Abstract
At the CC (β) chemokine receptor 2 (CCR2) and CCR5 loci, combinations of common single-nucleotide polymorphisms (SNPs) and a 32-bp deletion (Δ32) form nine stable haplotypes (designated A through G*2). The distribution of these CCR2-CCR5 haplotypes was examined among 703 participants in the Multicenter AIDS Cohort Study (MACS), the District of Columbia Gay (DCG) Study, and the San Francisco Men’s Health Study (SFMHS). Highly exposed and persistently seronegative (HEPS; n = 90) Caucasian men from MACS more frequently carried heterozygous G*2 (Δ32) genotypes (especially A/G*2) and less frequently carried the homozygous E/E genotype compared with 469 Caucasian seroconverters (SCs) from the same cohort (P = 0.004 to 0.042). Among 341 MACS Caucasian SCs with 6- to 12-month human immunodeficiency virus type 1 (HIV-1) seroconversion intervals and no potent antiretroviral therapy, mean plasma HIV-1 RNA level during the initial 42 months after seroconversion was higher in carriers of the E/E genotype and lower in those with the 64I-bearing haplotype F*2 or the Δ32-bearing haplotype G*2 (and especially genotypes A/G*2 and F*2/G*2). A multivariable model containing these CCR markers showed significant composite effects on HIV-1 RNA at each of four postconversion intervals (P = 0.0004 to 0.050). In other models using time to AIDS as the endpoint, the same markers showed more modest contributions (P = 0.08 to 0.24) to differential outcome during 11.5 years of follow-up. Broadly consistent findings in the larger MACS Caucasian SCs and the smaller groups of MACS African-American SCs and the DCG and SFMHS Caucasian SCs indicate that specific CCR2-CCR5 haplotypes or genotypes mediate initial acquisition of HIV-1 infection, early host-virus equilibration, and subsequent pathogenesis.
Polymorphisms in the human CC (β) chemokine receptor system are well known to modulate the natural history of human immunodeficiency virus type 1 (HIV-1) infection (reviewed in references 29, 49, and 53). Individual variants in the coding or promoter and regulatory regions of the chemokine receptors 5 (CCR5) and 2 (CCR2) have often been associated with various events in the pathogenesis of HIV-1/AIDS (2, 4, 12, 14, 27, 36, 38, 44, 46, 59, 68). There is strong evidence that homozygosity for the 32-bp deletion (Δ32) in the coding region of CCR5 nearly completely protects Caucasians from HIV-1 infection and that heterozygosity for Δ32 modestly retards disease progression (4, 12, 26, 27, 77). Overall, findings on CCR variants other than Δ32 in relation to HIV-1 transmission and pathogenesis have been difficult to interpret in the absence of clear confirmatory data and functional correlates.
The full extent of common CCR5 single-nucleotide polymorphisms (SNPs) and relationships among them had not been examined until recently (20, 52, 70). Thorough documentation of the extended CCR2 and CCR5 haplotypes has suggested that stable CCR haplotypes, alone or paired as genotypes, may influence the course of HIV-1 infection differentially according to their racial distribution (20, 21, 52); if so, variations in genotype frequencies among populations could have a sizeable differential impact on the course and the burden of disease (21). Conflicting observations on CCR2-64I (3, 14, 15, 20, 24, 27, 35, 51, 64, 68, 74); on SDF1 (27, 61, 73, 76), and on CX3CR1 effects (16, 45) may attest to similar population-specific effects of chemokine receptor and ligand genes other than CCR5.
The epidemiological importance of CCR polymorphisms has been inferred largely from population studies of late clinical events (AIDS or death). CCR receptor biology concurs broadly with the epidemiologic findings, but efforts to unravel the intricate receptor-virus relationships have not definitively shown how or when the polymorphisms alter in vivo HIV-1 pathogenesis. Differential regulation of CCR receptors could trigger the emergence of X4 viruses that predominantly use CXCR4 or others that use both CCR5 and CXCR4 (11). Thus, over time, earlier unequivocal effects may dwindle and previously less prominent factors may emerge as the virus alters its cell tropism and employs other mechanisms for adaptation to host immunity.
The plasma HIV-1 RNA concentration measured in the months after untreated infection accurately reflects the early host-virus equilibrium (10, 14, 32, 38, 47, 48), which in turn is highly predictive of the rate of later disease progression (5, 9, 28, 47, 57, 66) as well as HIV-1 transmissibility from infected to uninfected individuals regardless of the transmission mode (17, 18, 56, 58, 60, 65). Clinical and virological studies of the impact of chemokine receptor-ligand variants on viral dynamics have already suggested a role for Δ32 and 64I in determining this equilibrium, apparently even in the context of antiretroviral therapy (4, 22, 31, 53). Our own observations, based on comprehensive analyses of the relative effects of host genetic variations, now imply that several CCR haplotypes or genotypes can independently influence HIV-1 seroconversion, early HIV-1 RNA levels, and variability in the course of disease.
MATERIALS AND METHODS
Subject selection for analysis of acquisition of HIV-1 infection.
From approximately 3000 Multicenter AIDS Cohort Study (MACS) (30) participants who were seronegative at enrollment, the 100 most highly exposed and persistently HIV-1-seronegative (HEPS) men with quantifiable risk had been selected for a previous study (77). All HEPS men reported unprotected homosexual contact with the greatest number (up to 500+) of partners between 1984 and 1986. We excluded the seven HEPS men already shown to be protected by CCR5-Δ32 homozygosity and three additional men who were not Caucasian or who had no specimen available. The remaining 90 men in the Caucasian HEPS group were compared with two groups of Caucasian seroconverters (SCs): all 469 SCs with available specimens (1 to 500+ partners, median = 18) and a subset of 270 SCs reporting higher numbers of partners (50 to 500+, median =113) between 1984 and 1986.
Subject selection for analysis of virological and clinical outcomes of infection.
From 513 (469 Caucasians and 44 African-Americans) MACS HIV-1 SCs, 341 Caucasians, and 31 African-Americans were selected for viral load analyses according to the following criteria: (i) a seroconversion interval of ≤12 months; (ii) ≥1 measurement of serum or plasma RNA concentration available within the first 42 months after seroconversion (39, 47), (iii) a DNA sample available for genotyping, and (iv) adequate follow-up visits until onset of AIDS or death or 1 January 1996, when protease inhibitor-containing antiretroviral regimens began to gain wide application. AIDS by 1987 Centers for Disease Control (CDC) criteria (8) and death were used in separate analyses, but because findings with the two endpoints were quite similar, only selected analyses based on death are presented. SCs in the District of Columbia Gay (DCG) (n = 95) cohort (19) and the San Francisco Men’s Health Study (SFMHS) (n = 49) (75) contributed, respectively, 39 and 37 SCs who also met inclusion criteria. The MACS, DCG, and SFMHS SCs selected for genetic association analyses were highly comparable (Table 1).
