Abstract
Previous studies of the avian reovirus strain S1133 (ARV-S1133) S1 genome segment revealed that the open reading frame (ORF) encoding the ςC viral cell attachment protein initiates over 600 nucleotides distal from the 5′ end of the S1 mRNA and is preceded by two predicted small nonoverlapping ORFs. To more clearly define the translational properties of this unusual polycistronic RNA, we pursued a comparative analysis of the S1 genome segment of the related Nelson Bay reovirus (NBV). Sequence analysis indicated that the 3′-proximal ORF present on the NBV S1 genome segment also encodes a ςC homolog, as evidenced by the presence of an extended N-terminal heptad repeat characteristic of the coiled-coil region common to the cell attachment proteins of reoviruses. Most importantly, the NBV S1 genome segment contains two conserved ORFs upstream of the ςC coding region that are extended relative to the predicted ORFs of ARV-S1133 and are arranged in a sequential, partially overlapping fashion. Sequence analysis of the S1 genome segments of two additional strains of ARV indicated a similar overlapping tricistronic gene arrangement as predicted for the NBV S1 genome segment. Expression analysis of the ARV S1 genome segment indicated that all three ORFs are functional in vitro and in virus-infected cells. In addition to the previously described p10 and ςC gene products, the S1 genome segment encodes from the central ORF a 17-kDa basic protein (p17) of no known function. Optimizing the translation start site of the ARV p10 ORF lead to an approximately 15-fold increase in p10 expression with little or no effect on translation of the downstream ςC ORF. These results suggest that translation initiation complexes can bypass over 600 nucleotides and two functional overlapping upstream ORFs in order to access the distal ςC start site.
The orthoreoviruses consist of a number of diverse virus species, all of which have a similar capsid structure and a genome composed of 10 double-stranded RNA (dsRNA) segments (38). For the most part, the genome segments of reoviruses are monocistronic, each encoding a single unique polypeptide. The mammalian reovirus (MRV) S1 genome segment, however, is bicistronic and encodes two unrelated polypeptides from overlapping open reading frames (ORFs). The ς1 viral cell attachment protein is encoded by a large ORF that originates near the 5′ end of the mRNA and spans almost the complete length of the S1 mRNA (12, 37). A second small ORF, beginning at nucleotide position 71, is nested within the ς1 ORF in a +1 reading frame and encodes the small nonstructural ς1NS protein of MRV (14, 23).
Since the translation start site for ς1 is in a nonpreferred context (a pyrimidine at position −3), leaky scanning by the preinitiation complex most likely accounts for translation of the downstream ς1NS ORF from the MRV S1 mRNA (28). In addition to leaky scanning, there is evidence suggesting that translation of the ς1 protein from the bicistronic S1 mRNA is modulated by ribosomal pausing at the downstream ς1NS start site (7, 15).
As with MRV, the avian reovirus (ARV) cell attachment protein, termed ςC, is encoded by the S1 genome segment (33, 46, 47). However, the initiation codon for the ARV strain S1133 (ARV-S1133) ςC ORF occurs over 600 nucleotides distal from the 5′ end of the RNA, and this ORF is preceded by two small nonoverlapping ORFs that could potentially encode 9.8-kDa and 3.8-kDa polypeptides (46). We have recently shown that the first ORF encodes a p10 protein, responsible for the syncytium-inducing properties of the fusogenic reoviruses (26, 48). The presence of two potential ORFs, at least one of which is functional, upstream of the ςC ORF suggests that unconventional translation initiation mechanisms may regulate ARV ςC expression.
Nelson Bay reovirus (NBV) represents an additional species of orthoreovirus with extensive sequence divergence from both ARV and MRV (9). NBV was isolated from a flying fox in Nelson Bay, New South Wales, Australia, and is one of only two known reoviruses isolated from mammals that have the syncytium-inducing property typical of ARV isolates (13, 17, 26). In spite of its mammalian host, NBV is more closely related to ARV than to the prototypical nonfusogenic MRV isolates. However, amino acid sequence identities of approximately 30 to 60% between ARV and NBV are well below the greater than 90% identity exhibited by members within the ARV and MRV species groups (9). Consequently, NBV has been classified as a separate species within the genus Orthoreovirus.
Comparative analysis of polycistronic mRNAs derived from two reovirus species with divergent nucleotide sequences should facilitate analysis of the factors that influence translation initiation. We therefore undertook sequence and functional analyses of the S1 genome segments of NBV and two additional strains of ARV. Our results indicate that both the ARV and NBV S1 genome segments contain three conserved ORFs arranged in a sequential, partially overlapping fashion. This unusual tricistronic arrangement of ORFs influences the translational control mechanisms that regulate expression of this polycistronic eukaryotic mRNA, as indicated by a lack of correlation between the translation efficiencies of the 5′- and 3′-proximal ORFs.
MATERIALS AND METHODS
Virus strains and cells.
ARV strains 176 (ARV-176) and 138 (ARV-138) have been described previously (9). NBV was isolated from the heart blood of a flying fox (17) and was obtained from Terrence Dermody (Vanderbilt University). The ARV isolates were plaque purified and amplified to passage 4 at a multiplicity of infection (MOI) of 0.01 in a continuous quail cell line, QM5 (3), as previously described (8). NBV was similarly plaque purified and amplified in Vero cells.
Cloning and sequencing the S1 genome segment.
Viral dsRNA was isolated from concentrated virus stocks as previously described (10), and the RNA was poly(A) tailed and used as a template for cDNA production using oligo(dT)18-NotI primers and reverse transcription-PCR, with Superscript reverse transcriptase (Life Technologies Inc.) and Vent polymerase (New England Biolabs) using standard protocols (9). The amplified cDNA was digested with NotI and cloned into the NotI site of pBluescript II SK (Stratagene). Clones containing cDNA inserts the approximate size of the full-length S1 genome segment were sequenced using a Licor automated sequencer at the National Research Council-Dalhousie Core Sequencing Facility. All clones were sequenced completely in both directions.
The NBV S1 genome segment sequence was determined for two independently isolated cDNA clones in two different laboratories. The accession numbers for the new S1 genome segment sequences are AF218360 (NBV), AF218359 (ARV-138), and AF218358 (ARV-176). Sequences were compiled and analyzed using the University of Wisconsin GCG software, version 8 (6).
Antiserum production.
