Abstract
The A/teal/Hong Kong/W312/97 (H6N1) influenza virus and the human H5N1 and H9N2 influenza viruses possess similar genes encoding internal proteins, suggesting that H6N1 viruses could become novel human pathogens. The molecular epidemiology and evolution of H6 influenza viruses were characterized by antigenic and genetic analyses of 29 H6 influenza viruses isolated from 1975 to 1981 and 1997 to 2000. Two distinct groups were identified on the basis of their antigenic characteristics. Phylogenetic analysis revealed that all H6N1 viruses isolated from terrestrial poultry in 1999 and 2000 are closely related to A/teal/Hong Kong/W312/97 (H6N1), and the nucleotide sequences of these viruses and of A/Hong Kong/156/97 (H5N1) were more than 96% homologous. The hemagglutinin (HA) of the 1999 and 2000 terrestrial viruses does not have multiple basic amino acids at the site of cleavage of HA1 to HA2; however, a unique insertion of aspartic acid in HA1 between positions 144 and 145 (H3 numbering) was found. The neuraminidase of these terrestrial H6N1 viruses has a deletion of 19 amino acids characteristic of A/Hong Kong/156/97 (H5N1). Evolutionary analysis suggested that these H6N1 viruses coevolved with A/quail/Hong Kong/G1/97-like H9N2 viruses and became more adapted to terrestrial poultry. These terrestrial 1999 and 2000 A/teal/Hong Kong/W312/97 (H6N1)-like viruses, along with the H9N2 viruses, could have been involved in the genesis of the pathogenic H5N1 influenza viruses of 1997. The presence of H6N1 viruses in poultry markets in Hong Kong that possess seven of the eight genes of the A/Hong Kong/156/97 (H5N1) virus raises the following fundamental questions relevant to influenza pandemic preparedness: could the pathogenic H5N1 virus reemerge and could the H6N1 viruses directly cross the species barrier to mammals?
An H6N1 influenza virus was isolated from a green-winged teal during the H5N1 outbreak in Hong Kong Special Administrative Region (SAR) in 1997 (8). Subsequent characterization of the virus showed that seven of its eight genes are closely related to those of the pathogenic H5N1 influenza virus, which affected humans in 1997 (1, 22, 26, 32). This A/teal/Hong Kong (HK)/W312/97 (H6N1) influenza virus is the first known isolate with seven A/HK/156/97 (H5N1)-like gene segments. In addition to the H6N1 isolate, several other subtypes of influenza viruses were isolated in the live-poultry retail markets of Hong Kong. Among them, H9N2 influenza viruses, which were present in chickens, ducks, geese, and a variety of minor poultry, were the second most common subtype isolated in 1997 (23). One of the H9N2 isolates, A/quail/HK/G1/97 (H9N2), possessed six gene segments that are highly homologous to those of the human H5N1 virus (6). In 1999, A/quail/HK/G1/97 (H9N2)-like influenza viruses infected two humans in Hong Kong (10, 14). The incident highlights the potential for avian influenza viruses to cross the species barrier and infect humans without prior reassortment in an intermediate host, such as the pig (16). The common features shared by H5N1 and H9N2 influenza viruses isolated from humans are the genes encoding the proteins of the replicating complex, the matrix protein (M) gene and the nonstructural protein (NS) gene (10). How these six internal protein genes confer a virus’s ability to infect humans is not known, but influenza A viruses that possess these genes are of particular concern and pose a potential public health threat. The A/teal/HK/W312/97 (H6N1) virus possesses not only these six internal protein genes but also the N1 neuraminidase (NA) gene of the human H5N1 influenza virus (8). This virus essentially represents the reemergence of the H5N1 influenza viruses with a different hemagglutinin (HA). Therefore it is important to understand the molecular epidemiology and evolution of H6 influenza viruses in southeastern China.
The human H5N1 influenza virus has been proposed to be a reassortant of three precursor viruses. Both A/quail/HK/G1/97 (H9N2)-like and A/teal/HK/W312/97 (H6N1)-like viruses are possible gene donors in the genesis of the H5N1 influenza viruses (6, 8). The A/quail/HK/G1/97 (H9N2)-like influenza viruses have lower rates of amino acid change in the six internal proteins than do the pathogenic H5N1 influenza viruses, indicating that the introduction of H9N2 influenza viruses into terrestrial poultry is not a recent one (7). In other words, the H9N2 influenza viruses could have donated genes for the generation of the pathogenic H5N1 influenza viruses. However, the evolution of H6N1 influenza viruses has not been elucidated. A question remains: were the pathogenic H5N1 influenza viruses in Hong Kong reassortants of A/quail/HK/G1/97-like or A/teal/HK/W312/97-like viruses or were the H5N1 and A/teal/HK/W312/97-like viruses themselves the products of reassortment events?
Until recently, influenza viruses of the H6 subtype have received little attention. The H6 subtype was first isolated from a turkey in 1965, and other viruses that belong to the H6 subtype were subsequently isolated from shorebirds and wild ducks (3, 4, 17). The full-length nucleotide sequences of the H6 HA gene are most homologous to those of the H2 subtype (12). Previous surveillance in Hong Kong of poultry from southern China (1975 to early 1980s) revealed that H6 influenza viruses occurred in domestic ducks and geese and only one H6 influenza virus, viz., H6N4, was isolated from a chicken (19, 21). Of the viruses isolated during the H5N1 incident, four were H6 influenza viruses, and those were isolated from geese and a green-winged teal (this study; 8). These findings demonstrate the continuous circulation of H6 influenza viruses in poultry in southeastern China.
The antigenic and genetic characteristics of influenza viruses from different types of poultry were investigated to determine the molecular epidemiology and evolution of H6 influenza viruses in southeastern China. The results of the present study show the diversity of H6 viruses in poultry in this region. The presence of H6N1 viruses in poultry that possess seven of the eight genes of the human H5N1 virus raises the following question: will this pathogenic H5N1 virus reemerge (24) or will H6N1 viruses directly cross the species barrier into mammals?
MATERIALS AND METHODS
Virus sampling.
H6 influenza viruses isolated in Hong Kong and their abbreviations used in this study are listed in Table 1. Viruses isolated in Hong Kong during the 1970s were collected during an epidemiological survey conducted from 1975 to 1981 as described elsewhere (19, 21). Viral surveillance was not conducted from 1982 to early 1997. The viruses isolated in 1997 were collected during the H5N1 outbreak. After the H5N1 outbreak, the market system was changed: live domestic terrestrial poultry continue to be sold in the live-poultry retail markets, whereas aquatic poultry, viz., ducks and geese, are now slaughtered at a central slaughtering facility and the carcasses are sold at the live-poultry retail markets. Viruses isolated from terrestrial poultry were collected at 19 markets from April 1999 to April 2000 during routine surveillance as previously described (7). Briefly, an average of 50 fecal samples were collected from the trays positioned under the poultry cages in each market. The number of samples collected from each type of poultry was approximately proportional to the number of that poultry in the market. Terrestrial poultry present in the markets included chickens, chukars, guinea fowl, pheasants, pigeons, partridges, quail, and Silkie chickens. Fecal samples were collected in tissue culture medium M199 containing antibiotics and antifungal agents (7). Viruses from aquatic hosts were collected at the central slaughtering facility during routine surveillance by the Agriculture, Fisheries and Conservation Department and the Food and Environmental Hygiene Department of the Government of Hong Kong SAR. Cloacal swabs were collected from 14 ducks or geese from each consignment shipped to the facility. Viruses were isolated in embryonated chicken eggs, and the infected allantoic fluids were used for characterization studies.