TABLE 1.
General characteristics of HIV-1 SCs selected from the MACS, DCG cohort, and SFMHS
| HIV-1 SCs (n)a | Mean age (yr) | Median sero- conversion intervalb | No. of AIDS cases/ median AIDS- free time (yr) | No. of deaths/median survival | Mean HIV-1 RNA level (log10 copies/ml) in plasmac
|
||||
|---|---|---|---|---|---|---|---|---|---|
| <6 mo | 6–18 mo | 19–30 mo | 31–42 mo | 6–42 mo | |||||
| MACS | |||||||||
| Caucasians (341) | 34.4 | 6 | 163/8.4 | 129/10.1 | 4.3 (275) | 4.3 (281) | 4.3 (258) | 4.3 (219) | 4.3 (341) |
| African Americans (31) | 31.7 | 6 | 8/10.9 | 7/11.1 | 4.4 (29) | 4.4 (28) | 4.3 (22) | 4.3 (16) | 4.4 (31) |
| DCG (39) | 35.2 | 12 | 19/8.1 | 22/9.1 | 4 (7) | 4.3 (33) | 4.4 (23) | 4.1 (19) | 4.2 (39) |
| SFMHS (37) | 36.9 | 7 | 16/9.3 | 3.5 (23)* | 3.9 (24)*** | 3.6 (20)** | 3.9 (20) | 3.8 (37)**** | |
Total number of eligible SCs as defined by seroconversion interval and number of HIV-1 RNA measurements at any given interval (see text).
Seroconversion interval = difference (months) between date of last seronegative visit and date of first seropositive visit.
Range of time intervals from date of estimated seroconversion to median date of assay for HIV-1 RNA levels. Total numbers of measurements (subjects) are shown in parentheses. HIV RNA levels in SFMHS SCs differed from those in MACS SCs at these intervals according to pooled t tests: *, P < 0.05; **, < 0.01; ***, P < 0.001; ****, P < 0.0001.
CCR2 and CCR5 genotyping.
Our typing scheme differentiated the dimorphic variants at eight sites: the SNP encoding V64I in CCR2, six SNPs (A29G [=G58755A], G303A, T627C, C630T, A676G, and C927T) in or adjacent to the cis-regulatory or promoter region of CCR5, and the 32-bp deletion (Δ32) in CCR5. Haplotyping based on PCR with combinations of sequence-specific primers (SSP) (70) was supplemented with four additional SSP reactions in conjunction with T627C-specific primers to define the A29G variants. Typing for the rare SNP G208T among the 630C-676A subset was performed initially to differentiate A and B haplotypes but omitted later because the rare haplotype B had negligible potential impact on the results (20; J. Tang, unpublished observations). Combinations of variants at the typed sites form the eight haplotypes according to the nomenclature of the Tri-Service HIV-1 Natural History Study (TSS) (20). For comparability to the TSS, we applied its nomenclature, but omitted the HH prefix (e.g., HHA is referred to as A and HHG*2 as G*2) as adopted more recently (41).
Virological measurements.
For MACS participants, at least one measurement of plasma HIV-1 RNA concentration was available during each of four postseroconversion intervals: ≤6 months (median = 4.1), 6 to 18 months (median = 12.5), 19 to 30 months (median = 23.7), and 31 to 42 months (median = 36.2), as well as during the entire 6 to 42 months (median = 18.1). Plasma viral RNA concentrations were quantified by reverse transcription-PCR (RT-PCR) with the standard Amplicor assay (Roche Diagnostics, Nutley, N.J.) with a detection limit of 400 RNA copies per ml of plasma. Nine (∼3%) of the MACS SCs had undetectable viral RNA, for a total of 34 measurements; an arbitrary value of 300 was assigned for these visits. In the DCG and the SFMHS cohorts, plasma HIV-1 RNA level was measured by the RT-PCR Amplicor Monitor assays (sensitivity = 50 RNA copies/ml) (Roche Diagnostics). For undetectable levels in one SC in SFMHS and three in DCG, the arbitrary value was set at 50. Overall, HIV-1 RNA levels in the SFMHS SCs were 0.4 to 0.6 log lower (P < 0.001 to 0.12) than in the MACS and DCG SCs (Table 1).
Statistical analysis.
All HIV-1 RNA measurements (values transformed to log10) and survival analyses were censored at 1 January 1996. Multiple postseroconversion measurements taken within the various intervals used for analysis were averaged, and repeated measures analysis was applied to genotype comparisons. Loss to follow-up during successive postconversion study intervals was evaluated by comparing men with and men without later measurements for their initial mean RNA levels and subsequent time to AIDS.
Statistical routines available in SAS (Statistical Analysis Software, version 8.0; SAS Institute, Cary, N.C.) were applied throughout the analyses. General linear model (GLM) and logistic regression statistics were used to assess the association of individual genotypes (for frequencies ≥2% based on direct counting) with HIV-1 RNA levels, to perform multivariate comparisons, and to model overall relationships. Age was initially treated as a covariate in all GLM procedures; but adjustment for age proved unnecessary in the absence of clear differences among carriers of major CCR variants. Curves depicting the times from seroconversion to 1987 CDC-defined AIDS and death were generated by the Kaplan-Meier product-limit method. The univariate relative hazards (RH) of AIDS for specific genotypes were estimated by Cox proportional hazards models with subjects grouped by the genotypes under study. Both Wilcoxon and log-rank tests of significance are shown. The former gives greater weight to the earlier time period, when larger amounts of data are included and CCR5 serves as the predominant HIV-1 coreceptor, while the latter provides a common approach for making inferences by using Kaplan-Meier and Cox proportional hazards methods. Analyses of genetic effects emphasized their consistency, both internal (i.e., similarity between subpopulations and various endpoints from seroconversion to later disease) and external (i.e., similarity to findings in other highly comparable studies). Correction of P values for multiple comparisons was deemed generally inappropriate and even potentially misleading in this setting: much of the effort addressed previously reported or predicted relationships, and the multifaceted analyses introduced considerable uncertainty about the various numbers and degrees of independence of comparative tests either performed or implied (see Appendix).
RESULTS
Overall distribution of CCR2-CCR5 haplotypes.