Polyclonal monospecific antiserum was raised in rabbits against the C-terminal domain of p10 using a purified maltose-binding protein-p10 chimeric protein construct expressed in Escherichia coli (48). A similar approach was used to express the full-length p17 protein, and the chimeric maltose-binding protein-p17 protein was used for antiserum production in rabbits. The ςC protein was immunoprecipitated using polyclonal rabbit antiserum raised against purified virus particles that contained ςC but not the nonstructural p10 protein (8).
In vitro translation.
The full-length S1 cDNA clones in pBluescript were used for the production of capped mRNA transcripts. The plasmids were linearized on the 3′ side of the cDNA insert with HindIII, and runoff plus-strand transcripts synthesized in vitro using T7 RNA polymerase and 7mGpppG, as previously described (11). The quality and quantity of the capped RNA were assessed on denaturing agarose gels. The mRNA was translated in rabbit reticulocyte lysates (Promega Biotech), using 250 ng of transcript per 50 μl of translation reaction, for 90 min at 30°C in the presence of 5 μCi of [3H]leucine (Amersham). The reactions were diluted to 1 ml with radioimmunoprecipitation assay (RIPA) buffer, and the p10, p17, and ςC proteins were identified by immunoprecipitation using monospecific rabbit polyclonal antiserum as previously described (48). Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% polyacrylamide) and detected by fluorography.
Translation analysis in virus-infected and transfected cells.
QM5 cell monolayers in six-well cluster plates were infected at an MOI of 10 using ARV-176. Infected cells were incubated for 16 h before being pulse-labeled with 100 μCi of [3H]leucine (New England Nuclear) per ml for 1 h. The labeled cells were lysed with RIPA buffer, immunoprecipitated, resolved by SDS-PAGE, and processed for fluorography as previously described (10, 48).
Translation levels of the p10 and ςC ORFs of the ARV S1 genome segment were assessed in cells transfected with a pcDNA3 vector (Invitrogen) containing the authentic S1 genome segment cDNA or a cDNA construct containing an optimized p10 translation start site. Site-directed mutagenesis was used to convert the authentic p10 start site (CGTCGATGC) into an optimized start site (CCACCATGG) as previously reported (48). The authentic and optimized expression plasmids were used to transfect QM5 cell monolayers using Lipofectamine (Life Technologies Inc.), transfected monolayers were labeled with [35S]methionine, and radiolabeled cell lysates were immunoprecipitated with anti-p10 or anti-ARV serum as previously described (48). The precipitated products were resolved by SDS-PAGE and detected by fluorography (10, 48). The translation levels of p10 and ςC were quantified using the Image-Pro Plus software (version 4.0). Translation levels were obtained from three separate experiments using multiple exposures of the fluorograms to ensure that exposures were in the linear range.
RESULTS
Gene organization of NBV S1 genome segment.
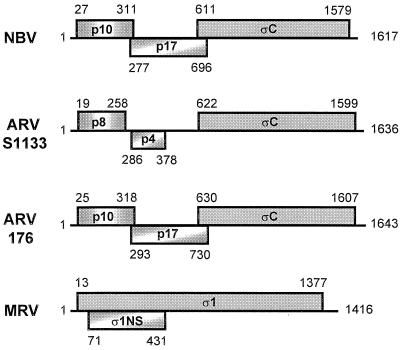
The cDNA and predicted amino acid sequences of the NBV S1 genome segment were determined from two independent cDNA clones. The cDNA nucleotide sequence contained the conserved 5′-terminal GCT triplet and 3′-terminal pentanucleotide sequence (TCATC-3′) characteristic of the other genome segments of the orthoreoviruses (9), indicating that the cDNA clones were full-length replicas of the dsRNA S1 genome segment of NBV. An examination of the coding potential of the NBV S1 genome segment revealed an unexpected gene arrangement. The cDNA sequence of the S1 genome segment of NBV contained three sequential ORFs, similar to the reported situation with ARV-S1133 (46). In the case of NBV, however, the two 5′-proximal ORFs are extended and encode predicted gene products of 95 or 140 amino acids (Fig. 1). As a result, these three ORFs are arranged in a sequential, partially overlapping fashion on the NBV S1 genome segment.
FIG. 1.
Gene arrangement of the polycistronic S1 genome segments of orthoreoviruses. The arrangement of predicted ORFs present in the ARV-176, ARV-S1133, NBV, and MRV S1 genome segments is depicted. The ORFs are shown as shaded boxes above and below a line representing the mRNA. The ORFs are identified by the name or approximate molecular mass (in kilodaltons) of the predicted gene product. Numbers indicate the first and last nucleotide numbers of each ORF (including the initiation codon, excluding the termination codon). The ARV-138 gene arrangement is identical to that depicted for ARV-176. The ARV-S1133 gene arrangement is based on the previously published sequence (46), while the MRV gene arrangement depicted is based on the published sequence of strain Dearing (12).
Identification of NBV cell attachment protein.
Although the S1 genome segments of ARV and MRV are known to encode homologous viral cell attachment proteins (ςC and ς1, respectively), such is not the case with baboon reovirus (BRV), in which no ςC/ς1 equivalent exists in the S-class genome segments (9). Therefore, we sought to identify the cell attachment protein of NBV by comparison of the sequences of the predicted NBV gene products to the previously published sequences of the ARV and MRV cell attachment proteins.
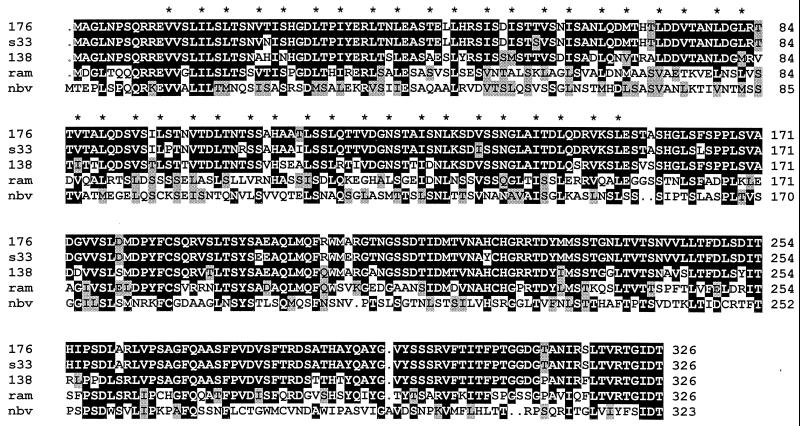
Sequence analysis of the NBV ORF3 gene product revealed a low level of amino acid sequence identity (24 to 27%) to the ςC proteins of the various strains of ARV. More importantly, a multiple sequence alignment (Fig. 2) revealed the presence of a conserved extended heptad repeat structure in the amino-proximal region of the ςC proteins, including the NBV ORF3 gene product. A heptad repeat motif present in the MRV ς1 cell attachment protein is responsible for the formation of a coiled-coil structure which imparts stability to the functional homotrimeric form of the protein (31, 49). A similar heptad repeat was previously identified in the ARV-S1133 ςC cell attachment protein that presumably contributes to the formation of ςC multimers (33, 46, 47). The presence of this characteristic motif in the predicted NBV ORF3 gene product indicates that this ORF encodes the NBV ςC viral cell attachment protein that also likely functions as a coiled-coil-stabilized trimer.
