Abstract
A human cytomegalovirus mutant (TNsubIE2P) was constructed with alanine substitutions of four residues (T27, S144, T233, and S234) previously shown to be phosphorylated in the immediate-early 2 (IE2) protein. This mutant grew as well as the wild type at both low and high multiplicities of infection. The mutant activated the major immediate-early, UL4, and UL44 promoters to similar levels, and with similar kinetics, as wild-type virus. However, the TNsubIE2P mutant virus transactivated an endogenous simian virus 40 early promoter 4 h earlier and to higher levels than the wild-type virus in infected human fibroblasts. The modification of the IE2 protein by SUMO-1 (i.e., its sumoylated state) was also examined.
The major immediate-early 2 (IE2) protein of human cytomegalovirus (HCMV) is essential for lytic replication in tissue culture (23). The predominant form of the protein is an 86-kDa nuclear phosphoprotein. The IE2 protein has been shown to be a promiscuous transcriptional activator, transactivating many early and late viral promoters as well as many cellular promoters (6, 9, 10, 12, 14, 16, 17, 19, 20, 22, 24, 30–32, 35, 36). The IE2 protein has also been shown to modulate the cell cycle (3–5, 26, 28, 33, 34) and block apoptosis (18, 37). Previously, it was shown that IE2 protein is phosphorylated by cellular kinases in vivo and in vitro (11). Specifically, the mitogen-activated protein (MAP) kinase ERK-2 was shown to phosphorylate threonines and serines in consensus MAP kinase motifs in the IE2 protein. Furthermore, it was demonstrated that MAP kinase-mediated phosphorylation of IE2 protein may reduce the protein’s transcriptional activation function. In transient-transfection assays, an IE2 protein phosphorylation site mutant, with alanine mutations at T27, S144, T233, and S234, activated transcription from a simple promoter (a Tef-1 site and a TATA element) 10- to 12-fold, compared to 4- to 5-fold activation by the wild-type protein (11).
In order to determine whether phosphorylation at these MAP kinase sites may play a role in regulating viral replication, especially the transcriptional activation of viral promoters, a mutant virus containing alanine substitution mutations at T27, S144, T233, and S234 was constructed.
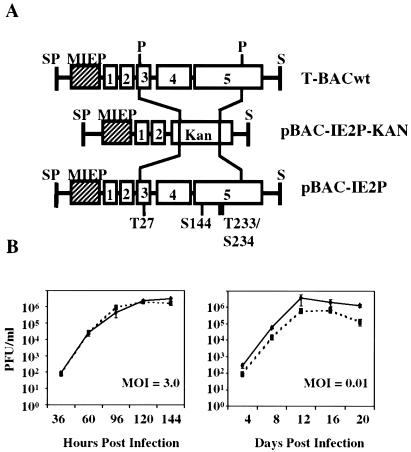
The four alanine substitutions were introduced into the bacterial artificial chromosome (BAC)-cloned Towne strain of HCMV (T-BACwt) (23), by allelic exchange (29), at positions T27, S144, T233, and S234. Figure 1A illustrates the method by which these mutations were introduced. The first clone (pBAC-IE2P-Kan) was made by insertion of a kanamycin resistance cassette into the major immediate-early locus, deleting a 2,476-bp fragment including the residues to be mutated. Clones of the kanamycin insertion mutant were verified by restriction enzyme analysis, and pBAC-IE2P-Kan was used for another round of allelic exchange in which the kanamycin cassette was replaced by a fragment of the major immediate-early locus containing the four alanine substitutions. The correct clone (pBAC-IE2P) was verified by restriction digest and sequencing of BAC DNA (data not shown). Five micrograms of purified pBAC-IE2P DNA and 2 μg of a pp71 expression plasmid (pCGN-71) (2) were used to transfect human foreskin fibroblasts (HFFs). The transfection produced a virus, TNsubIE2P, without the need for complementation. Virus was collected and used to infect more HFFs to create virus stocks. The stocks were quantitated for PFU by titration on HFFs. The presence of the mutations in the virus genome was confirmed by sequence analysis (data not shown).