Between 97 and 100% of the CCR2-CCR5 haplotypes were resolved unambiguously in the three cohorts (Table 2). The prevalence of eight CCR haplotypes detected in the three cohorts ranged from 0% (D) to 37% (C) (Table 2). Consistent with earlier studies (20, 21, 70), G*1 was the only haplotype with similar frequencies among Caucasians (2 to 4%) and African-Americans (3%). Haplotype D was largely restricted to African-Americans, and G*2 was far less common in African-Americans than in Caucasians (P < 0.001 for both comparisons). Overall, six haplotypes (A, C, E, F*2 [=CCR2-64I], G*1, and G*2 [=Δ32]) were relatively frequent (>2% for the haplotype or about 4% for the haplotype carrier) in Caucasians, whereas African-Americans had seven frequent haplotypes (A, C, D, E, F*1, F*2, and G*1). The distribution of CCR2-CCR5 genotypes (pairs of haplotypes in each individual) in each cohort and in the analyzed subgroup within each cohort closely fit Hardy-Weinberg equilibrium, i.e., common haplotypes produced common genotypes and accounted for the vast majority of the homozygous genotypes (e.g., E/E and C/C). The predicted exception was the G*2/G*2 genotype (due to its complete absence in MACS SCs (0% observed versus 0.8% expected). Haplotype and genotype frequencies for the subset of 341 Caucasian MACS SCs who met the criteria for the detailed studies were comparable to those for all 469 Caucasian SCs (Table 2). Likewise, distributions of CCR2-CCR5 haplotypes or genotypes in MACS African-American SCs and DCG and SFMHS Caucasian SCs included in the analysis closely resembled those in the total groups from which they were selected (Table 2).
TABLE 2.
Frequencies of CCR2-CCR5 haplotypes and genotypes in HEPS and HIV-1 SCs from the MACS, the DCG Study, and the SFMHS
| No. (%) with haplotype or genotype givena
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype or genotype | MACS
|
DCG
|
SFMHS
|
||||||
| Caucasians (n = 90 HEPS + 469 SCs)
|
African-Americans (n = 44 SCs)
|
Caucasians (n = 95 SCs)
|
Caucasians (n = 49 SCs)
|
||||||
| HEPS | All SCs | Selected SCs | All | Selected | All | Selected | All | Selected | |
| Haplotype | 2n = 180 | 2n = 938 | 2n = 682 | 2n = 88 | 2n = 62 | 2n = 190 | 2n = 78 | 2n = 98 | 2n = 74 |
| A | 17 (9) | 88 (9) | 61 (9) | 22 (25) | 14 (23) | 23 (12) | 12 (15) | 11 (9) | 5 (7) |
| C | 63 (35) | 332 (34) | 239 (35) | 12 (14) | 10 (16) | 71 (37) | 25 (32) | 35 (36) | 29 (39) |
| D | 2 (1) | 2 (<1) | 0 | 15 (17) | 10 (16) | 0 | 0 | 0 | 0 |
| E | 49 (27) | 296 (32) | 215 (32) | 15 (17) | 10 (16) | 61 (32) | 28 (36) | 31 (31) | 22 (30) |
| F*1 | 0 | 2 (<1) | 2 (<1) | 3 (3) | 2 (3) | 3 (2) | 0 | 0 | 0 |
| F*2(64I) | 16 (9) | 90 (10) | 69 (10) | 14 (16) | 11 (18) | 14 (7) | 2 (3) | 7 (7) | 7 (9) |
| G*1 | 12 (7) | 33 (4) | 24 (4) | 3 (3) | 2 (3) | 3 (2) | 2 (3) | 2 (2) | 2 (3) |
| G*2 (Δ32) | 21 (12) | 86 (9) | 66 (10) | 2 (2) | 1 (2) | 15 (8) | 9 (12) | 11 (11) | 8 (11) |
| Unresolved | 0 | 9 (1) | 6 (1) | 2 (2) | 2 (3) | 0 | 1 (1) | 1 (1) | |
| Genotypeb | n = 90 | n = 469 | n = 341 | n = 44 | n = 31 | n = 95 | n = 39 | n = 49 | n = 37 |
| A/A | 0 | 7 (1) | 5 (1) | 2 (5) | 2 (6) | 2 (2) | 2 (5) | 2 (4) | 1 (3) |
| A/C | 3 (3) | 26 (6) | 14 (4) | 1 (2) | 0 | 9 (9) | 4 (10) | 2 (4) | 1 (3) |
| A/E | 4 (4) | 27 (6) | 20 (6) | 3 (7) | 0 | 7 (7) | 2 (5) | 2 (4) | 1 (3) |
| A/F*2 | 4 (4) | 8 (2) | 5 (1) | 4 (9) | 3 (10) | 0 | 0 | 0 | 0 |
| A/G*2 | 6 (7) | 12 (3) | 11 (3) | 1 (2) | 1 (3) | 3 (3) | 2 (5) | 3 (6) | 1 (3) |
| C/C | 11 (12) | 58 (12) | 44 (13) | 1 (2) | 1 (3) | 13 (14) | 4 (10) | 3 (6) | 3 (8) |
| C/E | 22 (24) | 102 (22) | 72 (21) | 1 (2) | 1 (3) | 21 (22) | 10 (26) | 17 (34) | 14 (38) |
| C/F*2 | 4 (4) | 31 (7) | 26 (8) | 4 (9) | 4 (13) | 7 (7) | 0 | 3 (6) | 1 (3) |
| C/G*1 | 5 (6) | 14 (3) | 9 (3) | 1 (2) | 1 (3) | 0 | 0 | 0 | 0 |
| C/G*2 | 7 (8) | 36 (8) | 26 (8) | 0 | 0 | 6 (6) | 3 (8) | 5 (10) | 3 (8) |
| E/E | 5 (6) | 56 (12) | 39 (11) | 3 (7) | 3 (10) | 9 (9) | 5 (13) | 4 (8) | 4 (10) |
| E/F*2 | 3 (3) | 23 (5) | 19 (6) | 2 (5) | 2 (6) | 7 (7) | 2 (5) | 2 (4) | 0 |
| E/G*1 | 3 (3) | 11 (2) | 9 (3) | 0 | 0 | 2 (2) | 1 (3) | 0 | 0 |
| E/G*2 | 7 (8) | 21 (4) | 17 (5) | 0 | 0 | 5 (5) | 3 (8) | 2 (4) | 2 (5) |
| F*2/F*2 | 1 (1) | 6 (1) | 3 (1) | 0 | 0 | 0 | 0 | 0 | 0 |
| F*2/G*2 | 1 (1) | 12 (2) | 9 (3) | 1 (2) | 0 | 0 | 0 | 1 (2) | 1 (3) |
| Others | 4 (4) | 18 (4) | 13 (4) | 20 (45)c | 13 (42)c | 4 (4) | 1 (3) | 2 (4) | 5 (14) |
Criteria for All: CCR typing and clinical outcome data available; criteria for selected, criteria for All plus seroconversion ≤12 months and having ≥1 measurement of HIV-1 RNA level at various intervals (see text and Table 1).
