Abstract
Infection of animals with a molecular viral clone is critical to study the genetic determinants of viral replication and virulence in the host. Type 2 porcine circovirus (PCV2) has been incriminated as the cause of postweaning multisystemic wasting syndrome (PMWS), an emerging disease in pigs. We report here for the first time the construction and use of an infectious molecular DNA clone of PCV2 to characterize the disease and pathologic lesions associated with PCV2 infection by direct in vivo transfection of pigs with the molecular clone. The PCV2 molecular clone was generated by ligating two copies of the complete PCV2 genome in tandem into the pBluescript SK (pSK) vector and was shown to be infectious in vitro when transfected into PK-15 cells. Forty specific-pathogen-free pigs at 4 weeks of age were randomly assigned to four groups of 10 each. Group 1 pigs served as uninoculated controls. Pigs in group 2 were each inoculated intranasally with about 1.9 × 105 50% tissue culture infective doses of a homogeneous PCV2 live virus stock derived from the molecular clone. Pigs in group 3 were each injected intrahepatically with 200 μg of the cloned PCV2 plasmid DNA, and pigs in group 4 were each injected into the superficial iliac lymph nodes with 200 μg of the cloned PCV2 plasmid DNA. Animals injected with the cloned PCV2 plasmid DNA developed infection resembling that induced by intranasal inoculation with PCV2 live virus stock. Seroconversion to PCV2-specific antibody was detected in the majority of pigs from the three inoculated groups at 35 days postinoculation (DPI). Viremia, beginning at 14 DPI and lasting 2 to 4 weeks, was detected in the majority of the pigs from all three inoculated groups. There were no remarkable clinical signs of PMWS in control or any of the inoculated pigs. Gross lesions in pigs of the three inoculated groups were similar and were characterized by systemically enlarged, tan lymph nodes and lungs that failed to collapse. Histopathological lesions and PCV2-specific antigen were detected in numerous tissues and organs, including brain, lung, heart, kidney, tonsil, lymph nodes, spleen, ileum, and liver of infected pigs. This study more definitively characterizes the clinical course and pathologic lesions exclusively attributable to PCV2 infection. The data from this study indicate that the cloned PCV2 genomic DNA may replace infectious virus for future PCV2 pathogenesis and immunization studies. The data also suggest that PCV2, although essential for development of PMWS, may require other factors or agents to induce the full spectrum of clinical signs and lesions associated with advanced cases of PMWS.
Porcine circovirus (PCV) was originally isolated as a cell culture contaminant of a porcine kidney cell line (PK-15) (56, 60). PCV is a small, nonenveloped virus that contains a single-stranded circular DNA genome of about 1.76 kb. PCV is classified in the family of Circoviridae, which consists of three other animal circoviruses (chicken anemia virus [CAV], psittacine beak and feather disease virus, and the recently discovered columbid circovirus [CoCV] from pigeons) and three plant circoviruses (banana bunchy top virus, coconut foliar decay virus, and subterranean clover stunt virus) (11, 35, 37, 38, 39, 61). Members of the three previously recognized animal circoviruses (PCV, CAV, and psittacine beak and feather disease virus) do not share nucleotide sequence homology or antigenic determinants with each other (11, 61). The genome of the newly identified columbid circovirus shared about 40% nucleotide sequence identity with that of PCV (37). Recently, a novel human virus with a circular genome, designated TT virus (TTV), was identified from individuals associated with posttransfusion hepatitis (40, 45) and a human TTV-like minivirus (TLMV) was also identified from normal blood donors (12, 55). The genomic organization of both human TTV and TLMV is similar to that of the CAV (12, 40, 55). Although antibodies to PCV were found in various animal species, including humans, mice, cattle, and pigs (1, 15, 16, 26, 44, 58, 59), little is known regarding the pathogenesis of PCV in these animal species. Experimental infection of pigs with the PK-15 cell-derived PCV did not produce clinical disease, and, thus, this virus is not considered to be pathogenic to pigs (2, 57). The nonpathogenic PCV derived from the contaminated PK-15 cell line was designated PCV1.
Postweaning multisystemic wasting syndrome (PMWS) is an emerging disease in pigs first described in 1991 (25). PMWS primarily affects pigs between 5 and 18 weeks of age. Clinical PMWS signs include progressive weight loss, dyspnea, tachypnea, anemia, diarrhea, and jaundice. Mortality rate may vary from 1 to 2% up to 30% in complicated cases. Microscopic lesions characteristic of PMWS include granulomatous interstitial pneumonia, lymphadenopathy, hepatitis, and nephritis (9, 25). PMWS has now been recognized in pigs in Canada, the United States (3, 5, 9, 17, 24, 30, 33, 39, 41), many European countries (5, 9, 16, 28, 36, 48, 53, 62), and some countries in Asia (13, 46) and potentially has serious economic impact on the swine industry worldwide.
The causative agent of PMWS is believed to be a pathogenic strain of PCV designated PCV2 (3, 5, 7, 9, 17, 24, 39, 41). The complete genomic sequence of the PMWS-associated PCV2 has been determined (20, 24, 35, 38, 39, 41). Sequence analyses revealed that the PMWS-associated PCV2 shared only about 75% nucleotide sequence identity with the nonpathogenic PCV1. Experimental reproduction of clinical PMWS in gnotobiotic pigs and conventional pigs with tissue homogenates from pigs with naturally occurring PMWS and with cell culture-propagated PCV2 produced mixed results. Clinical PMWS was reproduced in gnotobiotic pigs and colostrum-deprived and caesarean-derived pigs coinfected with PCV2 and porcine parvovirus (PPV) (88, 32) and in PCV2-inoculated gnotobiotic pigs when their immune system was activated by keyhole limpet hemocyanin in incomplete Freund’s adjuvant (31). However, clinical PMWS was not reproduced in gnotobiotic pigs infected with PCV2 alone (4, 6, 8, 10, 18, 29, 31, 32, 47). The virus inocula used in these studies were either homogenates of tissues from pigs with naturally occurring PMWS or virus propagated in PK-15 cell cultures (4, 6, 8, 10, 18, 29, 31, 32, 47). Since tissue homogenates may contain other common swine agents, such as PPV and porcine reproductive and respiratory syndrome virus (PRRSV) (4, 8, 9, 19, 48), and since the ATCC PK-15 cell line used for PCV2 propagation was persistently infected with PCV1 (15), the clinical disease and pathologic lesions reproduced in those studies may not be solely attributable to PCV2 infection (4, 6, 8, 9, 19). Therefore, it will be advantageous to construct an infectious clone of PCV2 so that a biologically pure and homogeneous infectious virus stock can be generated for pathogenesis studies.
We report here for the first time that a molecular DNA clone of PCV2 is infectious when injected directly into the liver and lymph nodes of pigs. The course of clinical disease, virus distribution, and pathologic lesions associated with PCV2 infection was more definitively characterized by using this molecular DNA clone and a biologically pure and homogeneous infectious PCV2 virus stock derived from the molecular DNA clone.
MATERIALS AND METHODS
Source of PCV2.
The PCV2 isolate used in this study was from a spleen tissue sample of a pig with naturally occurring PMWS (PCV2 no. 40895) (20). Immunohistochemical staining (IHC) with PCV2 specific antibody confirmed the presence of PCV2 antigen in the tissue (data not shown). The spleen tissues were stored at −80°C until use.
Generation of a PK-15 cell line free of PCV1 contamination by end point dilution.
The PK-15 cell line purchased from the American Type Culture Collection was persistently infected with PCV1 (15). Since only a subpopulation of PK-15 cells was persistently infected (15), we therefore attempted to generate a PK-15 cell line that was free of PCV1 contamination by end point dilution. PK-15 cells were grown in minimum essential medium (MEM) with Earle’s salts and l-glutamine (Life Technologies, Inc., Grand Island, N.Y.) supplemented with 10% fetal bovine serum and 1× antibiotic (Life Technologies, Inc.). Confluent cell monolayers were trypsinized, and the cells were then counted and serially diluted to an end point with one cell per 0.2 ml. The end point dilution was plated in 96-well plates and allowed to grow into a monolayer starting from a single cell. Cells from each well were tested for PCV1 DNA using a PCR-restriction fragment length polymorphism (RFLP) assay capable of detecting and differentiating PCV1 and PCV2 (20). PK-15 cells from wells that tested negative for PCV1 by the PCR-RFLP assay were subsequently expanded. The PCV1-free PK-15 cell line used in this study was subcultured five additional passages and was found negative for PCV1 DNA by PCR at each passage.
Construction of PCV2 molecular DNA clone.