TABLE 1.
H6 influenza viruses characterized in this study
| Group | Isolate | Subtype | Abbreviation | Source |
|---|---|---|---|---|
| Contemporary terrestrial isolatesa | A/pheasant/HK/SH39/99 | H6N1 | Ph/HK/SH39/99 | Feces |
| A/quail/HK/1721-20/99 | H6N1 | Qa/HK/1721-20/99 | Feces | |
| A/quail/HK/1721-30/99 | H6N1 | Qa/HK/1721-30/99 | Feces | |
| A/quail/HK/SF550/00 | H6N1 | Qa/HK/SF550/00 | Feces | |
| A/quail/HK/SF595/00 | H6N1 | Qa/HK/SF595/00 | Feces | |
| A/pheasant/HK/NT261/00 | H6N1 | Ph/HK/NT261/00 | Feces | |
| A/chukarHK/FY295/00 | H6N1 | Cu/HK/FY295/00 | Feces | |
| Contemporary aquatic isolatesb | A/goose/HK/W217/97 | H6N9 | Gs/HK/W217/97 | Cloaca |
| A/goose/HK/W222/97 | H6N7 | Gs/HK/W222/97 | Cloaca | |
| A/goose/HK/W375/97 | H6N2 | Gs/HK/W375/97 | Cloaca | |
| A/duck/HK/323/98 | H6N2 | Dk/HK/323/98 | Chilled duck carcass | |
| A/duck/HK/324/98 | H6N2 | Dk/HK/324/98 | Chilled duck carcass | |
| A/duck/HK/1037-1/98 | H6N2 | Dk/HK/1037-1/98 | Cloaca | |
| A/duck/HK/1037-2/98 | H6N2 | Dk/HK/1037-2/98 | Cloaca | |
| A/duck/HK/1037-3/98 | H6N2 | Dk/HK/1037-3/98 | Cloaca | |
| A/duck/HK/1037-4/98 | H6N2 | Dk/HK/1037-4/98 | Cloaca | |
| A/duck/HK/3096/99 | H6N2 | Dk/HK/3096/99 | Cloaca | |
| A/duck/HK/3174/99 | H6N2 | Dk/HK/3174/99 | Cloaca | |
| A/duck/HK/3461/99 | H6N1 | Dk/HK/3461/99 | Cloaca | |
| A/duck/HK/3600/99 | H6N2 | Dk/HK/3600/99 | Cloaca | |
| 1970’s isolatesc | A/duck/HK/73/76 | H6N1 | Dk/HK/73/76 | Trachea |
| A/duck/HK/108/76 | H6N5 | Dk/HK/108/76 | Cloaca | |
| A/duck/HK/134/77 | H6N2 | Dk/HK/134/77 | Cloaca | |
| A/duck/HK/175/77 | H6N1 | Dk/HK/175/77 | Cloaca | |
| A/duck/HK/182/77 | H6N9 | Dk/HK/182/77 | Trachea | |
| A/duck/HK/202/77 | H6N1 | Dk/HK/202/77 | Cloaca | |
| A/duck/HK/221/77 | H6N8 | Dk/HK/221/77 | Cloaca | |
| A/goose/HK/17/77 | H6N4 | Gs/HK/17/77 | Cloaca | |
| A/chicken/HK/17/77 | H6N4 | Ck/HK/17/77 | Cloaca |
These samples were collected from live-poultry retail markets from April 1999 to April 2000.
Viruses isolated in 1997 were collected during the H5N1 outbreak, and those isolated after 1997 were collected from a central slaughtering facility for aquatic poultry.
These samples were collected from November 1975 to October 1979.
Antigenic analysis.
Hemagglutination inhibition (HI) and neuraminidase inhibition (NI) tests were performed with a panel of reference antisera (18).
Nucleotide sequence analysis.
Viral RNA was extracted from infected allantoic fluids by using an RNeasy RNA extraction kit (Qiagen, Hilden, Germany). Viral RNA was reverse transcribed using an oligodeoxynucleotide primer (5′AGCAAAAGCAGG) and Superscript II reverse transcriptase (Life Technologies Inc., Grand Island, N.Y.). PCR was performed with gene-specific primers (sequences are available upon request). Amplified products of the expected sizes were purified with a QIAquick PCR purification kit (Qiagen). Purified amplicons were sequenced by using the BigDye terminator cycle sequencing ready reaction (Applied Biosystems, Foster City, Calif.) and analyzed on an ABI377 automated sequencer (Applied Biosystems).
Phylogenetic analysis.
Published sequences used in phylogenetic comparison in this study were obtained from the Influenza Sequences Database (http://www.flu.lanl.gov). Editing, analysis, and alignment of sequence data were performed with the Lasergene package (version 4.0; DNASTAR, Madison, Wis.) and the Wisconsin Package (version 10.0; Genetics Computer Group, Madison, Wis.). Phylogenetic analysis was performed with Phylogenetic Analysis Using Parsimony (PAUP, version 4; D. Swofford, Sinauer Associates, Sunderland, Mass.) by PAUPSearch in the Wisconsin Package with 100 bootstrap replicates.
Evolutionary analysis.
A defined region of the eight gene segments of the H5N1, H6N1, and H9N2 influenza viruses was analyzed with the Molecular Evolutionary Genetics Analysis program (MEGA, version 2.0; S. Kumar, K. Tamura, I. Jakobsen, and M. Nei, Pennsylvania State University, University Park, Pa., and Arizona State University, Tempe, Ariz.). A reference virus was selected from each group of viruses analyzed. Reference viruses were compared pairwise with the members in their group, and the nonsynonymous substitutions were calculated using the Pamilo-Bianchi-Li method (9, 13). The rate of nonsynonymous substitutions was determined by dividing the average nonsynonymous substitutions by the number of years between the isolation of the reference virus and that of the member viruses. The rates of nonsynonymous substitutions for the virus groups were compared, and the P values were determined by the Student t test.
Nucleotide sequence accession numbers.
The sequence data obtained in this study have been submitted to the EMBL Nucleotide Sequence Database and are listed under accession no. AJ410495 through AJ410614.
RESULTS
H6 influenza viruses from poultry in Hong Kong.
Influenza virus surveillance studies carried out from 1975 to 1981 and 1997 to 2000 showed that H6 influenza viruses were present in aquatic and terrestrial poultry. The molecular epidemiology and evolution of these viruses were characterized using H6 isolates from different types of poultry (Table 1). The 1970s isolates were of various NA subtypes (N1, N2, N4, N5, N8, and N9). The contemporary aquatic isolates were of various NA subtypes, most being of the N2 subtype. All contemporary terrestrial isolates were of the N1 subtype.
In surveillance studies conducted during the 1970s, only ducks, geese, and chickens were sampled. The majority of the samples were collected from ducks (19, 21). One chicken, one goose, and seven duck H6 isolates were chosen for characterization in this study. During the H5N1 outbreak, emphasis was on the major poultry, viz., chickens, ducks, and geese, but minor poultry (quails, pheasants, pigeons, chukars, and guinea fowls) were also sampled. Three H6 influenza viruses were isolated from geese, and one was isolated from a green-winged teal (this study; 8). From 1998 to 1999, routine influenza virus surveillance of aquatic poultry at the central slaughtering facility resulted in the isolation of 10 H6 influenza viruses. From 1999 to 2000, in surveillance of the 19 live-poultry retail markets, seven H6 influenza viruses were isolated on six different sampling occasions in five of the markets from 162 quail, 153 pheasant, and 63 chukar fecal samples. These viruses represent the H6 isolates from terrestrial poultry in the present study.