Genotypes shown here are found in ≥5 (1%) MACS SCs.
Mostly consisting of genotypes involving haplotype D (i.e., A/D, C/D, D/D, D/E, and D/F*2).
Acquisition of HIV-1 infection.
Comparison of 90 MACS Caucasian HEPS with 469 SCs from the same cohort revealed two genetic differences: higher G*2-carrier and lower E/E genotype frequencies in HEPS than SCs (Table 3, model 1). Among men who seroconverted after exposure to relatively fewer (0 to 15) or more (16 to 500) partners, the frequencies of E/E and G*2 were similar. Multivariable odds ratios (ORs) and 95% confidence intervals (CIs) for seroconversion were similar for both comparison groups. A logistic regression model including both G*2 and E/E distinguished HEPS from all SCs (P = 0.009). Furthermore, the effect of the A/G*2 genotype (OR = 0.39, 95% CI = 0.14 to 1.08, model 2) accounted for much of the effect attributable to the G*2 carriers (OR = 0.54, 95% CI = 0.32 to 0.89, model 1). The frequencies of the A/G*2 (6.6%) and E/E (5.5%) genotypes in MACS HEPS also differed more from those observed in the DCG (3.2% for A/G*2 and 9.5% for E/E) than in the SFMHS SCs (6.1% for A/G*2 and 8.2% for E/E).
TABLE 3.
Association of CCR2-CCR5 genotypes with acquisition of HIV-1 infection in MACS based on the comparison of heavily exposed and persistently seronegative HEPS Caucasians with all or with heavily exposed Caucasian SCs
| CCR2-CCR5 genotype | No. (%) of HEPS (n = 90)a | No. (%) of SCsa
|
OR for analysis with all SCsa
|
OR for heavily exposed SCsa
|
|||
|---|---|---|---|---|---|---|---|
| All (n = 469) | Heavily exposed (n = 270) | Model 1b (PLR = 0.010) | Model 2b (PLR = 0.033) | Model 1b (PLR = 0.004) | Model 2b (PLR = 0.042) | ||
| G*2 (Δ32)/any | 21 (23.3) | 81 (17.2) | 50 (18.5) | 0.54 | 0.45 | ||
| A/G*2 (Δ32) | 6 (6.6) | 12 (2.5) | 3 (1.1) | 0.39 | 0.17 | ||
| Others | 64 (71.1) | 32 (68.8) | 188 (69.6) | Reference | Reference | Reference | Reference |
| 4 | |||||||
| E/E | 5 (5.5) | 56 (11.9) | 32 (11.9) | 1.95 | 2.21 | 1.87 | 2.15 |
As defined in the text, HEPS had up to 500+ homosexual partners between 1984 and 1986, while heavily exposed SCs had 50 to 500+ (median = 113) partners during the same period. OR and P values are based on LR by maximum-likelihood estimate.
Model 1 includes G*2/any, E/E, and others (reference); A/G*2 replaces G*2 in model 2.
Virological outcome in Caucasians.
In the 341 MACS participants who met the study criteria, the overall mean log10 RNA measurements remained rather constant (4.29 to 4.33) during the four early postseroconversion intervals examined (Table 4). The SCs (n = 72) tested at 6 months but not at 42 months had a 0.02-log lower mean RNA level and nearly identical AIDS-free times compared with participants tested at both intervals.
TABLE 4.
Mean HIV-1 RNA concentrations after seroconversion and relative hazard of AIDS by CCR2-CCR5 haplotypes and genotypes in MACS Caucasians
| Haplotype or genotype (n) | Mean plasma HIV-1 RNA concna at interval:
|
RH of AIDSc | ||||
|---|---|---|---|---|---|---|
| <6 mo (4.1)b | 6–18 mo (12.5)b | 19–30 mo (23.7)b | 31–42 mo (36.2)b | 6–42 mo (18.1)b | ||
| Overall mean (n = 341) | 4.33 | 4.29 | 4.33 | 4.30 | 4.31 | |
| Haplotype carriers | ||||||
| A (60) | 4.37 | 4.30 | 4.29 | 4.26 | 4.28 | 0.79 |
| C (239) | 4.38 | 4.29 | 4.38 | 4.29 | 4.32 | 1.07 |
| D (0) | ||||||
| E (218) | 4.28 | 4.36 | 4.40 | 4.29 | 4.35 | 1.56d |
| F*1 (2) | 4.63 | |||||
| F*2 (64I) (69) | 4.13 | 4.13 | 4.05 | 4.35 | 4.17 | 0.80 |
| G*1 (27) | 4.24 | 4.17 | 4.44 | 4.33 | 4.29 | 0.84 |
| G*2 (Δ32) (66) | 4.20 | 4.00 | 4.07 | 4.15 | 4.09 | 0.71 |
| Genotypes | ||||||
| A/A (5) | 4.78 | 4.50 | 4.38 | 4.39 | 4.41 | 0.75 |
| A/C (14) | 4.65 | 4.40 | 4.45 | 4.31 | 4.39 | 0.85 |
| A/E (20) | 4.37 | 4.41 | 4.47 | 4.23 | 4.38 | 1.11 |
| A/F*2 (5) | 3.87 | 4.29 | 4.12 | 4.72 | 4.37 | 1.86 |
| A/G*2 (11) | 3.89 | 3.70 | 3.54 | 3.82 | 3.69 | 0.26 |
| C/C (44) | 4.46 | 4.43 | 4.36 | 4.24 | 4.35 | 0.71 |
| C/E (73) | 4.30 | 4.33 | 4.50 | 4.20 | 4.35 | 1.69d |
| C/F*2 (26) | 4.31 | 4.21 | 4.11 | 4.41 | 4.23 | 0.70 |
| C/G*1 (10) | 4.38 | 4.04 | 4.51 | 4.66 | 4.31 | 0.47 |
| C/G*2 (26) | 4.40 | 4.08 | 4.36 | 4.36 | 4.26 | 1.26 |
| E/E (39) | 4.57 | 4.57 | 4.53 | 4.62 | 4.57 | 1.44 |
| E/F*2 (19) | 3.91 | 4.18 | 4.06 | 4.28 | 4.17 | 0.88 |
| E/G*1 (10) | 3.97 | 4.05 | 4.05 | 3.85 | 3.99 | 1.33 |
| E/G*2 (18) | 4.12 | 4.25 | 4.11 | 4.11 | 4.16 | 0.63 |
| F*2/F*2 (3) | 4.47 | 3.56 | 3.93 | 3.45 | 3.73 | 0.95 |
| F*2/G*2 (9) | 4.11 | 3.46 | 3.42 | 3.76 | 3.50 | 0.47 |
| G*2/non-A (55) | 4.24 | 4.06 | 4.10 | 4.17 | 4.11 | 0.86 |
| Others (8) | 4.50 | 4.47 | 4.49 | 4.59 | 4.51 | |
Expressed as mean log10 plasma HIV-1 RNA (copies per milliliter) based on ≥1 measurement available within interval.