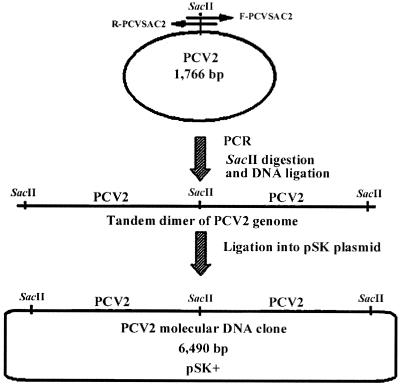
A pair of PCR primers was designed according to the published sequence of the PCV2 isolate 40895 (20): forward primer F-PCVSAC2 (5′-GAACCGCGGGCTGGCTGAACTTTTGAAAGT-3′) and reverse primer R-PCVSAC2 (5′-GCACCGCGGAAATTTCTGACAAACGTTACA-3′). This pair of primers amplifies the complete genome of PCV2 with an overlapping region containing the unique SacII restriction enzyme site (Fig. 1). Briefly, DNA was extracted using the QIAamp DNA Minikit (Qiagen, Inc., Valencia, Calif.) from a spleen tissue sample of a pig with naturally occurring PMWS (isolate 40895) (20). The extracted DNA was amplified by PCR with AmpliTaq Gold polymerase (Perkin-Elmer, Norwalk, Conn.). The PCR consisted of an initial enzyme activation step at 95°C for 9 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 48°C for 1 min, extension at 72°C for 3 min, and a final extension at 72°C for 7 min. The PCR product of expected size was separated by gel electrophoresis and purified with the glass milk procedure with a Geneclean Kit (Bio 101, Inc., La Jolla, Calif.).
FIG. 1.
Construction of an infectious PCV2 molecular DNA clone. The relative positions of the primer pair used to amplify the complete PCV2 genome are indicated by the arrows (reverse primer, R-PCVSAC2; forward primer, F-PCVSAC2). The PCV2 genomic DNA amplified by PCR is digested with the SacII restriction enzyme and purified. The purified and SacII-digested genomic DNA was ligated to form concatemers. Ligated concatemers were separated by gel electrophoresis; the tandem genome dimmer of PCV2 was purified and cloned into the pSK vector, which is predigested with the SacII enzyme to produce a molecular PCV2 DNA clone.
To construct a molecular DNA clone containing a tandem dimmer of the PCV2 genome, the PCR product containing the complete PCV2 genome was first ligated into the advanTAge plasmid vector (Clontech, Palo Alto, Calif.). Escherichia coli DH5α competent cells were transformed. The recombinant plasmids were verified by restriction enzyme digestion. The full-length PCV2 genomic DNA was excised from the advanTAge vector by digestion with the SacII restriction enzyme. The digested PCV2 genomic DNA was ligated with T4 DNA ligase at 37°C for only 10 min, which favors the production of tandem dimmers. The tandem dimmers were subsequently cloned into the pBluescript SK (pSK) vector (Stratagene, La Jolla, Calif.) (Fig. 1). Recombinant plasmids containing tandem dimmers of the PCV2 genome (referred to as the PCV2 molecular DNA clone) were confirmed by PCR, restriction enzyme digestion, and DNA sequencing. The DNA concentration of the recombinant plasmids was determined spectrophotometrically.
In vitro transfection with PCV2 molecular DNA clone and generation of a biologically pure and homogeneous PCV2 infectious virus stock.
To test the infectivity of the molecular DNA clone in vitro, PK-15 cells free of PCV1 contamination were grown in eight-well LabTek chamber slides. When the PK-15 cells reached about 85% confluency, cells were transfected with the molecular DNA clone using Lipofectamine Plus Reagents according to the protocol supplied by the manufacturer (Life Technologies, Inc). Mock-transfected cells with empty pSK vector were included as controls. Three days after transfection, the cells were fixed with a solution containing 80% acetone and 20% methanol at 4°C for 20 min and an immunofluorescence assay (IFA) using a PCV2-specific rabbit polyclonal antiserum was performed to determine the in vitro infectivity of the molecular DNA clone (see below).
To generate a biologically pure and homogeneous PCV2 infectious virus stock for the animal inoculation experiment, PK-15 cells free of PCV1 contamination were cultivated in T-25 culture flasks and transfected with the PCV2 molecular DNA clone. Briefly, PK-15 cells were grown to about 85% confluency in T-25 flasks. The cells were washed once with sterile phosphate-buffered saline (PBS) buffer before transfection. For each transfection reaction in a T-25 flask, 12 μg of the PCV2 plasmid DNA was mixed with 16 μl of Plus Reagent in 0.35 ml of MEM. A flask of mock-transfected cells with empty pSK vector was included as the negative control. After incubation at room temperature for 15 min, 50 μl of Lipofectamine Reagent diluted in 0.35 ml of MEM was added to the mixture and was incubated at room temperature for another 15 min. The transfection mixture was then added to a T-25 flask of PK-15 cells containing 2.5 ml of fresh MEM. After incubation at 37°C for 3 h, the media were replaced with fresh MEM containing 2% fetal bovine serum and 1× antibiotics. The transfected cells were harvested at 3 days posttransfection and stored at −80°C until use. The infectious titer of the virus stock was determined by IFA (see below).
Virus titration by IFA.
To determine the infectious titer of the homogeneous PCV2 virus stock, PK-15 cells were cultivated on eight-well LabTek chamber slides. The virus stock was serially diluted 10-fold in MEM, and each dilution was inoculated onto 10 wells of the monolayers of the PK-15 cells growing on the LabTek chamber slides. Wells of noninoculated cells were included as controls. The infected cells were fixed at 3 days postinoculation with a solution containing 80% acetone and 20% methanol at 4°C for 20 min. After washing of the cells with PBS buffer, the infected cells were incubated with a 1:1,000-diluted PCV2 specific rabbit polyclonal antibody (50) at 37°C for 1 h. The cells were then washed three times with PBS buffer and were incubated with a secondary fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.) at 37°C for 45 min. After the slides were washed three times with PBS buffer, they were mounted with fluoromount-G, coverslipped, and examined under a fluorescence microscope. The 50% tissue culture infective dose (TCID50) per ml was calculated. Initially, we transfected cells with a plasmid construct containing a single copy of PCV2 genome, but the infectious PCV2 titer from the single genome construct was much lower than the one containing the tandem genome (data not shown). Therefore, the plasmid construct containing the dimeric form of PCV2 genome was used for the in vitro and in vivo transfection experiments in this study.
In vivo transfection of pigs with PCV2 molecular DNA clone and experimental inoculation of pigs with homogeneous PCV2 infectious virus stock.
Forty specific-pathogen-free (SPF) swine of 4 weeks of age were randomly assigned into four rooms of 10 animals each. Prior to inoculation, the SPF pigs were tested for antibodies to PCV, PRRSV, PPV, and swine hepatitis E virus. Pigs in group 1 were uninoculated and served as negative controls. Pigs in group 2 were each inoculated intranasally with about 1.9 × 105 TCID50 of the PCV2 infectious virus stock derived from the PCV2 molecular DNA clone. Pigs in group 3 received direct intrahepatic injection of the recombinant plasmid DNA of the PCV2 molecular clone. Each pig was injected with a total of 200 μg of recombinant plasmid DNA, through an ultrasound-guided technique, into six different sites of the liver. Pigs in group 4 were each injected with a total of 200 μg of the recombinant PCV2 plasmid DNA into the superficial iliac lymph nodes, and each lymph node received two separate injections. The animals were monitored daily for clinical signs of disease. Serum samples were collected from each animal at 0, 7, 14, 21, 28, and 35 days postinoculation (DPI). At 21 DPI, five pigs were randomly selected from each group and necropsied. The remaining five animals in each group were necropsied at 35 DPI. Various tissues and organs were collected during necropsy and processed for histlogical examination and immunohistochemical staining (see below).
Clinical evaluation.
Pigs were weighed on 0 DPI and at the time of necropsy. Rectal temperatures and clinical respiratory disease scores, ranging from 0 to 6 (0 = normal; 6 = severe) (23), were recorded every other day from 0 to 35 DPI. Clinical observations, including evidence of central nervous system disease, liver disease (icterus), musculoskeletal disease, and changes in body condition, were also recorded daily. Clinical evaluation was performed by a team of two people.
Gross pathology and histopathology.