Antigenic analysis.
The isolates were characterized antigenically by HI tests (Table 2). The H6 influenza viruses isolated in southeastern China reacted with hyperimmune antisera raised against the viruses A/turkey/Massachusetts/1/65 (H6N2) and A/shearwater/Australia/1/72 (H6N5). While the H6 rabbit hyperimmune reference antiserum raised against A/teal/HK/W312/97 (H6N1) of the World Health Organization supplemental influenza reagent kit (Centers for Disease Control and Prevention, Atlanta, Ga.) reacted with all contemporary terrestrial isolates, it showed limited reactivity to contemporary aquatic isolates and reduced reactivity to the 1970s isolates. This rabbit hyperimmune reference antiserum has broader cross-reactivity than ferret antiserum raised against the same virus. The H6 influenza viruses can be divided into terrestrial and aquatic groups on the basis of their reaction with postinfection chicken antiserum, postinfection ferret antiserum, and monoclonal antibody. Postinfection chicken antiserum to Ph/HK/SH39/99 (H6N1) reacted with all contemporary terrestrial isolates but, with the exception of Tl/HK/W312/97 (H6N1) and Dk/HK/3461/99 (H6N1), did not react with contemporary aquatic and 1970s isolates. Postinfection chicken antiserum to Gs/HK/W217/97 (H6N2) reacted primarily with contemporary aquatic and 1970s isolates and showed low reactivity with contemporary terrestrial isolates. Postinfection ferret antiserum and monoclonal antibody against Tl/HK/W312/97 (H6N1) showed similar specificities and reacted only with contemporary terrestrial isolates, but postinfection chicken antiserum against Ck/HK/17/77 (H6N4) cannot differentiate the different groups of viruses. These results show the presence of two antigenically distinct groups of H6 influenza viruses in the terrestrial and aquatic poultry of this region.
TABLE 2.
Antigenic analysis of H6 influenza viruses by HIa
| Virus | Titerb for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Hyperimmune rabbit antisera against:
|
Postinfection chicken antisera against:
|
Postinfection ferret antiserum against Tl/HK/W312/97 | Monoclonal antibody against Tl/HK/W312/97 | |||||
| Tk/Mass/1/65 | Sh/Au/1/72 | WHO H6 reference | Ph/HK/SH39/99 | Gs/HK/W217/97 | Ck/HK/17/77 | |||
| Reference strains | ||||||||
| Tk/Mass/1/65 | 2,560 | 1,280 | 320 | 80 | 640 | 320 | < | < |
| Sh/Au/1/72 | 640 | 5,120 | 160 | 80 | 640 | 640 | < | < |
| WHO H6N1c | 1,280 | 1,280 | 320 | 320 | 80 | 320 | 160 | 5,120 |
| Contemporary terrestrial isolates | ||||||||
| Ph/HK/SH39/99 | 2,560 | 2,560 | 640 | 320 | 40 | 640 | 160 | 5,120 |
| Qa/HK/1721-20/99 | 1,280 | 2,560 | 320 | 320 | 40 | 320 | 160 | 5,120 |
| Qa/HK/1721-30/99 | 1,280 | 2,560 | 320 | 320 | 40 | 320 | 160 | 5,120 |
| Qa/HK/SF550/00 | 2,560 | 2,560 | 320 | 320 | 40 | 320 | 160 | 5,120 |
| Qa/HK/SF595/00 | 1,280 | 2,560 | 320 | 320 | 40 | 320 | 80 | 1,280 |
| Ph/HK/NT261/00 | 320 | 640 | 320 | 320 | 40 | 320 | 80 | 5,120 |
| Cu/HK/FY295/00 | 640 | 1,280 | 320 | 320 | 40 | 320 | 80 | 5,120 |
| Contemporary aquatic isolates | ||||||||
| Gs/HK/W217/97 | 640 | 640 | 80 | < | 640 | 640 | < | < |
| Gs/HK/W222/97 | 2,560 | 2,560 | 640 | < | 1,280 | 640 | < | < |
| Tl/HK/W312/97 | 1,280 | 2,560 | 320 | 320 | 80 | 320 | 160 | 5,120 |
| Gs/HK/W375/97 | 640 | 640 | < | < | 1,280 | < | < | < |
| Dk/HK/323/98 | 1,280 | 1,280 | < | < | 1,280 | 160 | < | < |
| Dk/HK/324/98 | 1,280 | 1,280 | < | < | 1,280 | 80 | < | < |
| Dk/HK/1037-1/98 | 160 | 160 | < | < | 640 | 40 | < | < |
| Dk/HK/1037-2/98 | 640 | 640 | < | < | 640 | 160 | < | < |
| Dk/HK/1037-3/98 | 640 | 640 | < | < | 640 | 160 | < | < |
| Dk/HK/1037-4/98 | 640 | 640 | < | < | 640 | 160 | < | < |
| Dk/HK/3096/99 | 320 | 640 | < | < | 320 | 160 | < | < |
| Dk/HK/3174/99 | 320 | 640 | 40 | < | 320 | 160 | < | < |
| Dk/HK/3461/99 | 640 | 2,560 | 80 | 80 | 160 | 1,280 | < | < |
| Dk/HK/3600/99 | 640 | 1,280 | 40 | < | 640 | 320 | < | < |
| 1970s isolates | ||||||||
| Dk/HK/73/76 | 1,280 | 5,120 | 80 | < | 320 | 640 | < | < |
| Dk/HK/108/76 | 640 | 2,560 | 40 | < | 160 | 320 | < | < |
| Dk/HK/134/77 | 2,560 | 5,120 | 80 | < | 160 | 2,560 | < | < |
| Dk/HK/175/77 | 320 | 2,560 | 40 | < | 160 | 320 | < | < |
| Dk/HK/182/77 | 640 | 2,560 | 160 | < | 640 | 1,280 | < | < |
| Dk/HK/202/77 | 640 | 2,560 | 80 | < | 320 | 640 | < | < |
| Dk/HK/221/77 | 640 | 2,560 | 160 | < | 320 | 1,280 | < | < |
| Gs/HK/17/77 | 1,280 | 5,120 | 160 | < | 320 | 1,280 | < | < |
| Ck/HK/17/77 | 1,280 | 5,120 | 160 | < | 320 | 640 | < | < |
Abbreviations: Tk/Mass/1/65, A/turkey/Massachusetts/1/65 (H6N2); Sh/Au/1/72, A/shearwater/Australia/1/72 (H6N5); Tl/HK/W312/97, A/teal/Hong Kong/W312/97 (H6N1); WHO, World Health Organization.
Titers in bold face are for homologous sera; <, no inhibition was detected at a serum dilution of 1:40.
Inactivated Tl/HK/W312/97 (H6N1) influenza virus.
Genetic analysis of the HA and NA genes.
