. J. Rosenberg, S. E. Straus, J. K. Dale, L. A. Middleton, A. Y. Lin, W. Strober, M. J. Lenardo, and J. M. Puck. 1995. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 81:935–946. [DOI] [PubMed] [Google Scholar]
- 8.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528–535. [PubMed] [Google Scholar]
- 9.Hsu, H., H. B. Shu, M. G. Pan, and D. V. Goeddel. 1996. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84:299–308. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, H., J. Xiong, and D. V. Goeddel. 1995. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell 81:495–504. [DOI] [PubMed] [Google Scholar]
- 11.Hu, S., C. Vincenz, M. Buller, and V. M. Dixit. 1997. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J. Biol. Chem. 272:9621–9624. [DOI] [PubMed] [Google Scholar]
- 12.Kischkel, F. C., D. A. Lawrence, A. Chuntharapai, P. Schow, K. J. Kim, and A. Ashkenazi. 2000. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 12:611–620. [DOI] [PubMed] [Google Scholar]
- 13.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104:487–501. [DOI] [PubMed] [Google Scholar]
- 14.Martin, D. A., L. Zheng, R. M. Siegel, B. Huang, G. H. Fisher, J. Wang, C. E. Jackson, J. M. Puck, J. Dale, S. E. Straus, M. E. Peter, P. H. Krammer, S. Fesik, and M. J. Lenardo. 1999. Defective CD95/APO-1/Fas signal complex formation in the human autoimmune lymphoproliferative syndrome, type Ia. Proc. Natl. Acad. Sci. USA 96:4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medema, J. P., C. Scaffidi, F. C. Kischkel, A. Shevchenko, M. Mann, P. H. Krammer, and M. E. Peter. 1997. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J. 16:2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muzio, M., A. M. Chinnaiyan, F. C. Kischkel, K. O’Rourke, A. Shevchenko, J. Ni, C. Scaffidi, J. D. Bretz, M. Zhang, R. Gentz, M. Mann, P. H. Krammer, M. E. Peter, and V. M. Dixit. 1996. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85:817–827. [DOI] [PubMed] [Google Scholar]
- 17.Pan, G., Bauer, J. H., Haridas, V., Wang, S., Liu, D., Yu, G., Vincenz, C., Aggarwal, B. B., Ni, J., Dixit, V. M. 1998. Identification and functional characterization of DR6, a novel death domain-containing TNF receptor. FEBS Lett. 431:351–356. [DOI] [PubMed] [Google Scholar]
- 18.Perez, D., and E. White. 1998. E1B 19K inhibits Fas-mediated apoptosis through FADD-dependent sequestration of FLICE. J. Cell Biol. 141:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peter, M. E., C. Scaffidi, J. P. Medema, F. Kischkel, and P. H. Krammer. 1999. The death receptors. Results Probl. Cell Differ. 23:25–63. [DOI] [PubMed] [Google Scholar]
- 20.Senkevich, T. G., J. J. Bugert, J. R. Sisler, E. V. Koonin, G. Darai, and B. Moss. 1996. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science 273:813–816. [DOI] [PubMed] [Google Scholar]
- 21.Shisler, J. L., and B. Moss. 2001. Molluscum contagiosum virus inhibitors of apoptosis: The MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology 282:14–25. [DOI] [PubMed] [Google Scholar]
- 22.Siegel, R. M., J. K. Frederiksen, D. A. Zacharias, F. K. Chan, M. Johnson, D. Lynch, R. Y. Tsien, and M. J. Lenardo. 2000. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 288:2354–2357. [DOI] [PubMed] [Google Scholar]
- 23.Siegel, R. M., D. A. Martin, L. Zheng, S. Y. Ng, J. Bertin, J. Cohen, and M. J. Lenardo. 1998. Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J. Cell Biol. 141:1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sprick, M. R., M. A. Weigand, E. Rieser, C. T. Rauch, P. Juo, J. Blenis, P. H. Krammer, and H. Walczak. 2000. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 12:599–609. [DOI] [PubMed] [Google Scholar]
- 25.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517–521. [DOI] [PubMed] [Google Scholar]
- 26.Tsukumo, S. I., and S. Yonehara. 1999. Requirement of cooperative functions of two repeated death effector domains in caspase-8 and in MC159 for induction and inhibition of apoptosis, respectively. Genes Cells. 4:541–549. [DOI] [PubMed] [Google Scholar]
- 27.Wang, G. H., J. Bertin, Y. Wang, D. A. Martin, J. Wang, K. J. Tomaselli, R. C. Armstrong, and J. I. Cohen. 1997. Bovine herpesvirus 4 BORFE2 protein inhibits Fas- and tumor necrosis factor receptor 1-induced apoptosis and contains death effector domains shared with other gamma-2 herpesviruses. J. Virol. 71:8928–8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, J., L. Zheng, A. Lobito, F. K. Chan, J. Dale, M. Sneller, X. Yao, J. M. Puck, S. E. Straus, and M. J. Lenardo. 1999. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell 98:47–58. [DOI] [PubMed] [Google Scholar]
- 29.Yeh, W. C., J. L. Pompa, M. E. McCurrach, H. B. Shu, A. J. Elia, A. Shahinian, M. Ng, A. Wakeham, W. Khoo, K. Mitchell, W. S. El-Deiry, S. W. Lowe, D. V. Goeddel, and T. W. Mak. 1998. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 279:1954–1958. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, J., D. Cado, A. Chen, N. H. Kabra, and A. Winoto. 1998. Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature 392:296–300 [DOI] [PubMed] [Google Scholar]