FIG. 2.
Multiple sequence alignment of the reovirus cell attachment proteins. The predicted amino acid sequences of the two ARV ςC proteins determined in this study (ARV-176 [176]) and ARV-138 [138]) and of the gene product of the 3′-proximal ORF of NBV (nbv) were aligned with the predicted sequences of the ςC cell attachment proteins of two previously sequenced ARV isolates (ARV-S1133 [s33] and ARV-Ram1 [ram]) using PILEUP and shaded using BOXSHADE. The asterisks denote the locations of the apolar residues present in the conserved heptad repeat structure. Identical residues present in three or more of the aligned sequences are indicated by black background shading, and grey shading denotes conservation of similar residues. Dots indicate insertions.
Conserved tricistronic gene arrangement in the ARV S1 genome segment.
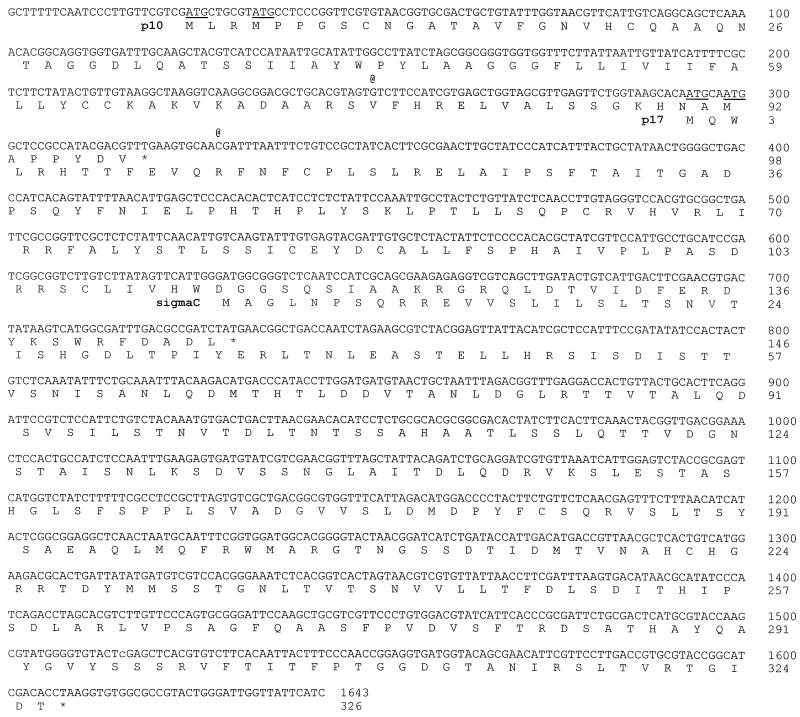
In view of the discrepancy between the NBV and ARV-S1133 S1 genome segment gene arrangements, we cloned and sequenced the S1 genome segments of two additional strains of ARV. The ARV-138 and ARV-176 cDNA sequences were 83% identical and contained the conserved terminal nucleotide signature sequences of the orthoreoviruses. The cDNA and predicted amino acid sequences of the ARV-176 S1 genome segment are presented in Fig. 3. Nucleotide sequence comparisons of ARV-176 with a published sequence of ARV-S1133 (accession number L39002) revealed a high degree of sequence identity, with only 20 nucleotide mismatches. Fourteen of the mismatches occur in the 3′-proximal ςC ORF, resulting in 10 predicted amino acid changes. The other six substitutions occur in the 5′-terminal 450 nucleotides preceding the ςC translation start site at nucleotide 630.
FIG. 3.
ARV-176 S1 genome segment cDNA sequence and predicted amino acid sequences of the encoded gene products. The complete cDNA plus-strand sequence of the ARV-176 S1 genome segment is presented, along with the amino acid sequences of the encoded p10, p17, and ςC proteins. Asterisks indicate the locations of the termination codons of the three ORFs. Potential start codons lying upstream of the ςC translation start site are underlined. The two @ symbols above the cDNA sequence denote the locations of single-base insertions, relative to the published ARV-S1133 sequence, that change the reading frame of the p10 and p17 ORFs (see text).
In addition to the limited number of nucleotide mismatches, four insertions and deletions were detected in the aligned cDNA sequences of ARV-176 and ARV-S1133. Two of the insertions-deletions occur at the ends of the cDNA sequence (a six-nucleotide 5′-terminal deletion [GCTTTT] and 3′-terminal addition of a single C residue in the ARV-S1133 sequence relative to ARV-176). In view of the conservation of our terminal nucleotide sequences with those of the other S-class genome segments (9), the terminal sequences of ARV-176 and ARV-138 are considered correct. Most importantly, the other two detected insertions in the ARV-176 and ARV-138 sequences, relative to the sequence of ARV-S1133, lie internally and contribute to reading frame shifts in the two small predicted 5′-proximal ORFs.
These insertions correspond to a G residue at position 250 and a C residue at position 329 (positions refer to the ARV-176 sequence depicted in Fig. 3). Similar insertions are also present in the S1 genome segment sequences of several other strains of ARV deposited in GenBank (ARV-1733, accession number AAB61606; ARV-Som, accession number L07069; and ARV-Ram, accession number L38502) (27). The two internal nucleotide insertions detected in the ARV-176 and ARV-138 S1 genome segment extend these upstream ORFs to encode predicted gene products of 98 and 146 amino acids, respectively, resulting in a sequential overlapping arrangement of ORFs similar to that present in the NBV S1 genome segment (see Fig. 1).
S1 genome segments are functionally tricistronic.