The full-length sequences of the HA genes of 29 H6 influenza viruses were determined to analyze the phylogenetic relationships among H6 influenza viruses isolated in southeastern China. These HA genes were 87 to 91% homologous to that of A/shearwater/Australia/1/72 (H6N5) (12). On the basis of this analysis, two distinct groups of H6 HA genes were identified. Each H6 HA gene of contemporary influenza viruses isolated from aquatic poultry and those isolated during the 1970s has an open reading frame of 1,701 bp that encodes a precursor polypeptide of 566 amino acids. In contrast, each H6 HA gene of contemporary influenza viruses isolated from terrestrial poultry has an open reading frame of 1,704 bp that encodes a precursor of 567 amino acids. The insertion of aspartic acid between positions 144 and 145 (H3 numbering) of the precursor polypeptides distinguishes the two groups of H6 HA genes and appears only in contemporary isolates from terrestrial hosts. All H6 isolates possess the sequence, PQIETR↓G at the cleavage site (arrow) between HA1 and HA2, and none contains the sequence of multiple basic amino acids which is found in highly pathogenic chicken influenza A viruses (25, 28). The consensus amino acid sequences revealed five potential N-linked glycosylation sites in HA1 (asparagine 26 or 27, 39, 182, 306, and 311) and two in HA2 (asparagine 498 and 557). Amino acid changes in Gs/HK/W222/97, Gs/HK/W375/97, and Dk/HK/134/77 (N182K), Dk/HK/175/77 and Dk/HK/3461/99 (N182S), and Dk/HK/1037-1/98 (N306I) led to the loss of potential glycosylation sites.
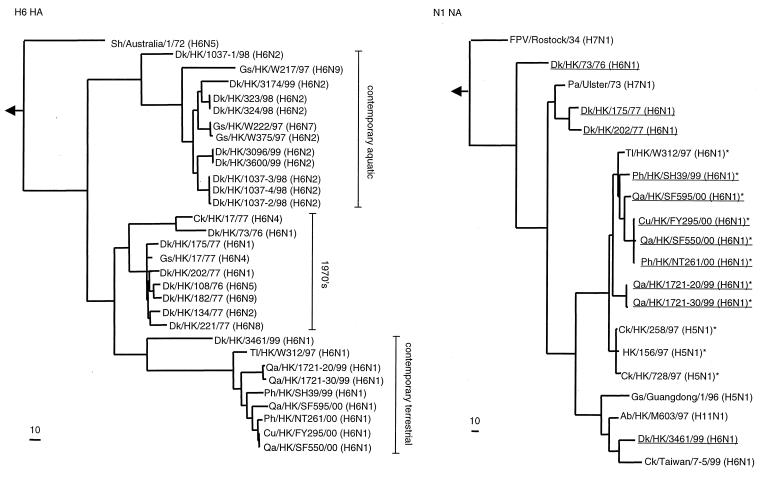
The H6 HA phylogenetic tree revealed that the H6 isolates are clustered into three branches: the 1970s, the contemporary aquatic, and the contemporary terrestrial branches (Fig. 1). All viruses in the contemporary terrestrial branch are exclusively of the N1 subtype. The Tl/HK/W312/97 (H6N1) virus was grouped with the H6 contemporary terrestrial isolates and diverge after the separation from duck virus Dk/HK/3461/99 (H6N1), suggesting that contemporary terrestrial isolates may have been derived from an aquatic ancestor. This observation was strengthened by the cross-reactivity that postinfection chicken antiserum to Ph/HK/SH39/99 (H6N1) demonstrated with Tl/HK/W312/97 (H6N1) and, to a limited extent, Dk/HK/3461/99 (H6N1). The contemporary terrestrial isolates and those of the 1970s branch showed a sister group relationship, indicating that these viruses may have a common ancestor. Thus the contemporary terrestrial branch may have been established in the 1970s or earlier and the ancestor could have been a virus present in aquatic hosts of the time.
FIG. 1.
Phylogenetic trees based on the sequences of the H6 HA and N1 NA genes of influenza viruses. Nucleotides 29 to 1730 (1,701 bp) of the H6 HA gene were used to construct the HA phylogenetic tree, which was rooted to A/mallard/Potsdam/178-4/83 (H2N2). Nucleotides 66 to 1373 (1,439 bp) of the N1 NA gene were used to construct the NA phylogenetic tree, which was rooted to A/WSN/33 (H1N1). H6 influenza viruses characterized in this study are underlined (NA phylogenetic tree). *, viruses with characteristic deletion of codons for 19 amino acids from the NA gene. The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. Gene sequences used in this study are listed under the following EMBL Nucleotide Sequence Database accession numbers: D90303, X5226, K02252, AF036357, AF057292, AF098548, AF144304, AF098551, and AF208598. Abbreviations: Sh, shearwater; Pa, parrot; Ab, aquatic bird.
Because the NA gene of Tl/HK/W312/97 (H6N1) is closely related to that of the pathogenic H5N1 influenza viruses (8), the N1 genes of all H6 isolates were sequenced. All N1 NAs of contemporary terrestrial isolates as well as that of the teal isolate clustered into one branch with those of the pathogenic H5N1 influenza viruses of 1997 (Fig. 1). Those from contemporary aquatic poultry formed another branch with those of the Gs/Guangdong/1/96 (H5N1), Ab/HK/M603/97 (H11N1), and Ck/Taiwan/7-5/99 (H6N1). The 1970s isolates showed an ancestral relationship to both contemporary groups. Alignment analysis revealed a 19-amino-acid deletion in the stalk region of the NAs from all contemporary terrestrial isolates. This deletion is characteristic of the HK/156/97 (H5N1)-like viruses (1, 26). Therefore, the N1 NAs of contemporary terrestrial H6N1 influenza viruses are closely related to those of the pathogenic H5N1 influenza viruses of 1997. These findings show the diversity of H6 influenza viruses in this region and show that all the terrestrial isolates possess N1 with high identity to the N1 of the HK/156/97-like viruses.
Genetic analysis of internal genes.
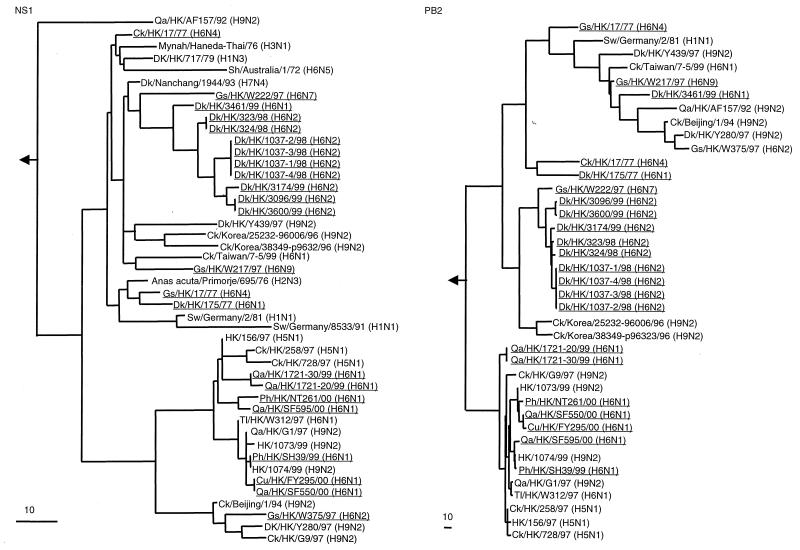
Surveillance in the Hong Kong live-poultry retail markets in 1997 revealed that the H6 influenza viruses were cocirculating with the H5N1 and H9N2 influenza viruses (23). This circulation increased the possibility of genetic exchange among these viruses. Therefore, the sequences of the NS and PB2 polymerase (PB2) genes of the H6 influenza viruses were characterized and the viruses with gene segments similar to those of the HK/156/97 (H5N1)-like viruses were identified. Phylogenetic analysis of the NS gene revealed that the H6 influenza viruses characterized in this study belonged to the A allele and were part of the Eurasian clade (Fig. 2). Contemporary aquatic and 1970s isolates clustered into a large group represented by the duck influenza viruses Dk/HK/439/97 (H9N2) and Dk/HK/717/79 (H1N3), respectively. As for the N1 NA phylogenetic tree, the 1970s isolates showed an ancestral relationship to the contemporary aquatic isolates. The NS genes of contemporary terrestrial isolates are phylogenetically similar to those of the human H5N1 (HK/156/97) and H9N2 (HK/1073/99 and HK/1074/99) influenza viruses.