Range (median) of interval (months) between estimated seroconversion and median date of HIV-1 RNA assay.
Based on Cox proportional hazards models.
For RH of AIDS in men carrying genotype of interest compared with all others in the cohort, P < 0.01.
In men with F*2 (64I), G*2 (Δ32), and F*2/G*2 (three markers all previously reported to have protective effects on progression to AIDS in MACS and other cohorts), mean log10 viral RNA levels were already lower (−0.20, −0.13, and −0.22, respectively) than the overall viral RNA during the first 6-month interval (Table 4). During the next three intervals, differences persisted and actually increased for F*2/G*2. SCs carrying the E/E genotype, previously associated with an accelerated course of infection in U.S. Caucasians and Argentinean children (20, 41), consistently showed higher mean log10 RNA levels than others (+0.26 to +0.42 log) not carrying F*2 or G*2 at each of the four visit intervals (Fig. 1) (P = 0.020 to 0.473). Carriage of E with other haplotypes, also previously associated with more frequent transmission and rapid progression (41), was not related to higher RNA level here (Table 4).
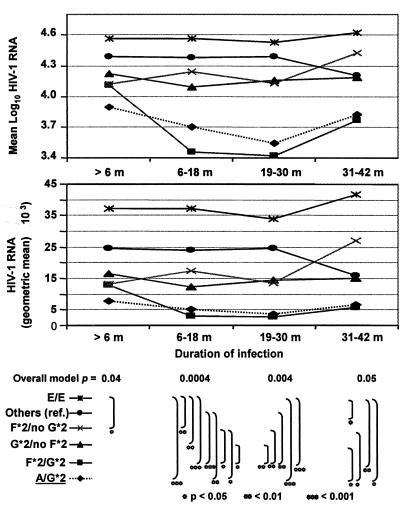
FIG. 1.
HIV-1 RNA concentrations (mean log10 and geometric mean) during initial 42 months following seroconversion. RNA levels are plotted at each of four successive intervals (>6, 6 to 18, 19 to 30, and 31 to 42 months) after seroconversion according to CCR2-CCR5 genotypes. At each point, a GLM test of the aggregate contribution of four major genotypes showed a statistically significant overall relationship (F test). Substantial differences were also seen in pairwise comparisons (t tests at three levels: *, ≤0.05; **, ≤0.01; and ***, ≤ 0.001). Mean viral RNA for carriers of A/G*2 (dotted line) is shown separately and as part of the G*2/no F*2 group.
During the earliest (<6 months) infection interval, multivariable analyses including carriers of the four haplotypes or genotypes noted above were independently responsible for much of the heterogeneity in the mean log10 HIV-1 RNA levels (P = 0.04) (Fig. 1). During that interval, mean levels in men bearing G*2 and F*2 separately and in combination were lower (−0.50 to −0.17) than those in the reference (“Others”) group, and levels in E/E homozygotes were higher (+0.18). Heterozygosity for E showed no appreciable difference. During the next (6 to 18 months) interval (Table 4 and Fig. 1), heterogeneity among the genotype groups compared (P = 0.0004) was largely attributed to nine F*2/G*2 carriers, whose mean viral RNA diverged from the level in those with E/E to a 1.11-log difference. The overall relationships were also quite consistent in the two subsequent 12-month intervals (P = 0.004 to 0.050). Within these intervals, HIV-1 RNA levels in E heterozygotes were again similar to those in the reference group without F*2 or G*2. The genotypes associated with the lowest (F*2/G*2) and the highest (E/E) mean RNA level were present in 3 and 11% of MACS subjects, respectively. During the entire 6- to 42-month period after seroconversion, the mean levels in men with F*2/G*2 remained approximately 1 log10 apart from levels in men with E/E (P = 0.06 to 0.001) (Fig. 1). The overall differences persisted when repeated measurements were taken into account for the entire period (P = 0.002).
As a subset of the G*2 carriers, men with the A/G*2 genotype had lower mean RNA levels (about −0.3 log10, P = 0.002) in all four intervals compared with any other G*2 carrier group besides F*2/G*2 (Table 4 and Fig. 1). The difference between A/G*2 and those with any other genotype (excluding F*2, G*2, or E/E) was −0.64 log10 (P < 0.0001). The stronger A/G*2 effect persisted in multivariable analysis (Table 5). On the other hand, neither the C/G*2 nor the C/E genotypes associated with disease acceleration in the Tri-Service Study (20) showed any appreciable effect on the mean viral RNA levels in the MACS participants.
TABLE 5.