The 5′-proximal ORFs of the ARV and NBV S1 genome segments encode predicted 95- to 98-amino-acid (10 kDa) proteins with 33% sequence identity. We have recently determined that these ORFs are functional and encode the membrane fusion proteins of these syncytium-inducing reoviruses (48). Previous studies have also shown that the ARV S1 genome segment expresses the ςC protein both in vitro and in vivo (46, 51). Consequently, the ARV S1 genome segment is at least bicistronic.
Our current sequence analysis revealed the presence of an additional ORF in both the ARV and NBV S1 genome segments that overlaps the stop codon of the p10 ORF by 26 to 35 nucleotides and the start codon of the ςC ORF by 86 to 101 nucleotides. The predicted 140- to 146-amino-acid (17 kDa) proteins encoded by the second ORFs of ARV and NBV possess 29% sequence identity (see Fig. 5). The maintenance of these ORFs in the S1 genome segments and the conservation of their predicted gene products in two divergent reovirus species implied that these centrally located ORFs are likely functional. To directly test this assumption, we generated an ARV p17-monospecific polyclonal antiserum raised against a maltose-binding protein-p17 chimeric protein and used this antiserum to detect ARV p17 expression in vitro and in virus-infected cells.
FIG. 5.
Multiple sequence alignment of the p17 and ς1NS proteins. The predicted amino acid sequences of the two ARV p17 proteins (ARV-176 [176]) and ARV-138 [138]) and of the NBV p17 protein (nbv) determined in this study were aligned with the predicted sequences of the p17 protein of a previously sequenced ARV isolate (ARV-Ram1 [ram]) and with the ς1NS protein of mammalian reovirus strain Dearing (mrv) using PILEUP and shaded using BOXSHADE. Identical residues present in three or more of the aligned sequences are indicated by black background shading, and grey shading denotes conservation of similar residues. Dots indicate insertions.
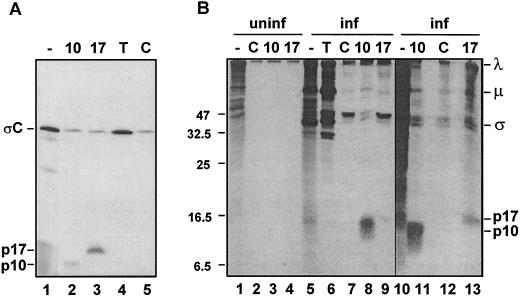
Immunoprecipitation analysis of the gene products generated by in vitro translation of the ARV S1 mRNA clearly revealed three unique gene products. As shown previously, polyclonal antisera raised against ARV structural proteins specifically recognized the predominant 32-kDa ςC polypeptide, while a polyclonal antibody specific for the product of the 5′ ORF detected the faint presence of a 10-kDa polypeptide corresponding to the nonstructural p10 fusion protein of ARV (Fig. 4A). Precipitation using antiserum raised against the gene product of the newly identified central ORF specifically recognized a 17-kDa polypeptide, indicating that the p17 ORF is functional. A low level of the ςC protein was nonspecifically precipitated by both the p10- and p17-specific antisera. This nonspecific precipitation, also evident when using preimmune control antiserum, was a reflection of the low level of p10 and p17 expression, which required using increased aliquots of the translation mixture for the precipitations. The unfavorable context of the p10 and p17 start sites, with pyrimidines located in the −3 and/or +4 positions (28), presumably contributes to the low-level expression of these gene products.
FIG. 4.
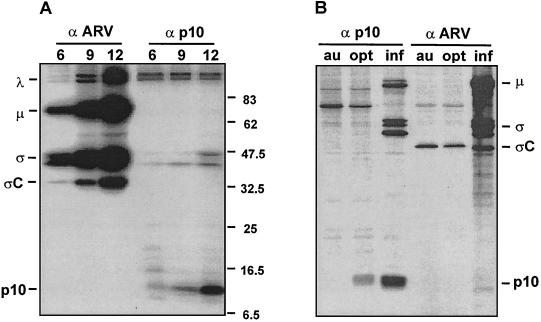
ARV S1 mRNA is functionally tricistronic in vitro and in vivo. (A) The full-length ARV-176 S1 cDNA clone was used for the production of capped mRNA by in vitro transcription. The transcripts were translated in rabbit reticulocyte lysates, and the [3H]leucine-labeled translation mixtures were fractionated by SDS-PAGE (15% acrylamide) either without immunoprecipitation (−) or after precipitation with the monospecific anti-p10 (10) or anti-p17 (17) serum, with antiserum raised against total virus structural proteins to detect ςC (T), or with control preimmune serum (C). Radiolabeled polypeptides were detected by fluorography. The locations of the ςC, p17, and p10 gene products are indicated on the left. (B) Uninfected (uninf., lanes 1 to 4) or ARV-infected (inf., lanes 5 to 13) QM5 cells were pulse-labeled with [3H]leucine for 1 h at 16 h postinfection. Cell lysates were fractionated by SDS-PAGE (15% acrylamide) either without immunoprecipitation (−) or after precipitation with the same antisera described for panel A. The locations of molecular size markers are indicated on the left (in kilodaltons), and the locations of the p10, p17, and major λ, μ, and ς virus structural proteins are indicated on the right.
The tricistronic nature of the ARV S1 genome segment was confirmed in vivo. Antisera raised against virus structural proteins detected the major λ-, μ-, and ς-size class proteins of ARV in the lysates obtained from virus-infected cells, but did not detect polypeptides corresponding to p10 or p17 (Fig. 4B). We have shown previously that p10 is a nonstructural protein and therefore is not recognized by antisera raised against purified virus particles (48). The p10 protein was clearly detected by p10-specific antiserum along with a background of virus structural proteins that were also nonspecifically precipitated by the preimmune rabbit serum. A faint band corresponding to p17 was detected using the p17-specific antiserum; this polypeptide band migrated slightly more slowly than p10 and was not present when uninfected cell lysates were immunoprecipitated with the same antiserum. Prolonged exposure of fluorograms (Fig. 4B, lanes 10 to 13) clearly revealed the presence of p17; the polypeptide band was not present in samples immunoprecipitated with either the p10 antiserum or control preimmune rabbit serum, confirming the identity of the polypeptide as p17. Therefore, the S1 genome segments of ARV, and by inference of NBV, are functionally tricistronic.
Sequence analysis of the p17 gene product.