FIG. 2.
Phylogenetic trees based on the sequences of the NS1 and PB2 genes of influenza viruses. H6 influenza viruses characterized in this study are underlined. The ranges of nucleotide sequences used to construct the phylogenetic trees are shown in Table 4. The NS1 phylogenetic tree is rooted to A/equine/Prague/1/56 (H7N7), and the PB2 phylogenetic tree is rooted to B/Lee/40. The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Vertical lines are for spacing branches and labels. Gene sequences used in this study are listed under the following EMBL Nucleotide Sequence Database accession numbers: AF156479, M17070, M60800, M55484, Z26865, U49492, L25830, AF156476, AF262212, U49493, AF250502, AF156482, AF156481, AF036360, AF098569, AF098571, AF250483, AF156477, AJ278649, AJ404735, AF156480, AF156472, AF156475, AF156440, AF156439, M55471, AF255624, AF156434, AF156437, AF156438, AF156436, AF156433, AF156430, AJ404630, AJ404630, AF036363, AF098577, AF098579, AF250476, and AF156435. Sw, swine.
The topology of the PB2 phylogenetic tree was similar to that of the NS phylogenetic tree: the 1970s, the contemporary aquatic, and the contemporary terrestrial isolates clustered into three different branches (Fig. 2). The 1970s isolates and two contemporary aquatic isolates formed an out-group that was distinct from other contemporary aquatic isolates. Whereas most of the contemporary aquatic isolates clustered together with two Korean H9N2 chicken influenza viruses. All contemporary terrestrial isolates were part of the same branch as the human H5N1 (HK/156/97) and H9N2 (HK/1073/99 and HK/1074/99) influenza viruses.
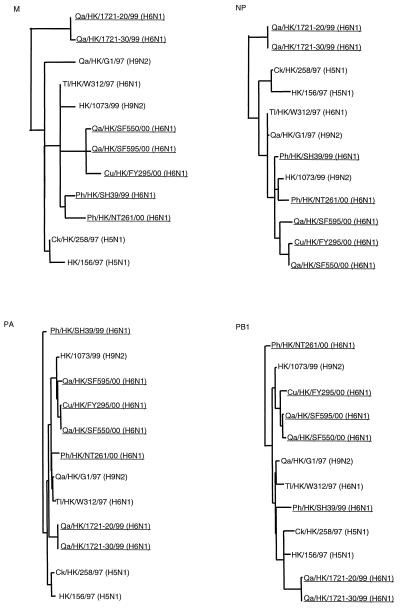
The phylogenetic analysis suggested that the N1 NA, NS, and PB2 genes of all contemporary terrestrial isolates are closely related to those of the H5N1 human influenza viruses. Therefore, the sequences of the M, nucleoprotein (NP), PA polymerase (PA), and PB1 polymerase (PB1) genes of the seven contemporary terrestrial H6N1 isolates were analyzed to explore this relationship further. The generalized phylogeny of these genes revealed that all seven contemporary terrestrial H6N1 isolates clustered with the H5N1 and H9N2 influenza viruses that infected humans (Fig. 3). Seven of the eight gene segments of all the contemporary terrestrial H6N1 influenza viruses are more than 96% homologous to those of human HK/156/97 (H5N1) (Table 3). In contrast, these seven gene segments of duck H6N1 influenza virus Dk/HK/3461/99 were less than 93% homologous to those of HK/156/97 although the Dk/HK/3461/99 HA gene belonged to the contemporary terrestrial branch of the H6 HA gene phylogenetic tree (Fig. 1). In addition, the homology among the gene segments of the influenza viruses of the contemporary aquatic group and the 1970s group and those of HK/156/97 was less than 93%. Thus, the molecular and phylogenetic results confirmed that H6N1 influenza viruses that have seven gene segments closely related to those of the pathogenic HK/156/97 (H5N1)-like viruses are still present in terrestrial poultry in southeastern China.
FIG. 3.
Generalized phylogeny of M1, NP, PA, and PB1 genes of influenza viruses. H6 influenza viruses characterized in this study are underlined. Nucleotide ranges used to construct the phylogenetic trees are shown in Table 4. Gene sequences used in this study are listed under the following EMBL Nucleotide Sequence Database accession numbers: AF098560, AF036358, AF156463, AF250482, AJ278646, AF057293, AF036359, AF156470, AF250480, AJ289871, AF098604, AF046095, AF156449, AF250478, AJ404637, AF098590, AF036362, AF156421, AF250477, and AJ404634.
TABLE 3.
Nucleotide homology between the gene segments of H6 viruses and human virus HK/156/97 (H5N1)
| Virus | % Homology to gene segments of HK/156/97a
|
||||||
|---|---|---|---|---|---|---|---|
| NS | M | NAb | NP | PA | PB1 | PB2 | |
| Tl/HK/W312/97 | 98.8 | 99.1 | 97.0 | 98.4 | 98.1 | 98.1 | 98.6 |
| Ph/HK/SH39/99 | 98.4 | 98.6 | 96.6 | 97.9 | 98.3 | 97.9 | 98.2 |
| Qa/HK/1721-20/99 | 98.5 | 98.9 | 96.7 | 97.6 | 97.8 | 98.1 | 98.1 |
| Qa/HK/1721-30/99 | 99.0 | 98.9 | 96.6 | 97.3 | 97.8 | 98.2 | 98.1 |
| Qa/Hk/SF550/00 | 98.2 | 98.4 | 96.0 | 97.6 | 97.2 | 98.0 | 97.5 |
| Qa/HK/SF595/00 | 98.8 | 98.4 | 96.4 | 97.4 | 97.2 | 98.0 | 98.0 |
| Ph/HK/NT261/00 | 98.4 | 98.4 | 96.1 | 97.8 | 97.5 | 98.2 | 97.5 |
| Cu/HK/FY295/00 | 98.2 | 98.1 | 96.1 | 97.5 | 97.1 | 97.9 | 97.4 |
| Dk/HK/3461/99 | 92.4 | 92.3 | 90.0 | 93.2 | 91.5 | 90.3 | 86.3 |
| Dk/HK/3096/99 | 92.4 | 91.3 | —c | 93.1 | NTd | 91.4 | 90.0 |
| Dk/HK/175/77 | 93.0 | NT | 90.2 | NT | NT | NT | 88.7 |
Calculation based on the nucleotide sequences of PB2 (nucleotides [nt] 988 to 2289), PB1 (nt 67 to 1428), PA (nt 25 to 1677), NP (nt 46 to 1398), NA (nt 66 to 1373), M (nt 50 to 784) and NS (nt 39 to 719) genes.
Dk/HK/3461/99 and Dk/HK/175/77 do not have the characteristic deletion of 19 amino acids in the stalk region.
—, different subtype.
NT, not tested.
Evolutionary analysis.