Models summarizing effects of contributing CCR2-CCR5 haplotypes or genotypes on HIV-1 RNA level during the first 3.5 years after seroconversion and on subsequent disease progression in MACS Caucasians
| CCR genotype (no. of subjects) | Mean HIV-1 RNA concna at interval:
|
Survival (time to AIDS) analysis
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| <6 mo (4.1)b | Pc | 6–18 mo (12.5)b | Pc | 19–30 mo (23.7)b | Pc | 31–42 mo (36.2)b | Pc | RHe of AIDS | Pc | |
| Model 1 | ||||||||||
| F*2, no G*2 (60) | 4.12 | 0.07 | 4.24 | 0.12 | 4.13 | 0.31 | 4.43 | 0.59 | 0.86 | 0.52 |
| G*2, no F*2 (57) | 4.22 | 0.24 | 4.09 | 0.01 | 4.16 | 0.08 | 4.18 | 0.61 | 0.77 | 0.24 |
| F*2/G*2 (9) | 4.11 | 0.32 | 3.46 | 0.001 | 3.42 | 0.001 | 3.75 | 0.10 | 0.45 | 0.18 |
| E/E (39) | 4.57 | 0.27 | 4.57 | 0.15 | 4.53 | 0.47 | 4.62 | 0.02 | 1.30 | 0.27 |
| Others/reference (158) | 4.39 | 4.38 | 4.39 | 4.20 | Reference | |||||
| Model Pd | 0.04 | 0.0004 | 0.004 | 0.05 | 0.23 (PLR) | 0.10f (Pw) | ||||
| Model 2 | ||||||||||
| F*2, no G*2 (60) | 4.12 | 0.10 | 4.24 | 0.20 | 4.13 | 0.42 | 4.43 | 0.61 | 0.88 | 0.56 |
| A/G*2 (11) | 3.89 | 0.14 | 3.70 | 0.004 | 3.54 | 0.05 | 3.82 | 0.03 | 0.26 | 0.06 |
| F*2/G*2 (9) | 4.11 | 0.35 | 3.46 | 0.001 | 3.42 | 0.0004 | 3.75 | 0.09 | 0.46 | 0.19 |
| E/E (39) | 4.57 | 0.21 | 4.57 | 0.08 | 4.53 | 0.35 | 4.62 | 0.02 | 1.32 | 0.24 |
| Others/Reference (213) | 4.37 | 4.34 | 4.36 | 4.22 | Reference | |||||
| Model Pd | 0.03 | 0.0001 | 0.003 | 0.01 | 0.08 (PLR) | 0.03f (Pw) | ||||
Expressed as mean log10 plasma HIV-1 RNA (copies/ml) based on ≥1 measurement available within each interval. The number of subjects differs between intervals, as shown in Table 1.
Range (median) of time interval between estimated date of seroconversion and median date of HIV RNA assay.
P values based on pairwise comparison between marker and reference (others) groups.
Overall P values based on multivariable analysis with GLMs.
RH according to cumulative proportion of those who develop AIDS over time with a CCR marker relative to those without it.
Log-rank (PLR) and Wilcoxon (PW) p values for AIDS-free time by Kaplan-Meier and Cox proportional hazards test (see text and Fig. 2).
The relative effects of all genotypes showing borderline or greater (P < 0.05) contributions to HIV-1 RNA levels were further evaluated with stepwise multivariate linear models (Table 5). When each mutually exclusive candidate genotype was examined in conjunction with the others, the individual effect was discernible, but weaker than that detected in sequential univariate analyses (data not shown), with the exception of the F*2/G*2 effect. Individual effects varied across intervals, but the overall models were quite consistent in demonstrating statistically significant discrimination of viral RNA level by CCR genotype (Table 5). In addition, model 2 (Table 5) improved the overall discrimination of mean RNA levels by substituting the A/G*2 genotype (n = 11) for any G*2 genotype (n = 66). None of the models containing the other genotype combinations showed better association with early mean RNA level (see Appendix).
Clinical outcome in MACS Caucasians.
The median times to 1987 CDC-defined AIDS (8.4 years) and death (10.1 years) in the MACS were typical for homosexual men not receiving potent antiretroviral therapy.
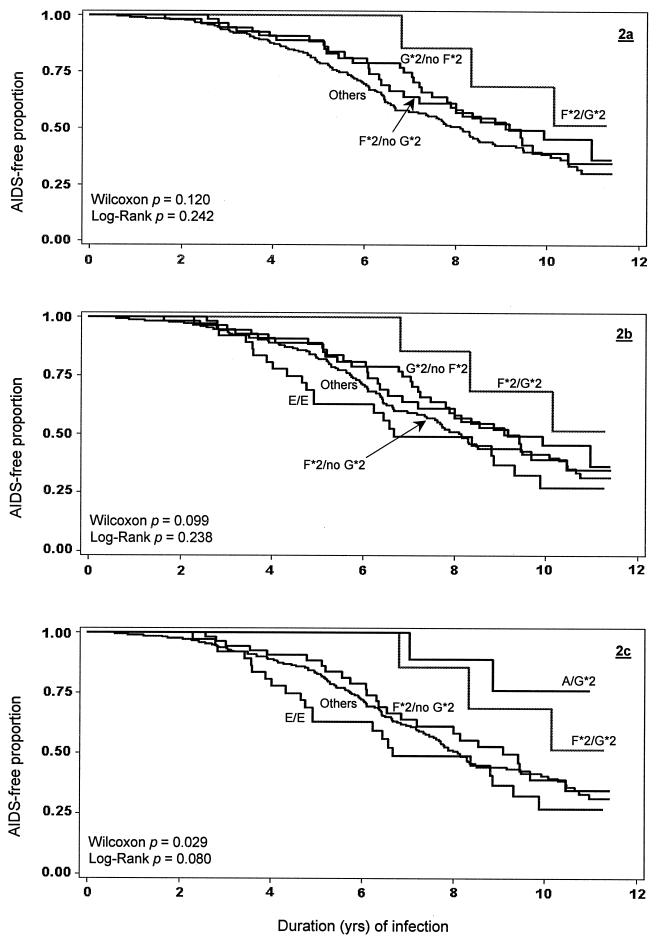
The series of Kaplan-Meier plots (Fig. 2a to c) illustrates how markers associated with lower or higher mean viral RNA levels contributed separately and jointly to AIDS-free time. CCR genotypes analyzed separately showed lower or higher RHs of AIDS with various degrees of significance (Table 4); the patterns of association between the contributory genotypes and time to AIDS (Fig. 2) largely paralleled those observed for viral RNA level. One notable exception to the pattern was the effect of E heterozygosity and particularly the C/E genotype: although they had no apparent effect on viral RNA level, carriage of either any E haplotype (RH = 1.56, 95% CI = 1.14 to 2.13, log-rank P = 0.006) or the C/E genotype (RH = 1.69, 95% CI = 1.19 to 2.38, log-rank P = 0.003) showed a strong association with more rapid progression to AIDS, supporting the observation in the Argentinean perinatal study (41). However, agreement with the Argentinean study was not complete: men with E/G*2 (RH = 0.63, 95% CI = 0.30 to 1.35, log-rank P = 0.23) more closely mirrored the overall protective effect of G*2 rather than the risk effect seen in perinatal disease.
FIG. 2.
Kaplan-Meier plots of time to AIDS following HIV-1 seroconversion. Time to AIDS is shown for the reference (Others) category and for men with different CCR2-CCR5 genotypes associated with contrasting levels of HIV-1 RNA levels during the initial 42 months of infection. The intermediate models (2a and 2b) in which all G*2/no F*2 genotypes were aggregated revealed a weaker overall effect compared with the final model (2c), which incorporated only A/G*2 rather than the entire G*2/no F*2 group. Models 2b and 2c shown here correspond to multivariable models 1 and 2 in Table 5.