FIG. 1.
(A) Generation of the TNsubIE2P phosphorylation mutant using a Towne HCMV BAC (T-BACwt). The schematic representation shows the major immediate-early locus, including exons 1 to 5 and the MIEP. Restriction enzyme sites: SP, SphI; P, PstI; S, SalI. Using allelic exchange in Escherichia coli, a 2,476-bp fragment flanked by PstI sites in the major immediate-early locus was replaced by a cassette conferring resistance to kanamycin (KAN). This clone, pBAC-IE2P-Kan, was then used in a second round of allelic exchange to replace the kanamycin cassette with the 2,476-bp fragment that was originally deleted, including four substitutions. Alanine was substituted at positions T27, S144, T233, and T234 in IE2 protein, resulting in the pBAC-IE2P mutant. (B) Growth curves of the HCMV Towne (solid lines) and TNsubIE2P (dashed lines) viruses. HFFs were infected at MOIs of 3.0 and 0.01 PFU/cell. Virus was collected at the indicated time points, and titers were determined on HFFs at 37°C.
The effects of the hypophosphorylated form of the IE2 protein (IE2P) on viral replication were first analyzed by the generation of viral growth curves. HFFs were infected with stocks of wild-type Towne (generated from the BAC clone T-BACwt) or the TNsubIE2P mutant viruses, whose titers had been determined by plaque formation, at multiplicities of infection (MOI) of 0.01 and 3.0 PFU/cell. The growth curves (Fig. 1B) show that at both high (3.0 PFU/cell) and low (0.01 PFU/cell) MOI, the TNsubIE2P virus grows as well as the wild-type virus. Therefore, the loss of these phosphorylation sites does not adversely affect viral growth in HFFs.
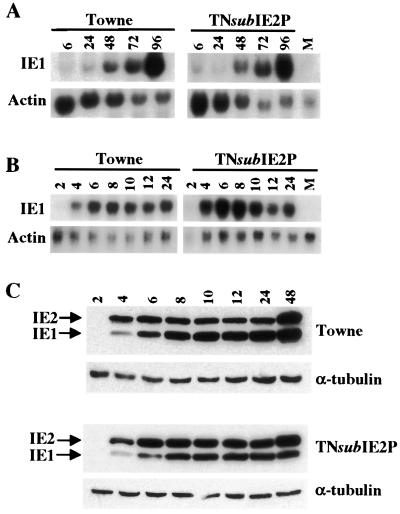
We next examined whether the loss of these phosphorylation sites had any effect on the ability of IE2 protein to transactivate viral promoters in the context of a lytic infection. Based on the transfection assay data (11), it was hypothesized that the TNsubIE2P mutant may transactivate viral promoters to a higher degree and earlier than wild-type Towne. We first examined the kinetics and transcription levels of the major immediate-early promoter (MIEP). A series of time courses were performed to determine transcription levels of the IE1 and IE2 transcripts. HFFs were infected with wild-type Towne virus and the TNsubIE2P mutant virus at an MOI of 3.0 PFU/cell. Cells were harvested at 6, 24, 48, 72, and 96 h postinfection, and RNA was collected by using the Trizol reagent (GIBCO). Northern transfers were then probed for IE1 and IE2 mRNAs. These transfers were stripped and reprobed with a 300-bp actin-specific probe as a control. It is important to note that actin mRNA levels are down regulated over time as a result of HCMV infection in HFFs. In Fig. 2A (and the similar analysis in Fig. 3A) it can be seen that actin mRNA levels vary essentially identically at comparable time points in the mutant and wild-type infections. Similarly, in Fig. 2A, IE1 mRNA levels are essentially identical at comparable time points in wild-type and mutant infections. Expression of IE1 mRNA is detectable in the mutant and wild-type infections at 6 and 24 h postinfection. The levels of transcript continue to increase through 96 h postinfection. Examining the actin and IE1 mRNA levels at each comparable time point for each infection indicates that there is no discernible difference in the synthesis of the IE1 transcript between the wild type and the TNsubIE2P mutant. Scanning the intensities of the IE1 and actin bands followed by plotting IE1 and actin levels against time provides overlapping curves (data not shown). Similar results were obtained when Northern blots were probed with an IE2-specific probe (data not shown).