Five pigs from each group were randomly selected for necropsies at 21 and 35 DPI. The necropsy team was blinded to infection status of the pigs at necropsy. Complete necropsies were performed on all pigs. An estimated percentage of the lung with grossly visible pneumonia was recorded for each pig based on a previously described scoring system (23). The scoring system is based on the approximate volume that each lung lobe contributes to the entire lung: the right cranial lobe, right middle lobe, cranial part of the left cranial lobe, and the caudal part of the left cranial lobe contribute 10% each of the total lung volume, the accessory lobe contributes 5%, and the right and left caudal lobes contribute 27.5% each. Other lesions, such as enlargement of lymph nodes, were noted separately. Sections for histopathological examination were taken from nasal turbinate, lungs (seven sections) (23), heart, brain, lymph nodes (tracheobronchial, iliac, mesenteric, and subinguinal), tonsil, thymus, liver, gall bladder, spleen, joints, small intestine, colon, pancreas, and kidney. The tissues were examined in a blinded fashion and given a subjective score for severity of lung, lymph node, and liver lesions. Lung scores ranged from 0 (normal) to 3 (severe lymphohistiocytic interstitial pneumonia). Liver scores ranged from 0 (normal) to 3 (severe lymphohistiocytic hepatitis). Lymph node scores were for an estimated amount of lymphoid depletion of follicles ranging from 0 (normal or no lymphoid depletion) to 3 (severe lymphoid depletion and histiocytic replacement of follicles).
Serology.
Blood was collected on arrival at 11 to 12 days of age and from all pigs at 0, 7, 14, 21, 28, and 35 DPI. Serum antibodies to PRRSV were assayed using Herd Check PRRSV enzyme-linked immunosorbent assay (IDEXX Laboratories, Westbrook, Mass.). Serum antibodies to PPV were detected by a hemagglutination inhibition (HI) assay (27). Serum antibodies to PCV2 were detected by a modified indirect enzyme-linked immunosorbent assay based on the recombinant ORF2 protein of PCV2 (42). Briefly, a partially purified PCV2 antigen was prepared from Hi Five cells (Invitrogen, Carlsbad, Calif.) infected with recombinant baculovirus containing the major capsid ORF2 protein of PCV2 (43). Cell lysates of Hi Five cells infected with wild-type baculovirus were prepared similarly and served as negative control antigen. The Immulon 2 HB polystyrene microtiter plates (Dynex Technologies Inc, Chantilly, Va.) were coated with optimal concentrations of positive and negative antigens at 4°C for 36 h. One hundred microliters of each serum sample diluted 1:100 in 5% milk diluent (Kirkegaard & Perry Laboratories, Inc.) was added into each well. The serum samples were tested in quadruplicate: two wells for negative control antigen and two parallel wells for PCV2 antigen. Positive control and negative control sera were included in each plate. The sera were incubated at 37°C for 30 min and then washed five times with 0.1 M PBS buffer containing 0.1% Tween 20. A peroxidase-labeled secondary anti-swine immunoglobulin G (Sigma Chemical Co., St. Louis, Mo.) was incubated at 37°C for 30 min. The plates were washed again and incubated with 2,2′-azino-di-(3-ethylbenzthiazoline-6-sulfonate) (Kirkegaard & Perry Laboratories, Inc.) at 37°C for 15 min for color development. The optical density (OD) was read at 405 nm. The corrected OD of each tested and control serum was calculated by subtraction of mean OD of the wells containing negative antigen from that of the parallel wells containing PCV2 antigen. The data were normalized by dividing the corrected OD of a tested serum sample by that of the positive control serum and were reported as ratios. The samples with serum sample/positive control serum ratios of ≤0.12, 0.12 to 0.2, and >0.2 were considered negative, equivocal, and positive, respectively.
PCR-RFLP analyses.
To measure PCV2 viremia in pigs transfected with PCV2 molecular DNA clone and in pigs infected with PCV2 infectious virus stock, serum samples collected on different DPI were tested for the presence of PCV2 DNA by a previously described PCR-RFLP assay (20). Viral DNA was extracted from 50 μl of each serum sample using the DNAzol reagent according to the protocol supplied by the manufacturer (Molecular Research Center, Cincinnati, Ohio). The extracted DNA was resuspended in DNase-, RNase-, and proteinase-free water and tested for PCV2 DNA by PCR-RFLP (20). PCR products from selected animals were sequenced to verify the origin of the virus infecting pigs.
IHC.
IHC detection of PCV2-specific antigen was performed on all tissues collected during necropsies on DPI 21 and 35. A rabbit polyclonal PCV2-specific antiserum was used for the IHC. The procedures have been previously described (50).
RESULTS
Generation of PK-15 cell line free of PCV1 contamination.
Four cell lines that were negative for PCV1 contamination were produced by end point dilution of the persistently infected PK-15 cells from the American Type Culture Collection. The cell lines remained negative for PCV1 by PCR after five additional passages. One of the cell lines was subsequently expanded and was shown to be able to support PCV2 replication when the cells were transfected with the PCV2 molecular DNA clone (Fig. 2) and were infected with PCV2 virus (data not shown). The cloned cells were further used for the in vitro transfection of the PCV2 molecular DNA clone to generate a biologically pure PCV2 infectious virus stock for the animal inoculation experiment.
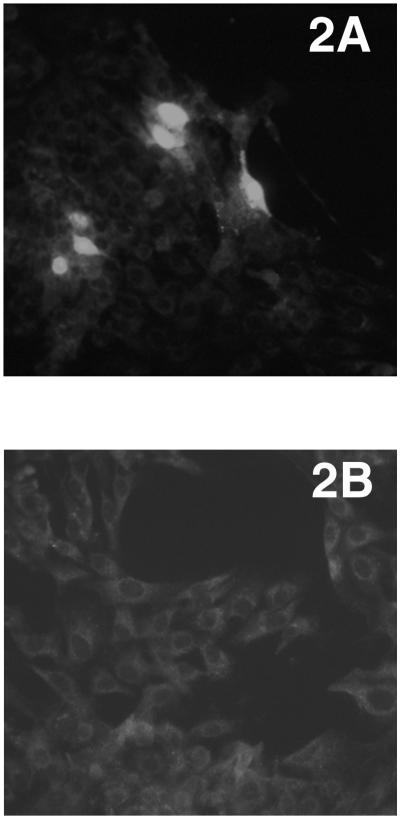
FIG. 2.
The cloned PCV2 plasmid DNA is infectious when transfected in vitro in PK-15 cells. (A) Detection of PCV2 antigen by IFA in PK-15 cells transfected with the cloned PCV2 plasmid DNA. Intense immunolabeling of PCV2 antigen was visualized in the nucleus and to a lesser degree in the cytoplasm of the transfected cells. (B) Mock-transfected PK-15 cells.
Construction of infectious PCV2 molecular DNA clone.
The complete genome of the PCV2 (isolate 40895) was amplified by PCR. Two copies of the complete PCV2 genome were ligated in tandem into the pSK vector to produce the PCV2 molecular DNA clone (Fig. 1). The infectivity of the PCV2 molecular DNA clone was determined by in vitro transfection of the PK-15 cells. IFA with PCV2 specific antibody confirmed that the molecular DNA clone was infectious in vitro and that about 10 to 15% of the PK-15 cells were transfected. PCV2 specific antigen was visualized by IFA in the nucleus and to a lesser degree in the cytoplasm of the transfected cells (Fig. 2). The cells mock transfected with the empty pSK vector remained negative for PCV2 antigen.
Generation of biologically pure and homogeneous PCV2 infectious virus stock.
The lack of a biologically pure form of PCV2 infectious virus stock has impeded the understanding of PCV2 pathogenesis and the etiological role of PCV2 in PMWS. In this study, we generated a biologically pure PCV2 infectious virus stock by transfection of PK-15 cells with the PCV2 molecular DNA clone. PCV2 virions produced by in vitro transfection were infectious, as the transfected cell lysates were successfully used to infect PK-15 cells. Thus, the PCV2 molecular DNA clone is capable of producing infectious PCV2 virions when transfected in vitro. The infectious titer of the homogeneous PCV2 virus stock prepared from transfected cells was determined to be 104.5 TCID50/ml, and this virus stock was used to inoculate pigs in group 2. Lysates of cells mock transfected with the empty pSK vector were unable to infect PK-15 cells.
PCV2 molecular DNA clone is infectious when injected directly into liver and superficial iliac lymph nodes of SPF pigs.
Serum samples were collected from all control and inoculated animals at 0, 7, 14, 21, 28, and 35 DPI and assayed for PCV2 viremia by detection of PCV2 DNA (20). PCV2 DNA was not detected in the group 1 uninoculated control pigs at any DPI (Table 1). Viremia was detected in 7 of 10 pigs from group 2 at 14 DPI, and 8 of 10 pigs had viremia by 35 DPI (Table 1). Viremia lasted only a few weeks, as the PCV2 DNA was not detectable at 28 and 35 DPI in all five remaining pigs from group 2. Of the pigs from group 3 that were intrahepatically injected with PCV2 molecular DNA clone, 8 of 10 were viremic at 14 DPI and 9 of 10 had had detectable viremia by 35 DPI (Table 1). Group 4 pigs were injected with PCV2 molecular DNA clone into the lymph nodes. Two of 10 pigs at 14 DPI and 8 of 10 pigs at 21 DPI from group 4 were viremic (Table 1). PCR products amplified from selected animals were sequenced. The sequence of the PCR products amplified from selected animals was identical to the corresponding region of the PCV2 molecular DNA clone (data not shown).