In contrast to the ARV and NBV S1 genome segments, which are tricistronic, the S1 genome segment of MRV is bicistronic and encodes a 120-amino-acid ς1NS protein in addition to the ς1 cell attachment protein. The ς1NS protein is a basic, nonstructural protein of the virus; it is nonessential for virus replication in cell culture, and it contributes to a virus-induced block in cell cycle progression at the G2/M phase (40, 43). Several similarities exist between p17 and ς1NS that might suggest a shared evolutionary past. Examination of the immunoprecipitation results indicates that an antiserum raised against ARV structural proteins failed to precipitate p17 (Fig. 4, lane 6), suggesting that p17, like ς1NS, may also be a nonstructural protein. This supposition is supported by several previous studies that failed to reveal a 17-kDa protein as a constituent of ARV particles (33, 36, 45). In addition, both ς1NS and p17 are basic proteins with isoelectric points of 8.3 to 8.9 for the ARV and NBV p17 proteins and 9.8 to 11.9 for the ς1NS proteins of the different serotypes of MRV. Furthermore, secondary-structure predictions for p17 and ς1NS predict that both are globular proteins consisting almost exclusively of predicted β-sheet and -turn structures, with little helix propensity (data not shown). Therefore, p17 and ς1NS are both small, basic, globular, and apparently nonstructural proteins of the virus.
Additional sequence analysis, however, failed to provide more compelling data in support of a homologous relationship between p17 and ς1NS. Alignments of the two proteins detected only 16% amino acid identity (the ARV and NBV p17 proteins possess 30% sequence identity), and there was no obvious clustering of similar amino acid residues into regions of higher conservation (Fig. 5). Although this level of sequence identity is approximately the same as would occur between two unrelated proteins aligned using the same procedure and may therefore not be significant, it should be noted that extensive sequence divergence exists between the homologous S-class genome segment gene products of MRV and those of ARV or NBV, with amino acid identities that range from 18 to 31% (9).
Analysis of the hydropathy profiles of the two proteins was also ambiguous (data not shown). This analysis was confounded by the dissimilar natures of the hydropathy profiles of the ARV and NBV p17 proteins, indicating that these clearly homologous proteins tolerate a fair degree of variability not only in their primary structure, but also in their distribution of similar amino acids. This variability in amino acid content between the ARV and NBV p17 proteins is compounded when comparing the less conserved ς1NS and p17 proteins, making it difficult to infer a clear homologous relationship between p17 and ς1NS based on the distribution of similar amino acids.
Both proteins were also examined for the presence of conserved structural or functional motifs. A potential transmembrane region was detected near the amino terminus of the ARV p17 proteins (residues 14 to 35) using the HMMTOP algorithm. This prediction was not, however, confirmed for ARV p17 using either the TMpred or PSORT algorithm. Moreover, a transmembrane domain was not detected in the NBV p17 protein, which is predominantly hydrophilic in this region, or in the hydrophobic amino terminus of ς1NS with any of the algorithms. The significance of the potential transmembrane domain in ARV p17 is unclear, but it does not appear to be conserved in either the NBV p17 protein or in ς1NS. The limited sequence similarity, absence of conserved functional motifs, and dissimilar hydropathy profiles failed to provide a clear indication of an evolutionary relationship between the ς1NS and p17 proteins. Therefore, in spite of some similar biochemical and biophysical properties, a conclusive determination of the homologous nature of the two proteins requires further functional characterization.
Translation analysis of the p10 and ςC ORFs in virus-infected cells.
The unusual sequential overlapping gene arrangement of the tricistronic S1 genome segments and the need to express at least one integral membrane protein, p10, and a soluble cytoplasmic protein, ςC, suggested that translation of the S1 genome segment ORFs might be temporally regulated. We therefore examined the time course of p10 and ςC expression in virus-infected cells. As shown in Fig. 6A, the major species of the ARV λ-, μ-, and ς-class proteins all displayed similar translation kinetics (gels were overexposed to reveal the early low-level translation of p10 and ςC). The appearance and duration of p10 and ςC translation followed an identical time course of expression as the major virus structural and nonstructural proteins, indicating no apparent temporal regulation of translation of the polycistronic S1 mRNA. Consequently, mechanisms explaining the translational regulation of the S1 mRNA would need to account for both the tricistronic nature of the mRNA and the need to simultaneously localize polysomes to two distinct subcellular compartments.
FIG. 6.
Translation analysis of the ARV S1 genome segment. (A) ARV-infected cells were pulse-labeled for 1 h using [35S]methionine at 6, 9, or 12 h postinfection. Infected cell lysates were immunoprecipitated using polyclonal antiserum specific for either ARV structural proteins, including ςC (α ARV) or the nonstructural p10 protein (α p10). Precipitates were fractionated by SDS-PAGE (15% acrylamide), and the labeled products were detected by autoradiography. The locations of molecular mass standards are indicated on the right (in kilodaltons). The locations of the major species of λ-, μ-, and ς-class ARV proteins are indicated on the left, along with the locations of the ςC and p10 gene products encoded by the polycistronic S1 genome segment. (B) QM5 cell monolayers were either infected with ARV-176 (inf) or transfected with plasmids expressing the S1 genome segment of ARV-176 containing either an authentic (au) or optimized (opt) p10 translation start site. Monolayers were labeled with [35S]methionine at 16 h postinfection or 36 h posttransfection, and the radiolabeled cell lysates were immunoprecipitated as described for panel A. Precipitates were fractionated by SDS-PAGE (15% acrylamide), and the labeled products were detected by autoradiography. The locations of the major species of μ- and ς-class ARV proteins are indicated on the right, along with the locations of the ςC and p10 gene products. Higher-molecular-weight species in lane 3 represent nonspecific trapping of radiolabeled virus particles by the precipitation reaction.
The tricistronic arrangement of ORFs on the ARV and NBV S1 genome segments indicated that translation initiation complexes would need to bypass two upstream functional ORFs and four methionine codons (see Fig. 3) to initiate translation at the 3′-proximal ςC ORF. As a first step toward defining the translation initiation mechanisms that might regulate ςC expression, we examined the consequences of increased translation of the p10 ORF on translation of the ςC ORF. The weak context of the ARV p10 start site with pyrimidines in both the −3 and +4 positions (28) was converted to an optimal initiation site (CCACCATGG) by site-directed mutagenesis. The translation levels of the p10 and ςC ORFs present in the authentic and optimized S1 constructs were then analyzed in transfected cells under quantitative conditions (see Materials and Methods).