It has been proposed that, when an influenza virus infects a new host, it undergoes a period of rapid evolution with an increased accumulation of amino acid substitutions (7). The rate of nonsynonymous substitutions in each gene segment of contemporary terrestrial H6N1 influenza viruses was calculated to determine the rate of evolution of the viruses (Table 4). H6N1 influenza viruses isolated in 2000 (group II) had a lower rate of nonsynonymous substitutions than did those isolated in 1999 (group I). This decrease in the rate of evolution indicates that H6N1 influenza viruses are becoming more adapted to terrestrial hosts. With the exceptions of the HA and NA genes of H6N1 influenza viruses isolated in 1999 (group I) and the HA gene of H9N2 influenza viruses, all genes were evolving slower than those of the H5N1 influenza viruses. The PB1, PA, and NS1 genes of H6N1 influenza viruses isolated in 1999 (group I) had higher rate of nonsynonymous substitutions than did those of the H9N2 influenza viruses; however, the NP and M genes of the H6N1 influenza viruses had lower rate of nonsynonymous substitutions than did those of the H9N2 influenza viruses. Despite the observed differences in the rates of nonsynonymous substitutions between these viruses, the P values indicated that five of the eight gene segments in the two compared groups were not significantly different. This finding suggests that these viruses coexisted and were introduced into their respective hosts at about the same time.
TABLE 4.
Rate of nonsynonymous substitutions of H6N1, H9N2, and H5N1 influenza viruses that circulate in Hong Kong
| Gene | Nucleotide range analyzede | Substitution rate (10−3/site/yr) ± SE for:
|
Pf for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Reference virus Tl/HK/W312/97 (H6N1) versus
|
Reference virus Qa/HK/G1/97 (H9N2) versus H9N2 viruses of 1999c | Reference virus Ck/HK/258/97 (H5N1) versus H5N1 viruses of 1997d | H6(I):H9 | H6(I):H5 | H6(II):H9 | H6(II):H5 | H5:H9 | |||
| H6N1 viruses of 1999 (I)a | H6N1 viruses of 2000 (II)b | |||||||||
| PB2 | 988–2289 | 1.8 ± 1.5 | 0.8 ± 0.7 | 1.8 ± 0.3 | 4.3 ± 1.5 | NS | <0.05 | <0.05 | <0.05 | <0.05 |
| PB1 | 67–1428 | 1.8 ± 0.3 | 1.0 ± 0.5 | 0.8 ± 0.4 | 4.8 ± 1.7 | <0.05 | <0.05 | NS | <0.05 | <0.05 |
| PA | 25–1677 | 1.4 ± 0.4 | 1.1 ± 0.2 | 1.3 ± 0.2 | 4.6 ± 1.7 | NS | <0.05 | NS | <0.05 | <0.05 |
| HA | 4.4 ± 1.4 | 2.9 ± 0.7 | 4.6 ± 0.9 | 4.2 ± 0.7 | NS | NS | <0.05 | <0.05 | NS | |
| NP | 46–1398 | 1.1 ± 0.9 | 0.4 ± 0.6 | 1.6 ± 0.5 | 2.7 ± 1.2 | NS | <0.05 | <0.05 | <0.05 | <0.05 |
| NA | 6.0 ± 1.3 | 5.2 ± 0.4 | 2.0 ± 0.2 | 4.6 ± 0.6 | NS | NS | <0.05 | NS | <0.05 | |
| M1 | 50–784 | 0.0 | 0.0 | 0.7 ± 0.7 | 3.0 ± 1.9 | <0.05 | <0.05 | <0.05 | <0.05 | <0.05 |
| NS1 | 39–719 | 7.0 ± 5.2 | 3.6 ± 3.3 | 0.2 ± 0.5 | 8.8 ± 2.6 | <0.05 | NS | <0.05 | <0.05 | <0.05 |
Reference virus compared to group I H6N1 viruses Ph/HK/SH39/99, Qa/HK/172120/99, and Qa/HK/172130/99.
Reference virus compared to group II H6N1 viruses Ph/HK/NT261/00, Qa/HK/SF550/00, Qa/HK/SF595/00, and Cu/HK/FY295/00.
Reference virus compared to H9N2 viruses Qa/HK/A17/99, Pg/HK/FY6/99, Ck/HK/NT16/99, Qa/HK/SSP10/99, and Ph/HK/SSP11/99.
Reference virus compared to H5N1 viruses Dk/HK/Y283/97, Dk/HK/P46/97, Gs/HK/W355/97, Ck/HK/786/97, and Ck/HK/915/97.
HA gene nucleotide ranges analyzed H5, 18 to 1046; H6, 29 to 1063; H9, 87 to 1076; NA gene nucleotide ranges analyzed: N1, 66 to 1373; N2, 20 to 1415.
Calculated for comparing the rates of nonsynonymous substitutions between different groups. H6(I):H9, H6N1 viruses of 1999 compared to H9N2 viruses; H6(I):H5, H6N1 viruses of 1999 compared to H5N1 viruses; H6(II):H9, H6N1 viruses of 2000 compared to H9N2 viruses; H6(II):H5, H6N1 viruses of 2000 compared to H5N1 viruses; H5:H9, H5N1 viruses compared to H9N2 viruses. If P was <0.05, there was a significant difference between the compared groups. If P was >0.05, there was no significant difference (NS) between the compared groups.
DISCUSSION
Two antigenically and genetically distinct groups of H6 influenza viruses circulate in domestic poultry in southeastern China: the aquatic poultry influenza viruses and the terrestrial poultry influenza viruses. Sequence analysis of the H6 HA gene revealed the insertion of an aspartic acid residue between positions 144 and 145 (H3 numbering) of the H6 HA in contemporary terrestrial isolates. The corresponding change in the gene sequence can be used as a genetic marker to distinguish terrestrial isolates from aquatic ones. The aspartic acid corresponded to proposed antigenic site A of the H3 HA molecule (29, 30). This antigenic site is on a protruding loop of amino acids 140 to 146, which projects from the H3 HA molecule surface. An insertion of an amino acid within this loop alters the loop shape and may change the antigenicity of the HA molecule. However, the Tl/HK/W312/97 (H6N1) virus does not possess this additional aspartic acid but showed an antigenicity similar to those of other contemporary terrestrial isolates in the HI tests. This suggests that antigenic site A on the H6 HA molecule does not solely determine the antigenicity of the H6 HA molecule.
Previous studies in the 1970s on the association of HA and NA genes in duck influenza viruses generally showed a nonrandomness of association (5). The H6 subtype was an exception in that, as it occurred in combination with all NA subtypes, N1 to N9, apart from N7, its association with N1 was nonrandom, occurring over fourfold more frequently than it might be expected to do. This pattern of H6 and N1 association is not apparent in contemporary aquatic influenza viruses but is found in contemporary terrestrial isolates (Table 1), a pattern that continues in ongoing surveillance studies (Y. Guam, unpublished data). No reason for this H6N1 association can be advanced, but the question of whether the insertion of aspartic acid between positions 144 and 145 (H3 numbering) may be a contributing factor to host range specificity might be raised. The deletion of 19 amino acids in the stalk region of the N1 of terrestrial H6N1 isolates is presumably a recent event associated with pathogenicity for chicken H5N1 influenza viruses (1).