TABLE 1.
Detection of viremia (PCV2 DNA) by PCR in sera of inoculated and control pigs
| Group | Inoculum | Route of inoculation | DPI
|
Total no. positive/no. tested | |||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 28 | 35 | ||||
| 1 | None | 0/10a | 0/10 | 0/10 | 0/10 | 0/5 | 0/5 | 0/10 | |
| 2 | PCV2 live virusb | Intranasal | 0/10 | 0/10 | 7/10 | 5/10 | 0/5 | 0/5 | 8/10 |
| 3 | PCV2 DNAc | Intrahepatic | 0/10 | 0/10 | 8/10 | 6/10 | 3/5 | 3/5 | 9/10 |
| 4 | PCV2 DNAc | Intralymphoid | 0/10 | 0/10 | 2/10 | 8/10 | 2/5 | 0/5 | 8/10 |
Ten pigs in each group; number positive/number tested.
A biologically pure and homogeneous PCV2 virus stock generated by transfection of PK-15 cells with the PCV2 molecular DNA clone.
Cloned PCV2 genomic DNA in pSK plasmid.
All inoculated pigs from groups 2, 3, and 4 were negative for PCV2 antibodies at 0 DPI. Two pigs in the uninoculated control group 1 had detectable PCV2 maternal antibody at 0 DPI. The maternal antibody in these two piglets waned by 7 DPI (Table 2). No seroconversion to PCV2 antibody was detected in any of the 10 uninoculated control pigs. Of the group 2 pigs intranasally inoculated with PCV2 infectious virus, one piglet seroconverted to PCV2 antibody at 21 DPI. By 35 DPI, four of the five remaining group 2 pigs had seroconverted (Table 2). Seroconversion in transfected animals from groups 3 and 4 first appeared at 28 DPI. By 35 DPI, five of five remaining pigs from group 3 and three of five remaining pigs from group 4 had seroconverted to the PCV2 antibody (Table 2).
TABLE 2.
Seroconversion to PCV2 specific antibodies in pigs inoculated with PCV2 live virus or directly injected with cloned PCV2 plasmid DNA
| Group | Inoculum | Route of inoculation | DPI
|
|||||
|---|---|---|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 28 | 35 | |||
| 1 | None | 2/10a | 0/10 | 0/10 | 0/10 | 0/5 | 0/5 | |
| 2 | PCV2 live virusb | Intranasal | 0/10 | 0/10 | 0/10 | 1/10 | 1/5 | 4/5 |
| 3 | PCV2 DNAc | Intrahepatic | 0/10 | 0/10 | 0/10 | 0/10 | 1/5 | 5/5 |
| 4 | PCV2 DNAc | Intralymphoid | 0/10 | 0/10 | 0/10 | 0/10 | 1/5 | 3/5 |
PCV2 antibody was measured with an enzyme-linked immunosorbent assay; number positive/number tested.
A biologically pure and homogeneous PCV2 virus stock generated by transfection of PK-15 cells with PCV2 molecular DNA clone.
Cloned PCV2 genomic DNA in pSK plasmid.
PPV antibodies were tested at 3 and 21 DPI for all pigs and at 35 DPI for the remaining pigs. As expected, maternal antibodies to the ubiquitous swine agent PPV were detected in the SPF piglets. The PPV HI antibody titers in all piglets but one decreased significantly from 3 DPI (an average titer of 1:2,665) to 21 DPI (an average titer of 1:246), indicating that the antibody detected in these piglets was passively derived. One piglet had a slightly increased PPV HI titer from 1:32 at 3 DPI to 1:64 at 21 DPI, which was likely due to testing variation. Serum samples collected from all pigs at 0, 21, and 35 DPI were further tested for PPV DNA with a published PCR assay (51). No PPV viremia was detected from any pigs at DPI 7, 21, or 35, further indicating that the pigs were not infected by PPV.
Clinical evaluation.
None of the control and inoculated pigs showed obvious signs of disease resembling those of clinical PMWS. There was no difference in weight gain or mean rectal temperatures between any of the four groups (data not shown). The group 1 control pigs remained normal throughout the study. There was mild transient respiratory disease observed in the majority of the pigs in PCV2 DNA-transfected and PCV2 virus-infected groups from 8 to 14 DPI. This was characterized by mild dyspnea (clinical respiratory scores of 1 to 2) of 1 to 2 days’ duration in individual pigs and 5 to 6 days’ duration for the group.
Gross lesions.
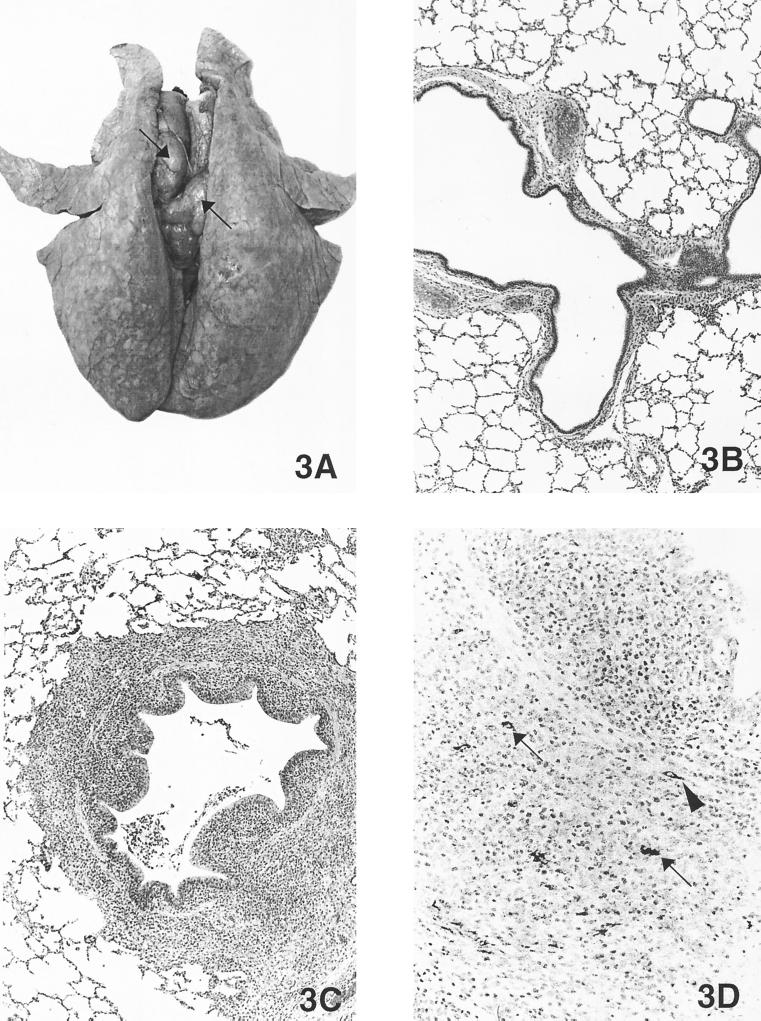
There were no gross lesions observed in the control pigs at necropsy. Pigs in the three inoculated groups had gross lesions limited to the lungs and lymph nodes (Table 3). The lesions were similar among pigs in the PCV2 plasmid DNA-transfected and PCV2 virus-infected groups. Lungs failed to collapse and had random, multifocal, moderately well demarcated areas of tan-to-purple consolidation involving 0 to 2% of the lung (Fig. 3) at 21 DPI and 0 to 13% of the lung at 35 DPI. Lymph nodes were systemically enlarged two to five times the normal size and were firm and tan (Fig. 3) at both 21 and 35 DPI in most of the pigs from all three PCV2-inoculated groups.
TABLE 3.
Gross lesions of lung and lymph nodes in control and PCV2-inoculated pigs
| Group | Inoculum | Route of inoculation | Results at:
|
|||
|---|---|---|---|---|---|---|
| 21 DPI in:
|
35 DPI in:
|
|||||
| Lymph nodes | Lung | Lymph nodes | Lung | |||
| 1 | None | 0/5a | 0/5 | 0/5 | 0/5 | |
| 2 | PCV2 live virusb | Intranasal | 5/5 | 1/5 (0–1)c | 5/5 | 4/5 (0–5) |
| 3 | PCV2 DNAd | Intrahepatic | 2/5 | 2/5 (0–2) | 5/5 | 2/5 (0–13) |
| 4 | PCV2 DNAd | Intralymphoid | 4/5 | 5/5 (0–1) | 3/5 | 1/5 (0–9) |
Five pigs from each group were necropsied at 21 DPI, and the remaining five pigs were necropsied at 35 DPI. Number positive/number tested.