In the final models evaluating the combined effects of the CCR markers on time to AIDS (Table 5), the impact of each individual marker on these outcomes was generally borderline significant when considered separately or when examined in light of the CCR effects on viral RNA level. As with the analysis of viremia, substitution of the A/G*2 genotype for the G*2 haplotype improved the discrimination of the RH estimate (Table 5).
Virologic and clinical outcomes in MACS African-Americans and in DCG and SFMHS Caucasians.
During postseroconversion intervals, African-American SCs with E/E genotype had a higher (+0.38) mean log10 HIV-1 RNA level compared with carriers of other genotypes, and the same individuals showed trends toward an accelerated course of infection in both univariate (RH = 4.0, P = 0.10) and multivariable (RH = 6.6, P = 0.06) analysis (data not shown). F*2 carriers, on the other hand, did not differ from others in their mean viral RNA. Genotypes containing F*2 showed no effect on time to AIDS individually or in a multivariable model. Absence of A/G*2 and F*2/G*2 in this ethnic group did not allow any further analyses.
Few of the 39 and 37 seroconverters from the DCG and the SFMHS, respectively, who met criteria for analysis carried the markers found to influence HIV-1 RNA level in the MACS: F*2 without G*2 (n = 2 and 3), G*2 without F*2 (n = 8 and 7), F*2/G*2 (n = 0 and 1), and E/E (n = 3 and 1). Overall mean log10 HIV-1 RNA concentrations at <6 and 6- to 42-month intervals were 4.10 and 4.20 for the DCG SCs and 3.50 and 3.80 for the SFMHS SCs, respectively. In the DCG participants during the 42-month period, there were nonsignificant trends toward lower mean log10 viral RNA levels in the presence of G*2 (−0.20) or F*2 (−0.07) and higher mean levels in the presence of E/E (+0.36 log). These findings were consistent with those in the MACS. Likewise, in SFMHS SCs during the <6- and 6- to 42-month intervals, respectively, there were consistent but not statistically significant reductions of log10 HIV-1 RNA in G*2 carriers (−0.25 and −0.39) and in F*2 carriers (−0.44 and −0.62). The effects of either G*2 (Δ32) or F*2 (64I) alone on disease progression in the DCG and SFMHS SCs were not based on large enough numbers to be meaningful in isolation, as noted previously (27).
Alternative analyses based on previous findings.
In earlier studies, CCR2 and CCR5 SNPs along with Δ32 have been grouped in different ways that did not target the entire CCR2-CCR5 haplotypes. We reanalyzed the genetic data from the MACS SCs according to earlier typing schemes and nomenclature. First, the equivalent (e) of F*2- and G*2-independent P1/P1 genotype was recaptured as P1/P1(e) (=E/E, E/F1, E/G1, F1/F1, F1/G1, and G1/G1). HIV-1 RNA at each of four infection intervals among the 49 P1/P1(e)-carrying MACS SCs (n = 341 in the selected cohort) were only slightly higher (+0.06 to +0.28 log, P > 0.10) than the F*2- or G*2-negative reference group, while time to AIDS was up to 1.5 years faster in P1/P1(e) carriers than in those with other genotypes excluding F*2 and G*2 (RH = 1.27, log-rank P = 0.29). The F*2/G*2-positive SCs showed much slower (3.5 to 4 years) progression to AIDS relative to the P1/P1(e) group. Compared with E/E homozygotes, the 10 MACS participants carrying E/G*1, the most common of the six possible non-E/E genotypes in the P1/P1(e) group, actually had considerably lower viral RNA levels, a rather clear indication that the risk was derived from E/E rather than all possible P1/P1 variants. Second, the MACS SCs carrying the 59029G/G equivalent [59029G/G(e) = all genotypes involving haplotypes A, B, C, and D; n = 63] differed from the F*2- or G*2-negative reference group by only 0.02 to 0.15 log in viral RNA level, whereas a delay in time to AIDS in the 59029G/G(e) carriers (adjusted RH = 0.65, 95% CI = 0.46 to 0.91, P = 0.013) was clearly independent of the genotypes involving F*2 and G*2 (adjusted RH = 0.60, 95% CI = 0.38 to 0.94, P = 0.027). The lack of association between the 59029G/G(e) genotype and early HIV-1 RNA levels in univariate analysis contrasted with its relationship to longer AIDS-free time, although it did not occur until about 6 years after seroconversion (data not shown).
DISCUSSION
The consistency in the effects of highly resolved CCR2-5 haplotype-genotype combinations on acquisition of infection, early virus concentration, and subsequent disease progression has not been addressed previously. This effort extends both the earlier epidemiologic comparison of HEPS and a subset of SCs in the MACS (77) and a more recent study of HEPS and SCs from several cohorts (43). Consistent with findings from in vitro experimental research on protection against infection by G*2 (Δ32) (25, 34, 55), the reduction in risk of seroconversion associated with G*2 heterozygosity closely parallels its favorable impact on viral RNA level and disease progression in infected individuals. Our high-resolution CCR genotyping further revealed that the A/G*2 and F*2/G*2 genotypes may account for much of the G*2 haplotype-related effects. Whether and how A/G*2 and F*2/G*2 differ from other G*2-bearing genotypes in regulating CCR5 expression remains to be clarified. The CCR effects on HIV-1 infection and disease progression in the MACS also applied to E/E homozygosity, as reported in an earlier analysis of perinatal transmission (41). The association of the rare 59356T/T genotype (=D/D haplotype pair) with increased vertical transmission of HIV-1 in African-Americans (36) could not be assessed here.
The effects of CCR2-CCR5 variants (the Δ32-linked G*2 haplotype, the 64I-linked F*2 haplotype, genotypic combination of F*2 and G*2, and the E/E genotype) on mean plasma HIV-1 RNA concentration was discernible within the first 6 months after seroconversion and increased as viral RNA level continued to drop in SCs carrying two protective genotypes (A/G*2 and F*2/G*2) (Fig. 1). The joint CCR effect was strong and consistent during three successive 1-year postconversion intervals in which virus concentration remained relatively stable. The implication is that these markers influence disease progression, in large part by modulating virus concentration soon after seroconversion.