Optimizing the p10 start site led to a substantial increase in p10 expression and in p10-induced syncytium formation, as reported previously (48). Interestingly, no concomitant decrease in ςC expression accompanied the increased translation of the p10 ORF (Fig. 6B). Virus-infected cells with increased levels of p10 and ςC expression, due to the more efficient infection of cells relative to transfection efficiencies, were used to ensure that the antibody concentrations used for immunoprecipitation of transfected cells were saturating and that precipitation of p10 and ςC was quantitative (Fig. 6B, lanes 3 and 6). In repeated experiments, the levels of p10 expression from the optimized construct ranged from 12- to 20-fold greater than the levels obtained from the authentic construct. In spite of this significant increase in p10 expression, ςC translation levels remained constant (±30% relative to the levels observed with the authentic construct). These results imply that ribosomes can bypass the upstream p10 ORF in order to access the distal ςC translation start site and suggest that ςC expression from the tricistronic S1 mRNA does not derive from a leaky scanning mechanism (28).
DISCUSSION
We recently demonstrated that the 5′-proximal ORFs of ARV and NBV encode homologous p10 proteins that are responsible for the unusual syncytium-inducing properties of these viruses (48). Our present analysis indicates that the 3′-proximal ORF of the NBV S1 genome segment encodes a homolog of the reovirus cell attachment protein. In spite of its mammalian origin, the NBV cell attachment protein is more closely related to the ςC cell attachment protein of ARV than to the homologous ς1 protein of the nonfusogenic mammalian reoviruses (Fig. 2). This observation is consistent with the closer evolutionary relationship previously noted in the other S-class gene products of ARV and NBV (9).
Interestingly, both the ARV and NBV ςC proteins are approximately one third smaller than the homologous ς1 protein of MRV (approximately 325 versus 450 residues). The conserved coiled-coil motifs in the ςC and ς1 proteins extend for approximately 150 residues (Fig. 2). Therefore, the majority of the sequences that are present in the ς1 protein of MRV but lacking in the ςC proteins reside in the carboxy-terminal half of the protein. This region of ς1 corresponds to two morphological regions, termed the neck and head (37), that contribute to the hemagglutinating and receptor-binding properties of the ς1 protein (4, 5, 56). The extensive deletions in the head and neck regions of the ςC proteins and the extremely low level of sequence conservation make it difficult to generate a meaningful alignment of the ςC proteins with ς1 in order to identify the conserved β-sheet structure and the Asn198, Arg202, and Pro 204 residues implicated in MRV serotype 3 ς1 sialic acid binding (4, 5). It seems likely, however, that the deletions in the head and neck of the ςC proteins would be expected to profoundly alter the structure of these regions and contribute to the lack of hemagglutinating activity displayed by the ςC proteins of the fusogenic reoviruses (18, 54).
Sequence analysis of NBV and of two strains of ARV corrected what appear to be sequencing or cloning errors that contributed to the premature termination of the two 5′-proximal predicted ORFs in the ARV-S1133 S1 cDNA sequence (46). This supposition was confirmed by examination of a second ARV-S1133 S1 sequence recently deposited in GenBank (accession number AF330703) that contains the G residue at position 249 and the C residue at position 329, which extend the p10 and p17 reading frames to comply with the gene arrangement predicted for ARV-176, ARV-138, and NBV. Our functional analysis indicates that the S1 genome segments of ARV and NBV encode a previously unidentified p17 gene product from the second of three functional ORFs. Whether the p17 protein is specific to the fusogenic reoviruses, similar to the situation with the p10 fusion proteins encoded by the first ORF, or is functionally homologous to the ς1NS protein of MRV is unclear. Additional sequence analysis of other fusogenic reoviruses such as baboon reovirus or the reptilian reoviruses (2, 13, 53) may help to clarify this point.
The sequential, partially overlapping, tricistronic arrangement of ORFs present in the ARV and NBV S1 genome segments is highly unusual for a polycistronic eukaryotic mRNA. The majority of mRNAs that encode more than one unique gene product are usually bicistronic and contain ORFs that are either nested within each other, as is the case for the bicistronic genome segments of MRV and the phytoreoviruses (14, 50), or overlap each other extensively. In the latter, partially overlapping arrangement, the start codons are generally located in relatively close proximity to each other, as exemplified by the polycistronic P/C mRNA of Sendai virus (29). There are, however, some exceptions to these general statements, such as the bicistronic E7/E1 mRNA of human papillomavirus, the bicistronic M2 mRNA of respiratory syncytial virus, and the bicistronic core/polymerase pregenomic mRNA of hepatitis B virus, all of which are similar to the situation reported here for the S1 genome segment but contain a sequential bicistronic arrangement of ORFs with minimal overlap (1, 39, 42).
The tricistronic sequential arrangement of ORFs in the S1 genome segment adds an additional level of novelty that has few parallels. One of the few examples of a similar tricistronic arrangement occurs in mRNA3 of infectious bronchitis virus, in which three sequential ORFs with minimal overlap encode the unique 3a, 3b, and 3c gene products. This unusual tricistronic sequential gene arrangement influences translation control mechanisms, as indicated by in vitro analysis suggesting that the downstream 3c ORF is translated via an internal ribosome entry mechanism (30, 32).
The sequential tricistronic gene arrangement on the ARV and NBV S1 genome segments raises a number of interesting issues regarding the mechanisms that regulate translation initiation on these polycistronic eukaryotic mRNAs. The absence of the viral RNA polymerase in in vitro translation mixtures and in transfected cells primed with full-length S1 mRNA indicates that RNA editing by the viral RNA polymerase is not required to generate a different mRNA for the expression of ςC, as occurs, for example, in the Sendai virus P/C mRNA (20, 52). It is also seems unlikely that leaky scanning alone could account for the expression of the majority of ςC polypeptides, since preinitiation complexes would need to scan over 600 nucleotides, two functional ORFs, and a total of four methionine codons in different reading frames, at least one of which naturally exists in a preferred context with purines at positions −3 and +4 (see Fig. 3), before initiating at the ςC start site. This situation applies to both the ARV and NBV S1 genome segments.
Our initial analysis of S1 translation confirmed the improbability of ςC translation initiation via leaky scanning, since optimizing the context of the p10 initiation site resulted in an approximately 15-fold increase in the expression of p10 with no deleterious effect on ςC expression (Fig. 6B). These results also suggest that ribosome termination, backscanning, and reinitiation is an unlikely means for initiation complexes to access the internal ςC translation start site, since increased production of upstream gene products would be expected to alter ςC expression (34, 44).