The host range of influenza A viruses is associated with differences in the specificity of the HA to attach to sialic acid (Sa)-containing receptors on susceptible cells (15). HAs of avian influenza viruses preferentially recognize Sa linked to galactose (Gal) by α2,3 linkages (Saα2,3Gal), whereas HAs of human influenza viruses preferentially recognize Saα2,6Gal linkages (11). Residues 226 and 228 in the HAs of the H2 and H3 subtypes of human influenza viruses may be important in determining receptor specificity (2, 27). In all H6 influenza viruses, except Gs/HK/W222/97 (H6N7) and Gs/HK/W375/97 (H6N2), the HA possesses a glutamine residue at position 226 and a glycine residue at position 228. This combination of amino acids binds preferentially to Saα2,3Gal and is essential for virus replication in the duck intestine (2, 27). The Q226L substitution in H6 HA of Gs/HK/W222/97 and Q226V in Gs/HK/W375/97 apparently did not impair the ability of these two viruses to replicate in the duck intestine, as both were isolated from the cloaca. However, if there is an additional substitution of G228S in Gs/HK/W222/97, it may enable this virus to recognize the mammalian receptor and significantly increase the potential of the virus to infect mammals (2).
Phylogenetic analysis of the HA genes indicated that the H6 influenza viruses isolated from domestic poultry in southeastern China can be classified into three distinct branches: the 1970s branch, the contemporary aquatic branch, and the contemporary terrestrial branch. The sequences of the H6 HA genes of viruses from terrestrial hosts originated from the same node of the H6 HA phylogenetic tree of the contemporary terrestrial branch and have an out-group relationship with aquatic isolate Dk/HK/3461/99 (H6N1). This finding suggests that the contemporary terrestrial H6N1 influenza viruses may have evolved from an aquatic bird virus. However, the HA gene of the H6N1 influenza viruses in the terrestrial branch is only about 91% homologous to that of Dk/HK/3461/99, indicating that this particular duck influenza virus did not directly contribute the HA gene to the contemporary terrestrial isolates. Further surveillance of influenza viruses in aquatic poultry may identify the precursor viruses of the contemporary terrestrial isolates.
The topology of the contemporary terrestrial branch of the H6 HA phylogenetic tree showed that the terrestrial isolates evolved in a sequential fashion and that contemporary terrestrial isolates evolved more rapidly than the Dk/HK/3461/99 (H6N1) virus. This trend in evolution is not seen in other branches of the H6 HA phylogenetic tree, e.g., the contemporary aquatic branch. Thus, it appears that the precursor virus of the contemporary terrestrial isolates was subjected to significant selective pressure once it infected terrestrial poultry and circulated in the wholesale and retail live-poultry markets.
The HA molecule is the prime determinant of host range of influenza viruses, but PB2, NP, and M2 are also thought to be involved. All contemporary terrestrial H6N1 viruses possessed avian virus-like amino acid E627, human virus-like amino acids T661 and I667 in the PB2 protein, and human virus-like amino acid M136 in the NP protein. These human virus-like amino acids are also found in the chicken H5N1 viruses of 1997 (33). The other human virus-like amino acids shared by the contemporary terrestrial H6N1 viruses and the chicken H5N1 viruses can be found in the M2 protein and are G16, V28, and F55. It is not known how these amino acids determine the host range of influenza viruses, but the presence of human virus-like amino acids in avian H6N1 viruses may enable the viruses to cross the host barrier and infect mammals.
The common features of the human H5N1 and the Qa/HK/G1/97 (H9N2) viruses are the six genes encoding the internal components (6). Whether this commonality is an important factor in the ability of these viruses to infect humans is not known, but influenza A viruses possessing this feature are of particular concern. The H6N1 influenza viruses from terrestrial poultry possess not only similar genes encoding internal proteins but also the N1 gene of HK/156/97 (H5N1) virus. The N1s of the H6N1 terrestrial isolates have the characteristic deletion of 19 amino acids in the stalk region. Thus, the terrestrial H6N1 influenza viruses may have the potential to infect humans. A 7-month surveillance study of the live-poultry retail markets conducted in 1999 resulted in the isolation of 16 H9N2 influenza viruses from quail, 3 from pheasants, and 2 from chukas (7). During the same surveillance period, only one H6N1 influenza virus was isolated, and that was from a pheasant. During the first 4 months of 2000, four H6N1 influenza viruses were isolated from quail, pheasants, and chukas. There appears to be an increasing prevalence of H6N1 influenza viruses from 1999 to the present (Guam, unpublished data), suggesting that they may be adapting to terrestrial poultry and establishing themselves in the live-poultry retail markets and perhaps in the wider region. The high prevalence of H9N2 influenza viruses in quail in the live-poultry retail markets in 1999 may be one of the factors that contributed to the human infection in that year (14). Thus, if the prevalence of H6N1 influenza viruses in the live-poultry retail markets reaches a level comparable to that of H9N2 influenza viruses, the chances of H6N1 influenza viruses infecting humans could increase.
Post-1997 surveillance reveals that Tl/HK/W312/97 (H6N1)-like viruses are not present in aquatic poultry; instead, these viruses are found in terrestrial poultry, suggesting that aquatic poultry are not a major host of them. The exception to this is the lone Dk/HK/3461/99 (H6N1) virus, with which the terrestrial H6N1 viruses have an out-group relationship. In this sense, the Tl/HK/W312/97 (H6N1) virus may be an anomaly since wild birds such as the teal, after capture, may be held on a farm prior to sale and may have acquired the virus from other poultry in the farm. Thus there is a possibility that the Tl/HK/W312/97 (H6N1) virus could have originated in part from terrestrial poultry and, as an isolate from an aquatic bird, is not the ideal representative for the terrestrial H6N1 group of viruses. The present study and ongoing surveillance indicate that terrestrial poultry, particularly the quail, is the major host for the Tl/HK/W312/97 (H6N1)-like viruses (Guam, unpublished data).
The presence of H6N1 influenza viruses bearing seven genes that are highly homologous to those of the pathogenic H5N1 viruses of 1997 highlights the potential of H6N1 viruses to reassort and generate pathogenic viruses. This is particularly important because it was hypothesized that the H6N1 virus may have been involved, along with the H9N2 viruses, in the genesis of the H5N1 viruses (8). It was believed that the H5N1 influenza viruses might be reassortants of three precursor influenza viruses. The HA gene could have originated from a Gs/Guangdong/1/96 (H5N1)-like virus whose HA gene is 99% homologous to the Hong Kong H5N1 influenza viruses in 1997 (31). The six genes encoding the internal proteins could have originated from a Tl/HK/W312/97 (H6N1)-like (8) or a Qa/HK/G1/97 (H9N2)-like virus (6), and the Tl/HK/W312/97-like virus could have provided the N1 gene. The hypothesis that H6N1 influenza viruses isolated from terrestrial poultry were involved in the genesis of the human H5N1 influenza viruses was investigated by comparing the rates of evolution of Tl/HK/W312/97 (H6N1)-like viruses with those of Qa/HK/G1/97 (H9N2)-like and Ck/HK/258/97 (H5N1)-like viruses. The genes of the Ck/HK/258/97 (H5N1)-like viruses appeared to be evolving more rapidly than those of the Qa/HK/G1/97 (H9N2)-like viruses, and this is consistent with the results of previous reports (7, 33). The rates of evolution of Tl/HK/W312/97 (H6N1)-like viruses characterized in this study and the Qa/HK/G1/97 (H9N2)-like viruses are not significantly different, suggesting that these viruses are coevolving. It was also suggested that the Qa/HK/G1/97 (H9N2)-like viruses were introduced into their terrestrial hosts earlier than the H5N1 viruses (7). The Tl/HK/W312/97 (H6N1)-like viruses could have also been introduced into their hosts before the H5N1 viruses. Together, these observations suggest that Tl/HK/W312/97 (H6N1)-like viruses and Qa/HK/G1/97 (H9N2)-like viruses were present in poultry before the H5N1 outbreak and that these viruses and the Gs/Guangdong/1/96 (H5N1)-like viruses were involved in the genesis of the pathogenic H5N1 influenza virus. On the other hand, the presence of all three precursors in the region means that there is the opportunity for the pathogenic H5N1 influenza virus of 1997 to reappear (24).