FIG. 2.
Expression of IE1 and IE2 transcripts and protein. HFFs were infected with wild-type Towne and mutant viruses at an MOI of 3.0 PFU/cell and incubated at 37°C until collection. (A) Northern analysis of IE1 transcription. Infected cells were collected at 6, 24, 48, 72, and 96 h postinfection, and total RNA was prepared. A total of 4 μg of RNA was used for each time point. The Northern filter was probed with a 32P-labeled DNA fragment corresponding to the first 300 bp of exon 4, which is unique to IE1. M, mock infected. The filter was stripped and probed with a 300-bp fragment of the actin gene as a loading control. (B) Early time course of IE1 transcription. Cells were infected and collected, and RNA was harvested at 2, 4, 6, 8, 10, 12, and 24 h postinfection. The Northern filter was probed with the same IE1-specific probe. M, mock infected. (C) Immunoblot of IE1 and IE2 proteins. Cells were infected at an MOI of 3.0 PFU/cell. Cells were collected at 2, 4, 6, 8, 10, 12, 24, and 48 h postinfection and lysed in radioimmunoprecipitation assay buffer. Protein concentrations were determined by a Bradford assay, and 40 μg of protein was used for each time point. The immunoblot was probed with a mouse anti-IE1 and -IE2 monoclonal antibody (MAb 810; Chemicon) and anti-α-tubulin.
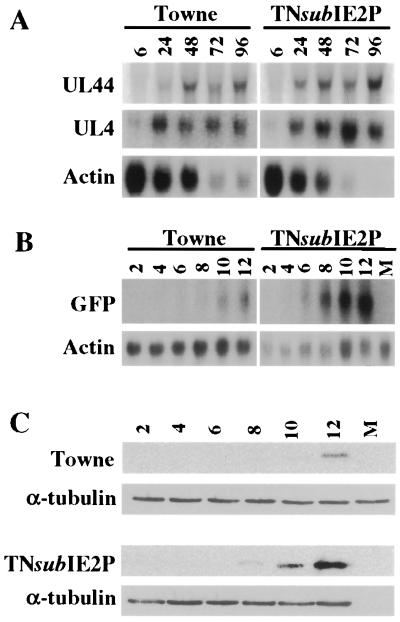
FIG. 3.
(A) Northern analysis of UL44 and UL4 early transcripts. Cells were infected and collected, and RNA was harvested at 6, 24, 48, 72, and 96 h postinfection. Northern filters were probed with a 32P-labeled DNA probe comprised of the first 300 bp of the UL4 and UL44 open reading frames. Filters were stripped and probed with actin as a loading control. (B) Northern analysis of GFP transcription. RNA was collected at 2, 4, 6, 8, 10, and 12 h postinfection. Northern filters were probed with a 32P-labeled DNA fragment of the GFP gene. (C) GFP expression in wild-type and TNsubIE2P viruses. Cells were collected at 2, 4, 6, 8, 10, and 12 h postinfection. Protein samples were prepared, and filters were probed with an anti-GFP monoclonal antibody (Clontech) and anti-α-tubulin.
To determine if the TNsubIE2P mutant was able to transactivate the MIEP at an earlier time after infection, a similar infection of HFFs was performed and RNA samples were prepared at 2, 4, 6, 8, 10, 12, and 24 h postinfection. Comparing the actin and IE1 mRNA levels at each comparable time point for each infections indicates (Fig. 2B) that the kinetics of MIEP activation is very similar between Towne and the TNsubIE2P mutant virus, even at very early time points.