A biologically pure and homogeneous PCV2 virus stock generated by transfection of PK-15 cells with the PCV2 molecular DNA clone.
Number with lesions/number tested (range of estimated percentage of the lung affected by grossly visible pneumonia lesions, 0 to 100%).
Cloned PCV2 genomic DNA in pSK plasmid.
FIG. 3.
(A) Lung from a pig inoculated by the intralymphoid route with PCV2 DNA and necropsied at 21 DPI. The lungs were rubbery, failed to collapse, and were mottled tan-red. Tracheobronchial lymph nodes were markedly enlarged and were tan (arrows). (B) Microscopic section of a normal lung from a control pig (magnification, ×21.25). (C) Microscopic section of the pig lung shown in Fig. 3A. Note the peribronchiolar lymphohistiocytic inflammation and mild necrotizing bronchiolitis (magnification, ×21.25). (D) Immunohistochemical staining of the lung shown in Fig. 3A. Note the PCV2 antigen in macrophages (arrows) and fibroblast-like cells (arrowhead) around airways (magnification, ×54.4).
Microscopic lesions.
Microscopic examination revealed no lesions in any tissues of the control pigs except for the livers. Eight of 10 control pigs had very mild multifocal lymphoplasmacytic inflammation predominately in the periportal regions of the liver, as is commonly observed in normal pigs and considered normal background (22).
Pigs from the two PCV2 plasmid DNA-transfected groups (intrahepatic and intralymphoid) and the PCV2 virus-infected group (intranasal) had similar lesions in brain, lung, heart, kidney, lymphoid tissues (tonsil, lymph nodes, and spleen), ileum, and liver (Table 4). Brain lesions were observed in 23 of 30 of the pigs from the three inoculated groups and were characterized as mild-to-moderate multifocal lymphoplasmacytic meningoencephalitis with perivascular cuffing and gliosis. Lung lesions were observed in 28 of 30 PCV2-inoculated pigs and were characterized as mild-to-moderate peribronchiolar lymphoplasmacytic and histiocytic bronchointerstitial pneumonia (Fig. 3C). One pig from PCV2 virus-infected group 2 necropsied at 21 DPI, and one pig each from the two PCV2 plasmid DNA-transfected groups necropsied at 35 DPI had ulcerative and proliferative bronchiolitis with fibroplasia and granulomatous inflammation in the lamina propria and peribronchiolar regions of bronchi. Mild multifocal lymphoplasmacytic myocarditis was also observed in 18 of 30 PCV2-inoculated pigs. Mild-to-moderate multifocal lymphoplasmacytic interstitial nephritis was observed in 14 of 30 of the PCV2-inoculated pigs. No lesions were observed in the thymuses. Mild-to-moderate lymphoid depletion (Fig. 4B) and histiocytic replacement of follicles were observed in the tonsil of 8 of 30, in the spleen of 7 of 30, and in the lymph nodes of 26 of 30 of the PCV2-inoculated pigs. Moderate granulomatous lymphadenitis with giant cells (Fig. 4C) was observed at 21 DPI in three pigs inoculated intranasally with PCV2 virus and in one pig at 35 DPI in each of the PCV2 plasmid DNA-transfected groups. Mild lymphoplasmacytic and histiocytic enterocolitis was observed in three of five pigs in the PCV2 virus-infected group, in three of five pigs in the intrahepatically PCV2 plasmid DNA-transfected group, and one of five pigs in the intralymphoidally PCV2 plasmid DNA-transfected group at 35 DPI. One pig in each of the PCV2 plasmid DNA-transfected groups had mild lymphoid depletion with histiocytic replacement and low numbers of giant cells in the Peyer’s patches. Mild-to-moderate lymphoplasmacytic hepatitis was observed in 29 of 30 of the PCV2-inoculated pigs. Low numbers of widely scattered individually necrotic hepatocytes surrounded by lymphohistiocytic inflammation were observed in one pig in each of the PCV2 plasmid DNA-transfected groups at 21 DPI. Lesions in other tissues were unremarkable.
TABLE 4.
Distribution of histopathological lesions in control and PCV2-inoculated pigs
| Group | Inoculum | Route of inoculation | DPIa | Results for:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lungb | Liverc | Lymph nodesd | Spleen | Thymus | Ileum | Brain | Heart | Kidney | Tonsil | ||||
| 1 | None | 21 | 0/5 (0.0) | 4/5 (0.8) | 0/5 (0.0) | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |
| 35 | 0/5 (0.0) | 4/5 (0.8) | 0/5 (0.0) | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | |||
| 2 | PCV2 virus | Intranasal | 21 | 5/5 (1.6) | 5/5 (1.2) | 3/5 (1.2) | 1/5 | 0/5 | 0/5 | 4/5 | 3/5 | 1/5 | 0/5 |
| 35 | 3/5 (0.6) | 4/5 (1.0) | 4/5 (0.8) | 3/5 | 0/5 | 3/5 | 4/5 | 0/5 | 1/5 | 3/5 | |||
| 3 | PCV2 DNA | Intrahepatic | 21 | 5/5 (1.0) | 5/5 (1.0) | 5/5 (1.0) | 1/5 | 0/5 | 0/5 | 5/5 | 4/5 | 1/5 | 0/5 |
| 35 | 5/5 (1.2) | 5/5 (1.0) | 4/5 (1.0) | 2/5 | 0/5 | 3/5 | 3/5 | 4/5 | 5/5 | 3/5 | |||
| 4 | PCV2 DNA | Intralymphoid | 21 | 5/5 (1.2) | 5/5 (1.0) | 5/5 (0.8) | 0/5 | 0/5 | 0/5 | 4/5 | 4/5 | 3/5 | 0/5 |
| 35 | 5/5 (1.0) | 5/5 (1.2) | 5/5 (1.4) | 0/5 | 0/5 | 1/5 | 3/5 | 3/5 | 3/5 | 2/5 | |||
Five animals from each group were necropsied at 21 DPI, and the remaining five animals from each group were necropsied at 35 DPI.
Number positive/number tested (average histological lung score: 0, normal; 1, mild interstitial pneumonia; 2, moderate; 3, severe).
Average histological liver score: 0, normal; 1, mild hepatitis; 2, moderate; 3, severe.
Average histological lymphoid (lymph nodes) depletion score: 0, normal; 1, mild; 2, moderate; 3, severe.
FIG. 4.
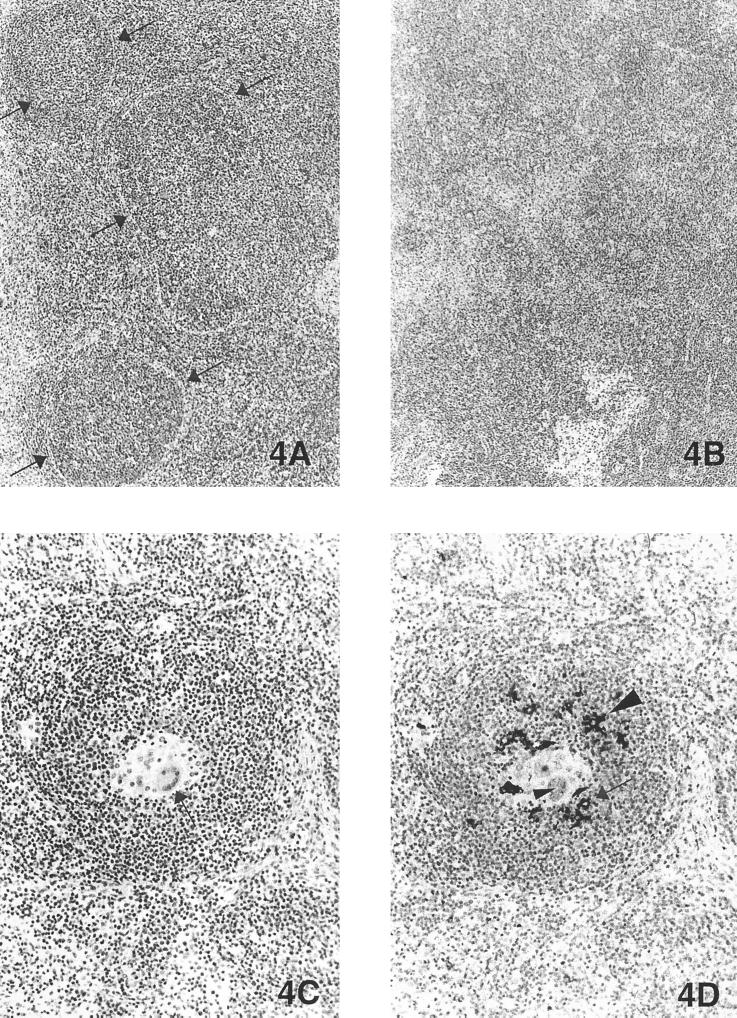
(A) Normal lymph node from a control pig. Note the well-defined lymphoid follicles (arrows) (magnification, ×21.25). (B) Microscopic section of the tracheobronchial lymph node, from the pig whose lung is shown in Fig. 3A, inoculated 21 days previously by the intralymphoid route with cloned PCV2 genomic DNA. Lymphoid follicles are poorly defined; there is mild-to-moderate lymphoid depletion and mild, multifocal, granulomatous inflammation (magnification, ×21.25). (C) Same lymph node as shown in Fig. 4B. Note the poorly defined follicle with macrophages and giant cells (arrow) replacing follicular lymphocytes (magnification, ×54.4). (D) Same lymph node as shown in Fig. 4B. Immunohistochemical detection of PCV2 antigen in macrophages (arrow) and giant cells (small arrowhead) and dendrite-like cells (large arrowhead) in the follicles (magnification, ×54.4).