The results of the present study show that antigenically distinct H6 influenza viruses are present in poultry in southeastern China. H6N1 influenza viruses which possess seven genes that are closely related to those of the human H5N1 influenza viruses are still circulating and exhibit increasing prevalence in the live-poultry retail markets (Guam, unpublished data). These findings highlight the need for further epidemiological and genetic studies of H6 influenza viruses in southeastern China, as has been the case with H9N2 influenza viruses (6, 7). Further surveillance studies will provide a better guide to the epidemiological patterns of these H6N1 influenza viruses in the hypothetical influenza epicenter of southern China (20) and hopefully will advance the understanding of the genetic parameters involved and contribute to baseline directions in influenza pandemic preparedness.
Acknowledgments
This work was supported by the Wellcome Trust grant 057476/Z/99/Z, Public Health Service research grants AI95357 and AI29680 from the National Institute of Allergy and Infectious Diseases, The University of Hong Kong Committee on Research and Conference Grants, Cancer Center Support CORE grant CA-21765, and the American Lebanese Syrian Associated Charities.
We gratefully acknowledge the continued support of the officers of the Agriculture, Fisheries and Conservation Department and the Food and Environmental Hygiene Department of the Hong Kong SAR Government. We also thank A. Hay for providing the postinfection ferret antiserum and K. Dyrting for supplying viruses. We thank P. Ghose for technical support.
References
- 1.Claas, E. C., A. D. Osterhaus, R. van Beek, J. C. de Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477. [DOI] [PubMed] [Google Scholar]
- 2.Connor, R. J., Y. Kawaoka, R. G. Webster, and J. C. Paulson. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23. [DOI] [PubMed] [Google Scholar]
- 3.Downie, J. C., and W. G. Laver. 1973. Isolation of a type A influenza virus from an Australian pelagic bird. Virology 51:259–269. [DOI] [PubMed] [Google Scholar]
- 4.Downie, J. C., R. G. Webster, G. C. Schild, W. R. Dowdle, and W. G. Laver. 1973. Characterization and ecology of a type A influenza virus isolated from a shearwater. Bull. W. H. O. 49:559–566. [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner, I. D., and K. F. Shortridge. 1979. Recombination as a mechanism in the evolution of influenza viruses: a two-year study of ducks in Hong Kong. Rev. Infect. Dis. 1:885–890. [DOI] [PubMed] [Google Scholar]
- 6.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 96:9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann, E., J. Stech, I. Leneva, S. Krauss, C. Scholtissek, P. S. Chin, M. Peiris, K. F. Shortridge, and R. G. Webster. 2000. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J. Virol. 74:6309–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, W. H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36:96–99. [DOI] [PubMed] [Google Scholar]
- 10.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matrosovich, M. N., A. S. Gambaryan, S. Teneberg, V. E. Piskarev, S. S. Yamnikova, D. K. Lvov, J. S. Robertson, and K. A. Karlsson. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224–234. [DOI] [PubMed] [Google Scholar]
- 12.Nobusawa, E., T. Aoyama, H. Kato, Y. Suzuki, Y. Tateno, and K. Nakajima. 1991. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182:475–485. [DOI] [PubMed] [Google Scholar]
- 13.Pamilo, P., and N. O. Bianchi. 1993. Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol. Biol. Evol. 10:271–281. [DOI] [PubMed] [Google Scholar]
- 14.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. S. Ip, R. W. M. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916–917. [DOI] [PubMed] [Google Scholar]
- 15.Rogers, G. N., J. C. Paulson, R. S. Daniels, J. J. Skehel, I. A. Wilson, and D. C. Wiley. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76–78. [DOI] [PubMed] [Google Scholar]
- 16.Scholtissek, C., H. Bürger, O. Kistner, and K. F. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287–294. [DOI] [PubMed] [Google Scholar]
- 17.Sharp, G. B., Y. Kawaoka, S. M. Wright, B. Turner, V. Hinshaw, and R. G. Webster. 1993. Wild ducks are the reservoir for only a limited number of influenza A subtypes. Epidemiol. Infect. 110:161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shortridge, K. F., W. K. Butterfield, R. G. Webster, and C. H. Campbell. 1977. Isolation and characterization of influenza A viruses from avian species in Hong Kong. Bull. W. H. O. 55:15–19. [PMC free article] [PubMed] [Google Scholar]
- 19.Shortridge, K. F. 1982. Avian influenza A viruses of southern China and Hong Kong: ecological aspects and implications for man. Bull. W. H. O. 60:129–135. [PMC free article] [PubMed] [Google Scholar]
- 20.Shortridge, K. F., and C. H. Stuart-Harris. 1982. An influenza epicentre? Lancet ii:812–813. [DOI] [PubMed] [Google Scholar]
- 21.Shortridge, K. F. 1992. Pandemic influenza: a zoonosis? Semin. Respir. Infect. 7:11–25. [PubMed] [Google Scholar]
- 22.Shortridge, K. F., N. N. Zhou, Y. Guan, P. Gao, T. Ito, Y. Kawaoka, S. Kodihalli, S. Krauss, D. Markwell, K. G. Murti, M. Norwood, D. Senne, L. Sims, A. Takada, and R. G. Webster. 1998. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology 252:331–342. [DOI] [PubMed] [Google Scholar]
- 23.Shortridge, K. F. 1999. Poultry and the influenza H5N1 outbreak in Hong Kong, 1997: abridged chronology and virus isolation. Vaccine 17:S26–S29. [DOI] [PubMed] [Google Scholar]
- 24.Shortridge, K. F., M. Peiris, Y. Guan, K. Dyrting, T. Ellis, and L. Sims. H5N1 virus: beaten but is it vanquished? In B. Dodet (ed.), Emergence and control of zoonotic ortho- and paramyxovirus diseases, in press. John Libbey Eurotext, Paris, France.
- 25.Stieneke-Grober, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396. [DOI] [PubMed] [Google Scholar]
- 27.Vines, A., K. Wells, M. Matrosovich, M. R. Castrucci, T. Ito, and Y. Kawaoka. 1998. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J. Virol. 72:7626–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webster, R. G., and R. Rott. 1987. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell 50:665–666. [DOI] [PubMed] [Google Scholar]
- 29.Wiley, D. C., I. A. Wilson, and J. J. Skehel. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366–373. [DOI] [PubMed] [Google Scholar]
- 31.Xu, X., K. Subbarao, N. J. Cox, and Y. Guo. 1999. Genetic characterization of the pathogenic influenza A/goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261:15–19. [DOI] [PubMed] [Google Scholar]
- 32.Yuen, K. Y., P. K. Chan, M. Peiris, D. N. Tsang, T. L. Que, K. F. Shortridge, P. T. Cheung, W. K. To, E. T. Ho, R. Sung, and A. F. Cheng. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, N. N., K. F. Shortridge, E. C. J. Claas, S. L. Krauss, and R. G. Webster. 1999. Rapid evolution of H5N1 influenza viruses in chickens in Hong Kong. J. Virol. 73:3366–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]