To further confirm that immediate-early gene expression was not altered by the IE2P mutations, HFFs were infected with each virus at an MOI of 3.0 PFU/cell and cell extracts were prepared at 2, 4, 6, 8, 10, 12, 24, and 48 h postinfection for Western analysis. Figure 2C shows an immunoblot probed with an anti-IE1 and -IE2 monoclonal antibody (MAb 810; Chemicon). The same immunoblot was reprobed with anti-α-tubulin as a loading control. The Western analysis results confirm the Northern data showing that the IE1 and IE2 proteins are produced in relatively equivalent amounts and with the same kinetics during the infection by wild-type and mutant viruses.
The cumulative data show that the TNsubIE2P mutations do not cause increased transcriptional activity of immediate-early genes. Previous studies have shown that IE2 can negatively regulate the MIEP through binding to specific sites in the MIEP (21, 27); our data suggest that the TNsubIE2P mutations do not significantly alter this level of regulation.
We next examined the activation of several HCMV early promoters previously shown to be activated by IE2 (7, 14, 15, 31). We hypothesized that the IE2 phosphorylation mutant may be able to activate these promoters with earlier kinetics than the wild-type IE2 protein. HFFs were infected at an MOI of 3.0 PFU/cell, and RNA was collected at 6, 24, 48, 72, and 96 h postinfection. Northern transfers probed for the early genes UL44 and UL4 are shown in Fig. 3A. Comparing the actin mRNA levels and the viral mRNA levels as described for Fig. 2, we conclude that the levels and kinetics of appearance of transcripts for both of these early genes are very similar in both wild-type and IE2 mutant infections. A second Northern time course was performed by using time points between 6 and 24 h postinfection and, again, revealed no difference between the two viruses (data not shown).
The viruses produced from the parental BAC, T-BACwt, and pBAC-IE2P contain an inserted heterologous gene cassette in the Us region of the viral genome (23). The cassette consists of the simian virus 40 early promoter and the green fluorescent protein (GFP) coding region. By monitoring the expression of GFP we investigated whether the endogenous, non-HCMV promoter could be transactivated more efficiently during infection by TNsubIE2P than by the wild type. Figure 3B shows a Northern transfer probed for GFP mRNA by using RNA sampled from HFF-infected cells at an MOI of 3.0 PFU/cell for 2, 4, 6, 8, 10, and 12 h. GFP mRNA was first detectable in the wild-type infection at 10 h postinfection. However, the TNsubIE2P mutant first produced detectable levels of GFP mRNA at 6 h postinfection; in addition, greater levels of GFP mRNA accumulated in the mutant infections. An immunoblot (Fig. 3C) probed with an anti-GFP monoclonal antibody (Clontech) confirmed that GFP expression is significantly increased in the TNsubIE2P infection compared to wild-type infection. The immunoblot was also probed for α-tubulin as a loading control. GFP protein was detectable at 12 h postinfection in the wild-type infection; however, in the mutant infection it was detectable by 8 h and it accumulated to higher levels than the wild-type. These data suggest that the mutation of the four specific MAP kinase sites in IE2P allows increased transcriptional activation of a heterologous promoter.