Two alternative possibilities could explain ςC translation, one cap independent and the other cap dependent. Internal ribosome entry sites (IRES) present in a number of viral and cellular mRNAs function in a cap-independent manner to mediate access of the translation machinery to internal start codons (21, 24, 25, 41). Since IRES elements consist of an extensive series of stem-loop structures, they are generally contained within relatively long 5′ noncoding regions (19, 22). The short 5′ noncoding region present on the S1 genome segments of ARV and NBV, in conjunction with the sequential overlapping gene arrangement, would require that an IRES element be located almost entirely within one of the functional ORFs preceding the ςC start site. Secondary-structure predictions of the 5′-terminal 600 nucleotides of the ARV and NBV S1 mRNAs, using either the entire 600 nucleotide sequence or a floating window of 250 nucleotides failed to reveal a conserved predicted secondary structure (data not shown), suggesting that a typical IRES element is not present upstream of the ςC start site.
Conversely, ribosomal shunting functions in a cap-dependent fashion to modulate the translation of downstream ORFs via limited 5′-terminal scanning before transfer of the preinitiation complex to the downstream start site, thereby bypassing continuous scanning of the upstream regions (16, 29, 42, 55). As in the case of IRES-mediated translation of ςC, cis-acting signals that might function as shunt donor and acceptor sites would need to be located within the coding regions for p10 and/or p17. Determining the cap dependency of ςC translation and substitution-deletion analysis of the S1 mRNA should offer insights into the means by which ribosomes access the internal start site of ςC.
A final unusual feature relating to translation of the tricistronic S1 genome segments of ARV and NBV involves the need to generate at least one nonstructural transmembrane protein (p10) in addition to the soluble ςC protein from a single species of mRNA. The ARV p10 protein assumes a type I transmembrane topology (48). Evidence suggests that a central transmembrane domain serves as an internal signal-anchor sequence for the cotranslational, signal recognition particle-dependent insertion of p10 into the membrane of the endoplasmic reticulum. Conversely, the ςC protein is an essential structural protein of the virus which needs to be localized to the cytoplasmic sites of virus assembly and therefore is presumably translated on cytoplasmic rather than on membrane-bound polysomes. Our temporal analysis of p10 and ςC expression (Fig. 6A) indicates that these two distinct populations of polysomal S1 mRNA exist concurrently in virus-infected cells.
It is not clear whether the choice of translation initiation site would lead to dedicated use of that polysome population for expression of either p10 or ςC, or whether a single S1 mRNA shuttles between the membrane-bound and cytosolic ribosome populations following each consecutive round of translation initiation, as occurs with togaviruses (35). As we now know, expression of the third p17 ORF also needs to be considered and adds yet another level of complexity to the mechanisms regulating translation initiation on this unusual tricistronic viral mRNA.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council (NSERC) of Canada to R.D. M.S. and S.D. were funded by the Killam Foundation and by NSERC Studentships. D.O. was supported by an NSERC Studentship. Z.Y. was supported by a Sir John and Lady Higgins Research Scholarship at the University of Melbourne.
The authors acknowledge Deborah Kool for making available the sequences of the ARV-Som and ARV-Ram S1 genes, which were generated as part of her Ph.D. studies at the University of Melbourne.
REFERENCES
- 1.Ahmadian, G., J. S. Randhawa, and A. J. Easton. 2000. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 19:2681–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahne, W., I. Thomsen, and J. Winton. 1987. Isolation of a reovirus from the snake Python regius. Arch. Virol. 94:135–139. [DOI] [PubMed] [Google Scholar]
- 3.Antin, P. B., and C. P. Ordahl. 1991. Isolation and characterization of an avian myogenic cell line. Dev. Biol. 143:111–121. [DOI] [PubMed] [Google Scholar]
- 4.Chappell, J. D., V. L. Gunn, J. D. Wetzel, G. S. Baer, and T. S. Dermody. 1997. Mutations in type 3 reovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein sigma1. J. Virol. 71:1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chappell, J. D., J. L. Duong, B. W. Wright, and T. S. Dermody. 2000. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 74:8472–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doohan, J. P., and C. E. Samuel. 1992. Biosynthesis of reovirus-specified polypeptides: ribosome pausing during the translation of reovirus S1 mRNA. Virology 186:409–425. [DOI] [PubMed] [Google Scholar]
- 8.Duncan, R. 1996. The low pH-dependent entry of avian reovirus is accompanied by two specific cleavages of the major outer capsid protein μ2C. Virology 219:179–189. [DOI] [PubMed] [Google Scholar]
- 9.Duncan, R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260:316–328. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, R., and K. Sullivan. 1998. Characterization of two avian reoviruses that exhibit strain-specific quantitative differences in their syncytium-inducing and pathogenic capabilities. Virology 250:263–272. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, R., E. Nagy, P. J. Krell, and P. Dobos. 1987. Synthesis of the infectious pancreatic necrosis virus polyprotein, detection of a virus-encoded protease and the fine structure mapping of the genome segment A coding regions. J. Virol. 61:3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan, R., D. Home, L. W. Cashdollar, W. L. Joklik, and P. W. K. Lee. 1990. Identification of conserved domains in the cell attachment proteins of the three serotypes of reovirus. Virology 174:399–409. [DOI] [PubMed] [Google Scholar]
- 13.Duncan, R., F. A. Murphy, and R. Mirkovic. 1995. Characterization of a novel syncytium-inducing baboon reovirus. Virology 212:752–756. [DOI] [PubMed] [Google Scholar]
- 14.Ernst, H., and A. J. Shatkin. 1985. Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc. Natl. Acad. Sci. USA 82:48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajardo, J. E., and A. J. Shatkin. 1990. Translation of bicistronic viral mRNA in transfected cells: regulation at the level of elongation. Proc. Natl. Acad. Sci. USA 87:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Futterer, J., Z. Kiss-Laszlo, and T. Hohn. 1993. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73:789–802. [DOI] [PubMed] [Google Scholar]
- 17.Gard, G., and R. W. Compans. 1970. Structure and cytopathic effects of Nelson Bay virus. J. Virol. 6:100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glass, S. E., S. A. Naqi, C. F. Hall, and K. M. Kerr. 1973. Isolation and characterization of a virus associated with arthritis of chickens. Avian Dis. 17:415–424. [PubMed] [Google Scholar]
- 19.Gray, N. K., and M. Wickens. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399–458. [DOI] [PubMed] [Google Scholar]
- 20.Hausmann, S., D. Garcin, C. Delenda, and D. Kolakofsky. 1999. The versatility of paramyxovirus RNA polymerase stuttering. J. Virol. 73:5568–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huez, I., L. Creancier, S. Audigier, M. C. Gensac, A. C. Prats, and H. Prats. 1998. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 18:6178–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs, B. L., and C. E. Samuel. 1985. The reovirus S1 mRNA encodes two primary translation products. Virology 143:63–74. [DOI] [PubMed] [Google Scholar]
- 24.Jendrach, M., V. Thiel, and S. Siddell. 1999. Characterization of an internal ribosome entry site within mRNA 5 of murine hepatitis virus. Arch. Virol. 144:921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4:1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamura, H., F. Shiinizu, M. Maeda, and H. Tsubahara. 1965. Avian reovirus: its properties and serological classification. Natl. Inst. Anim. Health Q. 5:115–124. [PubMed] [Google Scholar]
- 27.Kool, D. A. 1993. Relation of gene sequence to serotype in avian rotaviruses and reoviruses. Ph.D. thesis. University of Melbourne, Melbourne, Australia.