Microscopic lesions in the lung, liver, and lymph nodes were scored according to published scoring systems (Table 4) (22–23). There were no acceptable scoring systems for other tissues and organs. The average scores of lesions in lung and lymph nodes in pigs of the three PCV2-inoculated groups were statistically significantly different from those in group 1 control pigs. The average scores of the liver lesions in pigs of the three PCV2-inoculated groups were not statistically significantly different from those of control pigs.
Detection and tissue distribution of PCV2 antigen.
IHC staining of PCV2 antigen was done on brain, lungs, turbinate, heart, kidneys, tonsil, lymph nodes, spleen, thymus, ileum, liver, gall bladder, and pancreas of all pigs necropsied at 21 and 35 DPI. All tissues from the control pigs were negative for PCV2 antigen. Tissue distribution of PCV2 antigen in the three PCV2-inoculated groups was similar (Table 5). In the brain, PCV2 antigen was found predominately in mononuclear cells, fibroblast-like cells, and endothelial cells in the meninges and choroid plexus and less often in endothelial cells and perivascular mononuclear cells in the cerebrum and cerebellum. In the lungs, PCV2 antigen was detected within alveolar and septal macrophages and in fibroblast-like cells in the lamina propria of airways (Fig. 3D). In the heart, PCV2 antigen was detected in widely scattered macrophages and endothelial cells. In kidneys, PCV2 antigen was detected within tubular epithelial cells and mononuclear cells in the interstitium. In the lymphoid tissues (lymph nodes, spleen, tonsil, and Peyer’s patches), PCV2 antigen was detected primarily within macrophages and dendrite-like cells and giant cells within follicles (Fig. 4D). PCV2 antigen was also detected within macrophages in the lamina propria of the small intestine. In the liver, PCV2 antigen was detected within mononuclear cells and Kupffer cells. PCV2 antigen was not detected in turbinate, thymus, or gall bladder.
TABLE 5.
Detection and distribution of PCV2-specific antigen by immunohistochemistry in control and PCV2-inoculated pigs
| Group | Inoculum | Route of inoculation | DPIa | Results for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lung | Liver | Lymph nodes | Spleen | Thymus | Ileum | Brain | Heart | Kidney | Tonsil | |||||
| 1 | None | 21 | 0/5b | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | ||
| 35 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | ||||
| 2 | PCV2 virus | Intranasal | 21 | 4/5 | 5/5 | 5/5 | 3/5 | 0/5 | 3/5 | 3/5 | 1/5 | 1/5 | 2/5 | |
| 35 | 1/5 | 2/5 | 3/5 | 2/5 | 0/5 | 0/5 | 2/5 | 0/5 | 0/5 | 0/5 | ||||
| 3 | PCV2 DNA | Intrahepatic | 21 | 5/5 | 5/5 | 5/5 | 5/5 | 0/5 | 0/5 | 5/5 | 1/5 | 0/5 | 2/5 | |
| 35 | 4/5 | 4/5 | 3/5 | 4/5 | 0/5 | 3/5 | 4/5 | 2/5 | 2/5 | 3/5 | ||||
| 4 | PCV2 DNA | Intralymphoid | 21 | 4/5 | 4/5 | 5/5 | 4/5 | 0/5 | 3/5 | 3/5 | 0/5 | 0/5 | 3/5 | |
| 35 | 3/5 | 4/5 | 5/5 | 4/5 | 0/5 | 2/5 | 3/5 | 1/5 | 0/5 | 4/5 | ||||
Five animals from each group were necropsied at 21 DPI, and the remaining five animals from each group were necropsied at 35 DPI.
Number positive/number tested.
DISCUSSION
PMWS is a complex disease syndrome in swine, and multiple factors may be involved in the clinical presentation of PMWS. Increasing data indicate that PCV2 is the causative agent of PMWS (3, 5, 7, 9, 17, 24, 39, 41). However, the difficulty in producing a biologically pure form of PCV2 due to the presence of other common swine agents in the tissue homogenates of diseased pigs has impeded a definitive characterization of the clinical disease and pathologic lesions solely attributable to PCV2 infection.
We report here for the first time that the cloned PCV2 genomic DNA is infectious when directly injected into the livers and superficial iliac lymph nodes of SPF pigs. Animals directly injected with the cloned PCV2 plasmid DNA developed an infection and disease resembling those induced by infection via the intranasal route of inoculation with a homogeneous PCV2 infectious virus stock. It is known that PCV2 replicates in the lymph nodes, lungs, and liver during natural infection (9, 25, 29, 31, 62). It will be interesting to know if other routes of injection of the cloned PCV2 plasmid DNA, such as the intramuscular route, can also initiate an infection. Through the use of this PCV2 molecular DNA clone, the clinical disease, pathologic lesions, and virus distribution exclusively attributable to PCV2 infection were more definitively characterized. Viremia, beginning at 14 DPI and lasting about 2 to 4 weeks, was detected in the majority of the PCV2-inoculated animals. Similarly, the majority of inoculated pigs necropsied at 35 DPI seroconverted to PCV2 antibodies. PCV2 antigen was detected in various tissues and organs in inoculated pigs. Gross lesions were limited to the lungs and lymph nodes and were characterized by systemically enlarged, tan lymph nodes and lungs that failed to collapse and mild, multifocal, tan foci of consolidation. Histopathological lesions in multiple tissues and organs similar to those of PMWS were reproduced with the PCV2 molecular DNA clone as well as with the infectious virus prepared in vitro from the molecular DNA clone. However, we failed to reproduce characteristic clinical PMWS with the cloned PCV2 plasmid DNA or with a biologically pure PCV2 infectious virus stock. PCV2 is clearly responsible for the PMWS-like histopathological lesions reproduced in this study, but whether or not PCV2 is the sole cause of clinical PMWS remains questionable. Clinical PMWS was reproduced only in gnotobiotic pigs coinfected with PCV2 and PPV (8) and in PCV2-inoculated gnotobiotic pigs when their immune system was activated by keyhole limpet hemocyanin in incomplete Freund’s adjuvant (31). Clearly, more studies are needed to determine the etiological significance of PCV2 in clinical PMWS and its interrelationship with PRRSV, PPV, other infectious swine agents, and immune stimulants.
Will et al. (63) first demonstrated the feasibility of using cloned hepatitis B virus DNA to infect chimpanzees by direct in vivo injection. This approach has since been used to study viral replication and pathogenesis of several other viruses (14, 21, 34, 49, 52, 54, 63, 64). The construction of an infectious PCV2 molecular DNA clone and the demonstration of infection by direct injection of cloned PCV2 plasmid DNA into the liver and lymph nodes of pigs in this study should be very advantageous for future PCV2 studies. This in vivo transfection system will enable us to study the structural and functional relationship of PCV2 genes using recombinant plasmids constructed in vitro to test different regions or genes of PCV2 for their roles in virus replication and pathogenesis in the host. The replication and pathogenesis of PCV2 can be studied in vivo without having to produce infectious virus stocks by propagating PCV2 in cell cultures. This is advantageous, as serial cell culture passages may select for viral variants (1). Another advantage of using cloned PCV2 genomic DNA, instead of live virus, for animal studies is its relative ease for quantitation of the inoculation dose. The amount of the cloned PCV2 DNA used for animal inoculation can be easily determined by a spectrophotometer, whereas the dose of live PCV2 virus requires infectivity titration in cell cultures and confirmation of infection by IFA. Direct injection of animals with cloned PCV2 plasmid DNA eliminates the problems associated with the presence of other indigenous swine agents in tissue homogenate inocula in animal studies. From the vaccine development point of view, the relatively easy storage and stability of DNA and the economy of large-scale recombinant PCV2 plasmid DNA production should provide an attractive means of delivering genetically engineered, attenuated PCV2 vaccines to pigs. However, the intrahepatic and intralymphoid routes of inoculation used in this study are not practical for vaccine delivery; thus, future studies are warranted to determine if pigs can be infected by the intramuscular route of injection with the PCV2 DNA clone. The intramuscular and intradermal routes have been successfully used to infect animals with cloned genomic DNA of other viruses (34, 52, 64).