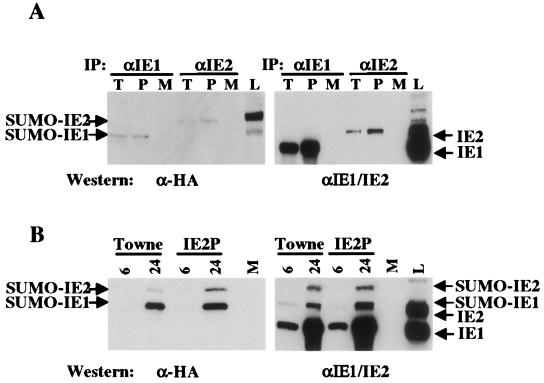
Finally, the modification of sites in IE2P by SUMO-1 (i.e., its sumoylated state) was examined to determine if it differed from the wild-type IE2 protein. IE2 has two sumoylation sites (lysines 175 and 180) that have been shown to function in SUMO modification both in vitro and in vivo (1, 13, 25). It has been shown that in other sumoylated proteins, such as IκBα, phosphorylation inhibits sumoylation (8). Therefore, we determined whether the decreased phosphorylation of IE2 could lead to an increase in sumoylation of the protein. HFFs were transfected with a hemagglutinin (HA)-SUMO expression plasmid (pHA-SUMO). Cells were infected 24 h later at an MOI of 3.0 PFU/cell with wild-type or TNsubIE2P mutant virus. Subsequently, cells were harvested, and 100 μg of total cell lysate were used in immunoprecipitation experiments. Figure 4A shows two panels of the same blot. The left panel shows the results of an immunoprecipitation experiment using monoclonal antibodies specific for IE1 (1B12) and IE2 (3H9). The blot was then probed with an anti-HA monoclonal antibody that detects any protein conjugated to the HA-SUMO modification. SUMO-modified IE1 (90 kDa) and IE2 (105 kDa) can be detected in both wild-type (lanes T) and TNsubIE2P mutant (lanes P) infections. The sumoylated forms of IE1 and IE2 are present. The right panel shows the same transfer stripped and probed with an anti-IE1/IE2 antibody (MAb 810). The nonconjugated forms of IE1 (72 kDa) and IE2 (86 kDa) were detected. The total lysate lane (lane L) shows all four forms detected by the anti-IE1/IE2 antibody (MAb 810).
FIG. 4.
Immunoprecipitation and Western analysis of pHA-SUMO-transfected and then infected cells. (A) HFFs were transfected with 5 μg of pHA-SUMO. Cells were infected 24 h later at an MOI of 3.0 PFU/cell with Towne and TNsubIE2P viruses. Infected cells were harvested 24 h postinfection and lysed in radioimmunoprecipitation assay buffer. Total protein (100 μg) was immunoprecipitated with an anti-IE1 (1B12) or anti-IE2 (3H9) monoclonal antibody. The pellet was resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis running buffer and then run on a sodium dodecyl sulfate–10% acrylamide gel. The gel was transferred overnight, and the immunoblot was probed with an anti-HA monoclonal antibody (BABCO). The panel on the left shows the immunoblot on the right, stripped and probed with an anti-IE1/IE2 antibody (MAb 810). T, wild-type virus; P, TNsubIE2P mutant virus; M, mock-infected lysate; L, total infected-cell lysate that was not subjected to immunoprecipitation. (B) HFFs were transfected and infected as described above and collected at 6 and 24 h postinfection. Lysates were immunoprecipitated with an anti-IE1/IE2 antibody (MAb 810) and then probed with an anti-HA antibody as described above. The panel on the right shows the immunoblot on the left, stripped and probed with the anti-IE1/IE2 antibody. IE2P indicates the TNsubIE2P virus.
Figure 4B shows results of the same experiment as Fig. 4A, with the anti-IE1/IE2 MAb 810 used for immunoprecipitation. At 24 h postinfection, both SUMO-modified forms of IE1 and IE2 were present in the wild-type and mutant infected-cell lysates. This blot was stripped and probed with the same antibody used for the immunoprecipitation, MAb 810. In the right panel of Fig. 4B all forms of IE1 and IE2 are present in both infections. There is no significant difference in the sumoylated forms of IE2 between the TNsubIE2P mutant and wild-type viruses.
We have demonstrated that an HCMV mutant, TNsubIE2P, lacking MAP kinase phosphorylation sites at T27, S144, T233, and S234 is viable and able to grow as well as the wild type in HFFs. The activation of immediate-early and early promoters by the TNsubIE2P mutant exhibits kinetics and levels similar to those of the wild-type virus infection. Nevertheless, an endogenous simian virus 40 early promoter was activated earlier and to a significantly higher level in cells infected by the mutant virus. This finding corroborates previously published data showing that in transient-transcription assays, the hypophosphorylated form of IE2 protein activated heterologous promoters to a significantly greater level than wild-type IE2 protein. That the hypophosphorylated protein had little effect on viral promoter activation was an unexpected result which suggests that the phosphorylation of these sites in IE2 may affect transcription from heterologous and viral promoters differently. It remains to be tested whether phosphorylation of IE2 may control separate transcriptional activation domains which can affect promoters differentially during the infection. Finally, these results are seen during lytic infections of HFFs, a cell type used because of its permissivity for HCMV. It is possible that the mutations may have a more significant effects in cells and tissues naturally infected by HCMV.