- 28.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latorre, P., D. Kolakofsky, and J. Curran. 1998. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 18:5021–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le, S.-Y., N. Sonenberg, and J. V. Maizel, Jr. 1994. Distinct structural elements and internal entry of ribosomes in mRNA3 encoded by infectious bronchitis virus. Virology 198:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leone, G., R. Duncan, D. C. W. Mah, A. Price, L. W. Cashdollar, and P. W. K. Lee. 1991. The N-terminal heptad repeat region of reovirus cell attachment protein ς1 is responsible for ς1 oligomer stability and possesses intrinsic oligomerization function. Virology 182:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, D. X., and S. C. Inglis. 1992. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J. Virol. 66:6143–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Costas, J., A. Grande, R. Varela, C. Garcia-Martinez, and J. Benavente. 1997. Protein architecture of avian reovirus S1133 and identification of the cell attachment protein. J. Virol 71:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy, J. E. G. 1998. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 62:1492–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melancon, P., and H. Garoff. 1987. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J. Virol. 61:1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni, Y., R. F. Ramig, and M. C. Kemp. 1993. Identification of proteins encoded by avian reoviruses and evidence for posttranslational modification. Virology 193:466–469. [DOI] [PubMed] [Google Scholar]
- 37.Nibert, M. L., T. S. Dermody, and B. N. Fields. 1990. Structure of the reovirus cell-attachment protein: a model for the domain organization of ςl. J. Virol. 64:2976–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1996. Reoviruses and their replication, p.1557–1623. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 39.Ou, J. H., H. Bao, C. Shih, and S. M. Tahara. 1990. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J. Virol. 64:4578–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poggioli, G. J., C. Keefer, J. L. Connolly, T. S. Dermody, and K. L. Tyler. 2000. Reovirus-induced G2/M cell cycle arrest requires sigma1s and occurs in the absence of apoptosis. J. Virol. 74:9562–9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole, T. L., C. Wang, R. A. Popp, N. D. Potgeiter, A. Siddiqui, and M. S. Collett. 1995. Pestivirus translation initiation occurs by internal ribosome entry. Virology 206:750–754. [DOI] [PubMed] [Google Scholar]
- 42.Remm, M., A. Remm, and M. Ustav. 1999. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 73:3062–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodgers, S. E., J. L. Connolly, J. D. Chappell, and T. S. Dermody. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a ς1s-null mutant. J. Virol. 72:8597–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samuel, C. E. 1989. Polycistronic animal virus mRNAs. Prog. Nucleic Acids Res. 37:127–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnitzer, T. J., T. Ramos, and V. Gouvea. 1982. Avian reovirus polypeptides: analysis of intracellular virus-specified products, virions, top component and cores. J. Virol. 43:1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapouri, M. R. S., M. Kane, M. Letarte, J. Bergeron, M. Arella, and A. Silim. 1995. Cloning, sequencing and expression of the S1 gene of avian reovirus. J. Gen. Virol. 76:1515–1520. [DOI] [PubMed] [Google Scholar]
- 47.Shapouri, M. R. S., M. Arella, M., and A. Silim. 1996. Evidence for the multimeric nature and cell binding ability of avian reovirus sigma 3 protein. J. Gen. Virol 77:1203–1210. [DOI] [PubMed] [Google Scholar]
- 48.Shmulevitz, M., and R. Duncan. 2000. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the nonenveloped fusogenic reoviruses. EMBO J. 19:902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strong, J. E., G. Leone, R. Duncan, R. K. Sharma, and P. W. K. Lee. 1991. Biochemical and biophysical characterization of the reovirus cell attachment protein ς1: evidence that it is a homotrimer. Virology 184:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki, N., M. Sugawara, D. L. Nuss, and Y. Matsuura. 1996. Polycistronic (tri- or bicistronic) phytoreoviral segments translatable in both plant and insect cells. J. Virol. 70:8155–8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varela, R., and J. Benavente. 1994. Protein coding assignment of avian reovirus strain S1133. J. Virol. 68:6775–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vidal, S., J. Curran, and D. Kolakofsky. 1990. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 64:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vieler, E., W. Baumgartner, W. Herbst, and G. Kohler. 1994. Characterization of a reovirus isolate from a rattlesnake, Crotalus viridis, with neurological dysfunction. Arch. Virol. 138:341–344. [DOI] [PubMed] [Google Scholar]
- 54.Wilcox, G. E., and R. W. Compans. 1983. Characterization of Nelson Bay virus and virus-induced cell fusion, p.391–403. In R. W. Compans and D. H. L. Bishop (ed.), Double-stranded RNA viruses. Elsevier Science Publishing Co., Inc., Amsterdam, The Netherlands.
- 55.Yeuh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557–1567. [DOI] [PubMed] [Google Scholar]
- 56.Yeung, M. C., D. Lim, Duncan, R., M. S. Shahrabadi, and P. W. K. Lee. 1989. The cell attachment proteins of type 1 and type 3 reovirus are differentially susceptible to trypsin and chymotrypsin. Virology 170:62–70. [DOI] [PubMed] [Google Scholar]