Acknowledgments
This study was supported by grants from Fort Dodge Animal Health Inc. (to X.J.M.) and from the Iowa Livestock Health Advisory Council (to P.G.H.).
We thank Prem Paul for his support and use of lab facilities, Stephen Boyle for his helpful discussion and support, and Elizabeth Riedesel for her expert help in the ultrasound-guided technique for direct injection of PCV2 DNA into the liver and lymph nodes of pigs. We also thank Jim Fosse and Megan O’Dea for photography.
REFERENCES
- 1.Allan, G. M., D. P. Mackie, J. McNair, B. M. Adair, and M. S. McNulty. 1994. Production, preliminary characterization and applications of monoclonal antibodies to porcine circovirus. Vet. Immunol. Immunopathol. 43:357–371. [DOI] [PubMed] [Google Scholar]
- 2.Allan, G. M., F. McNeilly, J. P. Cassidy, G. A. Reilly, B. Adair, W. A. Ellis, and M. S. McNulty. 1995. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig fetal material. Vet. Microbiol. 44:49–64. [DOI] [PubMed] [Google Scholar]
- 3.Allan, G. M., B. Meehan, D. Todd, S. Kennedy, F. McNeilly, J. Ellis, E. G. Clark, J. Harding, E. Espuna, A. Botner, and C. Charreyre. 1998. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet. Rec. 142:467–468. [PubMed] [Google Scholar]
- 4.Allan, G. M., F. McNeilly, J. Ellis, S. Krakowka, B. Meeham, I. McNair, I. Walker, and S. Kennedy. 2000. Experimental infection of colostrums deprived piglets with porcine circovirus 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch. Virol. 145:2421–2429. [DOI] [PubMed] [Google Scholar]
- 5.Allan, G. M., F. McNeilly, S. Kennedy, B. Daft, E. G. Clarke, J. A. Ellis, D. M. Haines, B. M. Meehan, and B. M. Adair. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 10:3–10. [DOI] [PubMed] [Google Scholar]
- 6.Allan, G. M., F. McNeilly, B. M. Meehan, J. A. Ellis, T. J. Conner, I. McNair, S. Krakowka, and S. Kennedy. 2000. A sequential study of experimental infection of pigs with porcine circovirus and porcine parvovirus: immunostaining of cryostat sections and virus isolation. J. Vet. Med. B47:81–94. [DOI] [PubMed] [Google Scholar]
- 7.Allan, G. M., F. McNeilly, B. M. Meehan, S. Kennedy, D. P. Mackie, J. A. Ellis, E. G. Clark, E. Espuna, N. Saubi, P. Riera, A. Botner, and C. E. Charreyre. 1999. Isolation and characterization of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet. Microbiol. 66:115–123. [DOI] [PubMed] [Google Scholar]
- 8.Allan, G. M., S. Kennedy, F. McNeilly, J. C. Foster, J. A. Ellis, S. J. Krakowka, B. M. Meehan, and B. M. Adair. 1999. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 121:1–11. [DOI] [PubMed] [Google Scholar]
- 9.Allan, G. M., and J. A. Ellis. 2000. Porcine circoviruses: a review. J. Vet. Diagn. Investig. 12:3–14. [DOI] [PubMed] [Google Scholar]
- 10.Balasch, M., J. Segales, C. Rosell, M. Domingo, A. Mankertz, A. Urniza, and J. Plana-Duran. 1999. Experimental inoculation of conventional pigs with tissue homogenates from pigs with post-weaning multisystemic wasting syndrome. J. Comp. Pathol. 121:139–148. [DOI] [PubMed] [Google Scholar]
- 11.Bassami, M. R., D. Berryman, G. E. Wilcox, and S. R. Raidal. 1998. Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anemia virus. Virology 249:453–459. [DOI] [PubMed] [Google Scholar]
- 12.Biagini, P., P. Gallian, H. Attoui, M. Touinssi, J.-F. Cantaloube, P. de Micco, and X. de Lamballerie. 2001. Genetic analysis of full-length genomes and subgenomic sequences of TT virus-like mini virus human isolates. J. Gen. Virol. 82:379–383. [DOI] [PubMed] [Google Scholar]
- 13.Choi, C., C. Chae, and E. G. Clark. 2000. Porcine postweaning multisystemic wasting syndrome in Korean pig: detection of porcine circovirus 2 infection by immunohistochemistry and polymerase chain reaction. J. Vet. Diagn. Investig. 12:151–153. [DOI] [PubMed] [Google Scholar]
- 14.Dubensky, T. W., B. A. Campbell, and L. P. Villarreal. 1984. Direct transfection of viral and plasmid DNA into the liver or spleen of mice. Proc. Natl. Acad. Sci. USA 81:7529–7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulac, G. C., and A. Afshar. 1989. Porcine circovirus antigens in PK-15 cell line (ATCC CCL-33) and evidence of antibodies to circovirus in Canadian pigs. Can. J. Vet. Res. 53:431–433. [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards, S., and J. J. Sands. 1994. Evidence of circovirus infection in British pigs. Vet. Rec. 134:680–681. [DOI] [PubMed] [Google Scholar]
- 17.Ellis, J., L. Hassard, E. Clark, J. Harding, G. Allan, P. Willson, J. Strakappe, K. Martin, F. McNeilly, B. Meehan, D. Todd, and D. Haines. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis, J., S. Krakowka, M. Lairmore, D. Haines, A. Bratanich, E. Clark, G. Allan, C. Konoby, L. Hassard, B. Meehan, K. Martin, J. Harding, S. Kennedy, and F. McNeilly. 1999. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J. Vet. Diagn. Investig. 11:3–14. [DOI] [PubMed] [Google Scholar]
- 19.Ellis, J. A., A. Bratanich, E. G. Clark, G. Allan, B. Meehan, D. M. Haines, J. Harding, K. H. West, S. Krakowka, C. Konoby, L. Hassard, K. Martin, and F. McNeilly. 2000. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J. Vet. Diagn. Investig. 12:21–27. [DOI] [PubMed] [Google Scholar]
- 20.Fenaux, M., P. G. Halbur, M. Gill, T. E. Toth, and X. J. Meng. 2000. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J. Clin. Microbiol. 38:2494–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girones, R., P. J. Cote, W. E. Hornbuckle, B. C. Tennant, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1989. Complete nucleotide sequence of a molecular clone of woodchuck hepatitis virus that is infectious in the natural host. Proc. Natl. Acad. Sci. USA 86:1846–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halbur, P. G., C. Kasorndorkbua, C. Gilbert, D. Guenette, M. B. Potters, R. H. Purcell, S. U. Emerson, T. E. Toth, and X. J. Meng. 2001. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 39:918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halbur, P. G., P. S. Paul, M. L. Frey, J. Landgraf, K. Eernisse, X. J. Meng, M. A. Lum, J. J. Andrews, and J. A. Rathje. 1995. Comparison of the pathogenicity of two U. S. porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet. Pathol. 32:648–660. [DOI] [PubMed] [Google Scholar]
- 24.Hamel, A. L., L. L. Lin, and G. P. Nayar. 1998. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72:5262–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding, J. C., and E. G. Clark. 1997. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS). Swine Health Prod. 5:201–203. [Google Scholar]
- 26.Hines, R. K., and P. D. Lukert. 1995. Porcine circovirus: a serological survey of swine in the United States. Swine Health Prod. 3:71–73. [Google Scholar]
- 27.Joo, H. S., C. R. Donaldson-Wood, and R. H. Johnson. 1976. A standardized haemagglutination inhibition test for porcine parvovirus antibody. Aust. Vet. J. 52:422–424. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy, S., G. Allan, F. McNeilly, B. M. Adair, A. Hughes, and P. Spillane. 1998. Porcine circovirus infection in Northern Ireland. Vet. Rec. 142:495–496. [PubMed] [Google Scholar]
- 29.Kennedy, S., D. Moffett, F. McNeilly, B. Meehan, J. Ellis, S. Krakowka, and G. M. Allan. 2000. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J. Comp. Pathol. 122:9–24. [DOI] [PubMed] [Google Scholar]
- 30.Kiupel, M., G. W. Stevenson, S. K. Mittal, E. G. Clark, and D. M. Haines. 1998. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet. Pathol. 35:303–307. [DOI] [PubMed] [Google Scholar]