Acknowledgments
J. Heider thanks W. Bresnahan and F. Goodrum for discussion and critical reading of the manuscript and M. Nevels for the pHA-SUMO expression vector.
This work was supported by a grant from the NIH (CA85786) to T. Shenk.
REFERENCES
- 1.Ahn, J. H., Y. Xu, W. J. Jang, M. J. Matunis, and G. S. Hayward. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonin, L. R., and J. K. McDougall. 1997. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J. Virol. 71:5861–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., T. Albrecht, and E. A. Thompson. 1998. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J. Biol. Chem. 273:22075–22082. [DOI] [PubMed] [Google Scholar]
- 5.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caswell, R., L. Bryant, and J. Sinclair. 1996. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J. Virol. 70:4028–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., and M. F. Stinski. 2000. Activation of transcription of the human cytomegalovirus early UL4 promoter by the Ets transcription factor binding element. J. Virol. 74:9845–9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233–239. [DOI] [PubMed] [Google Scholar]
- 9.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagemeier, C., S. M. Walker, P. J. Sissons, and J. H. Sinclair. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 73:2385–2393. [DOI] [PubMed] [Google Scholar]
- 11.Harel, N. Y., and J. C. Alwine. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J. Virol. 72:5481–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermiston, T. W., C. L. Malone, P. R. Witte, and M. F. Stinski. 1987. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J. Virol. 61:3214–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2–p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, L., C. L. Malone, and M. F. Stinski. 1994. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J. Virol. 68:2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, L., and M. F. Stinski. 1995. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J. Virol. 69:7612–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerry, J. A., M. A. Priddy, and R. M. Stenberg. 1994. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J. Virol. 68:4167–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klucher, K. M., M. Sommer, J. T. Kadonaga, and D. H. Spector. 1993. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol. Cell. Biol. 13:1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukac, D. M., and J. C. Alwine. 1999. Effects of human cytomegalovirus major immediate-early proteins in controlling the cell cycle and inhibiting apoptosis: studies with ts13 cells. J. Virol. 73:2825–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc. Natl. Acad. Sci. USA 90:707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monick, M. M., L. J. Geist, M. F. Stinski, and G. W. Hunninghake. 1992. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am. J. Respir. Cell Mol. Biol. 7:251–256. [DOI] [PubMed] [Google Scholar]
- 25.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinclair, J., J. Baillie, L. Bryant, and R. Caswell. 2000. Human cytomegalovirus mediates cell cycle progression through G1 into early S phase in terminally differentiated cells. J. Gen. Virol. 81 Pt 6:1553–1565. [DOI] [PubMed] [Google Scholar]
- 29.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staprans, S. I., D. K. Rabert, and D. H. Spector. 1988. Identification of sequence requirements and trans-acting functions necessary for regulated expression of a human cytomegalovirus early gene. J. Virol. 62:3463–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiebusch, L., and C. Hagemeier. 2001. The human cytomegalovirus immediate early 2 protein dissociates cellular DNA synthesis from cyclin-dependent kinase activation. EMBO J. 20:1086–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo, Y. D., C. J. Chiou, K. S. Choi, Y. Yi, S. Michelson, S. Kim, G. S. Hayward, and S. J. Kim. 1996. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J. Virol. 70:7062–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yurochko, A. D., T. F. Kowalik, S. M. Huong, and E. S. Huang. 1995. Human cytomegalovirus upregulates NF-κ B activity by transactivating the NF-κB p105/p50 and p65 promoters. J. Virol. 69:5391–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]