- 31.Krakowka, S., J. A. Ellis, F. McNeilly, S. Ringler, D. M. Rings, and G. Allan. 2001. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet. Pathol. 38:31–42. [DOI] [PubMed] [Google Scholar]
- 32.Krakowka, S., J. A. Ellis, B. Meehan, S. Kennedy, F. McNeilly, and G. Allan. 2000. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet. Pathol. 37:254–263. [DOI] [PubMed] [Google Scholar]
- 33.Larochelle, R., M. Morin, M. Antaya, and R. Magar. 1999. Identification and incidence of porcine circovirus in routine field cases in Quebec as determined by PCR. Vet. Rec. 145:140–142. [DOI] [PubMed] [Google Scholar]
- 34.Letvin, N. L., C. I. Lord, N. W. King, M. S. Wyand, K. V. Myrick, and W. A. Haseltine. 1991. Risks of handling HIV. Nature 349:573. [DOI] [PubMed] [Google Scholar]
- 35.Mankertz, J., H. J. Buhk, G. Blaess, and A. Mankertz. 1998. Transcription analysis of porcine circovirus (PCV). Virus Genes 16:267–276. [DOI] [PubMed] [Google Scholar]
- 36.Mankertz, A., M. Domingo, J. M. Folch, P. LeCann, A. Jestin, J. Segales, B. Chmielewicz, J. Plana-Duran, and D. Soike. 2000. Characterization of PCV-2 isolates from Spain, Germany and France. Virus Res. 66:65–77. [DOI] [PubMed] [Google Scholar]
- 37.Mankertz, A., K. Hattermann, B. Ehlers, and D. Soike. 2000. Cloning and sequencing of columbid circovirus (CoCV), a new circovirus from pigeons. Arch. Virol. 145:2469–2479. [DOI] [PubMed] [Google Scholar]
- 38.Meehan, B. M., J. L. Creelan, M. S. McNulty, and D. Todd. 1997. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J. Gen. Virol. 78:221–227. [DOI] [PubMed] [Google Scholar]
- 39.Meehan, B. M., F. McNeilly, D. Todd, S. Kennedy, V. A. Jewhurst, J. A. Ellis, L. E. Hassard, E. G. Clark, D. M. Haines, and G. M. Allan. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79:2171–2179. [DOI] [PubMed] [Google Scholar]
- 40.Miyata, H., H. Tsunoda, A. Kazi, A. Yamada, M. A. Khan, J. Murakami, T. Kamahora, K. Shiraki, and S. Hino. 1999. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J. Virol. 73:3582–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morozov, I., T. Sirinarumitr, S. D. Sorden, P. G. Halbur, M. K. Morgan, K. J. Yoon, and P. S. Paul. 1998. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 36:2535–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nawagitgul, P., P. A. Harms, I. Morozov, B. J. Thacker, S. D. Sorden, C. Lekcharoensuk, and P. S. Paul. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assay for detection of antibodies to PCV. Clin. Diagn. Lab. Immunol., in press. [DOI] [PMC free article] [PubMed]
- 43.Nawagitgul, P., I. Morozov, S. R. Bolin, P. A. Harms, S. D. Sorden, and P. S. Paul. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81:2281–2287. [DOI] [PubMed] [Google Scholar]
- 44.Nayar, G. P., A. L. Hamel, L. Lin, C. Sachvie, E. Grudeski, and G. Spearman. 1999. Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can. Vet. J. 40:277–278. [PMC free article] [PubMed] [Google Scholar]
- 45.Nishizawa, T., H. Okamoto, K. Konishi, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92–97. [DOI] [PubMed] [Google Scholar]
- 46.Onuki, A., K. Abe, K. Togashi, K. Kawashima, A. Taneichi, and H. Tsunemitsu. 1999. Detection of porcine circovirus from lesions of a pig with wasting disease in Japan. J. Vet. Med. Sci. 61:1119–1123. [DOI] [PubMed] [Google Scholar]
- 47.Pogranichnyy, R. M., K.-J. Yoon, P. A. Harms, S. L. Swenson, J. J. Zimmerman, and S. D. Sorden. 2000. Characterization of immune response of young pigs to porcine circovirus type 2 infection. Viral Immunol. 13:143–153. [DOI] [PubMed] [Google Scholar]
- 48.Rosell, C., J. Segales, J. A. Ramos-Vara, J. M. Folch, G. M. Rodriguez-Arrioja, C. O. Duran, M. Balasch, J. Plana-Duran, and M. Domingo. 2000. Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet. Rec. 146:40–43. [DOI] [PubMed] [Google Scholar]
- 49.Seeger, C., D. Ganem, and H. E. Varmus. 1984. The cloned genome of ground squirrel hepatitis virus is infectious in the animal. Proc. Natl. Acad. Sci. USA 81:5849–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorden, S. D., P. A. Harms, P. Nawagitgul, D. Cavanaugh, and P. S. Paul. 1999. Development of a polyclonal-antibody-based immunohistochemical method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. J. Vet. Diagn. Investig. 11:528–530. [DOI] [PubMed] [Google Scholar]
- 51.Soucie, J. M., D. D. Erdman, B. L. Anderson, T. J. Torok, M. El-Jamil, E. Barnhart, M. Tepper, H. N. Burrill, A. M. Pickett, and W. L. Mengeling. 2000. Investigation of porcine parvovirus among persons with hemophilia receiving Hyate:C porcine factor VIII concentrate. Transfusion 40:708–711. [DOI] [PubMed] [Google Scholar]
- 52.Sparger, E. E., H. Louie, A. M. Ziomeck, and P. A. Luciw. 1997. Infection of cats by injection with DNA of feline immunodeficiency virus molecular clone. Virology 238:157–160. [DOI] [PubMed] [Google Scholar]
- 53.Spillane, P., S. Kennedy, B. Meehan, and G. Allan. 1998. Porcine circovirus infection in the Republic of Ireland. Vet. Rec. 143:511–512. [PubMed] [Google Scholar]
- 54.Sprengel, R., H. E. Varmus, and D. Ganem. 1987. Homologous recombination between hepadnaviral genomes following in vivo DNA transfection: implications for studies of viral infectivity. Virology 159:454–456. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi, K., Y. Iwasa, M. Hijikata, and S. Mishiro. 2000. Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch. Virol. 145:979–993. [DOI] [PubMed] [Google Scholar]
- 56.Tischer, I., H. Gelderblom, W. Vettermann, and M. A. Koch. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64–66. [DOI] [PubMed] [Google Scholar]
- 57.Tischer, I., W. Mields, D. Wolff, M. Vagt, and W. Griem. 1986. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 91:271–276. [DOI] [PubMed] [Google Scholar]
- 58.Tischer, I., L. Bode, D. Peters, S. Pociuli, and B. Germann. 1995. Distribution of antibodies to porcine circovirus in swine populations of different breeding farms. Arch. Virol. 140:737–743. [DOI] [PubMed] [Google Scholar]
- 59.Tischer, I., L. Bode, J. Apodaca, H. Timm, D. Peters, R. Rasch, S. Pociuli, and E. Gerike. 1995. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch. Virol. 140:1427–1439. [DOI] [PubMed] [Google Scholar]
- 60.Tischer, I., R. Rasch, and G. Tochtermann. 1974. Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines. Zentbl. Bakteriol. Hyg. A 226:153–167. [PubMed] [Google Scholar]
- 61.Todd, D., F. D. Niagro, B. W. Ritchie, W. Curran, G. M. Allan, P. D. Lukert, K. S. Latimer, W. L. Steffens, and M. S. McNulty. 1991. Comparison of three animal viruses with circular single-stranded DNA genomes. Arch. Virol. 117:129–135. [DOI] [PubMed] [Google Scholar]
- 62.Wellenberg, G. J., S. Pesh, F. W. Berndsen, P. J. G. M. Steverink, W. Hunneman, T. J. K. Van der Vorst, N. H. M. T. Peperkamp, V. F. Ohlinger, R. Schippers, J. T. Van Oirschot, and M. F. de Jong. 2000. Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in the Netherlands. Vet. Q. 22:167–172. [DOI] [PubMed] [Google Scholar]
- 63.Will, H., R. Cattaneo, H.-G. Koch, G. Darai, H. Schaller, H. Schellekens, P. M. C. A. van Eerd, and F. Deinhardt. 1982. Cloned HBV DNA causes hepatitis in chimpanzees. Nature 299:740–742. [DOI] [PubMed] [Google Scholar]
- 64.Willems, L., D. Portetelle, P. Kerkhofs, G. Chen, A. Burney, M. Mammerickx, and R. Kettmann. 1992. In vivo transfection of bovine leukemia provirus into sheep. Virology 189:775–777. [DOI] [PubMed] [Google Scholar]