These findings confirmed the previously observed influence on viral RNA level of G*2 and F*2 haplotypes and their combination, in some cases, even under potent anti-retroviral treatment (4, 6, 14, 22, 27, 54, 62, 72). In the TSS, the F*2 effect was reported for African-Americans, but not Caucasians, in whom F*2 is less frequent; the paucity of subjects in the TSS with the F*2/G*2 genotype may explain the inability to detect its effect in Caucasians (20). The strong association of A/G*2, not reported in or implied by any previous work, might reflect CCR haplotypic interactions (20) deserving further experimental evaluation in populations frequently carrying this particular CCR genotype.
The E/E genotype association with accelerated disease progression in Caucasians here and elsewhere (20) was predicted (70). In addition, the E/E genotype represents the major portion of an unfavorable genotype previously defined as P1/P1 (44), which is mutually exclusive of the reportedly favorable 59029G/G (=303G/G here) (46). The consistency of the E/E association is noteworthy: it occurred more frequently in MACS Caucasian SCs than seronegatives, in MACS Caucasian SCs with higher early HIV-1 RNA levels, in MACS African-Americans with higher viral RNA and shorter AIDS-free time, and in Rwandans with rapid disease progression (R. A. Kaslow et al., 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., abstr. 45B, 2001). Such consistency across ethnic groups within the MACS and Rwandan studies extends to additional cohorts (20, 41), suggesting that E/E confers similar effects against distinct genetic backgrounds. Distinguishing the E/E genotype from the remainder of the P1/P1 genotype combinations largely resolves the apparent discrepancy in the associations of P1/P1 with rapid progression between Caucasians (recessive) and African-Americans (dominant) (2, 44). In individuals of African ancestry, higher frequencies of the non-E/E variants encompassed by P1/P1 (20, 70) dilute the effect of the less frequent E/E. Thus, the E/E genotype may warrant special attention because of its relative prevalence in major ethnic groups (20, 41, 52).
Some of the CCR effects appeared to be either time-dependent or outcome (endpoint)-specific. For example, without substantial impact on the early viral RNA levels, carriage of haplotype E and especially the genotype C/E was associated with accelerated disease progression in MACS Caucasians, as seen in Argentinean children (41). Likewise, association of 59029G/G with delayed onset of AIDS was not preceded by any substantial decrease in RNA levels. Associations of host genetic polymorphism with early virus concentration can be readily tested in other cohorts with adequate virological measurement in the absence of lengthy follow-up. However, assessment of both genetic determinants and viral RNA as simultaneous, independent predictors of long-term outcome will require models specifically developed to account for those factors taken separately and jointly. Exploratory work on such models has been undertaken (71). HIV-1 genotypes and phenotypes (1, 13, 50), along with CCR and other disease-modifying host genetic factors, will eventually contribute to a more complete model of host-virus relationships.
Initial discoveries of the role of CCR polymorphism in HIV-1 infection were largely based on late clinical outcomes. However, if CCR2-CCR5 variants tightly regulate viral entry into host cells, they are likely to influence early viral equilibrium at least as directly as the later immunopathologic processes; in the MACS, the effects of several CCR variants on early HIV-1 RNA levels were indeed clearer if not stronger than those on later disease progression. The influence of host genetic variation on HIV-1 RNA levels is probably more useful than it is on other outcome measures because of the dual impact of viral RNA levels on subsequent disease progression rates (5, 28, 47, 57, 66) and HIV-1 transmissibility from infected to uninfected individuals (17, 18, 56, 58, 60, 65).
Our work could not confirm other reported associations of CCR haplotypes and genotypes with clinical outcomes in HIV-1 infection. For example, the C/G*2 genotype appeared protective in Tri-Service Study adults and Argentinean children with fully resolved CCR2-CCR5 haplotypes (20, 41), but equally fine resolution and similar distribution of genotypes in the MACS yielded only transient association of C/G*2 with relatively low RNA level and no predisposition to slower progression. Such inconsistencies among cohorts reflect the pervasive difficulty of identifying complex genetic contributions in various populations. Complete agreement for every major genotype in every population should not be expected, considering that polymorphisms in other systems including genes coding for HLA (7, 23, 33, 40, 63, 69) and perhaps RANTES (37), interleukin 10 (67), CX3CR1 (16), or MIP-1 (21), or other yet unrecognized genes will inevitably be incorporated into a comprehensive multifactorial and potentially time-dependent model of AIDS pathogenesis.
In summary, contributions of CCR2-CCR5 haplotypes in general and their genotypes in particular to host-HIV-1 equilibration can be detected early in the course of infection. While some of the CCR effects can be highly consistent across population boundaries (20, 27, 41) or at multiple time points, the compound and clear effects of CCR markers on early viral RNA may not immediately translate into a significant impact on later disease outcomes or vice versa. Further attention to CCR variants showing consistent influences on HIV-1 transmission and viral RNA level may extend interest in pathogenetic models into the clinical realm. More specifically, evidence that acquisition of virus may be impeded by G*2 heterozygosity (this study and references 42 and 43) and enhanced by homozygosity for either the D (36) or the E (here and 41) haplotype in both ethnic Africans and Caucasians may encourage research on coreceptor polymorphisms in the interest of prevention as well as treatment. Reported detection of CCR5-G*2 (Δ32)-mediated effects during potent antiretroviral therapy (4, 22, 54, 72) have already implied that genetic typing may inform clinical decision making, particularly for patients with atypical responses to therapeutic agents.
Acknowledgments
We thank C. A. Rivers, A. D. Myracle, K. Schultz, and N. Michael for valuable contribution to various aspects of our studies. The Multicenter AIDS Cohort Study (MACS) involves the following additional principal investigators: John Phair (Northwestern University School of Medicine), Roger Detels (UCLA Schools of Public Health and Medicine), Charles R. Rinaldo (University of Pittsburgh Graduate School of Public Health), Alvaro Munoz (Johns Hopkins University School of Hygiene and Public Health), and Carl Dieffenbach (Project officer, NIAID).
This work was supported by NIAID grants R01 AI41951 (R.A.K.) and R01 AI36661.
APPENDIX
Supplementary analyses of HIV-1 RNA concentration categorized in tertiles (Table A1) also documented the joint effects of CCR genotypes consistently in univariate and multivariate models (Table A2). Here the test of significance of the association of A/G*2, newly identified as an apparent contributing genotype, was adjusted for the number of evaluable G*2-containing genotypes in 341 MACS seroconverters.
TABLE A1.
Means (log10) of HIV-1 RNA concentrations for each tertile of VL, aggregated by repeated measures technique and by mean of means
| VL by tertile (n) | Mean VLa | SD | P | GMb |
|---|---|---|---|---|
| Repeated measures (6–42 mo) | ||||
| Tertile 1, low VL (113) | 3.33 | 0.69 | Reference | 2,138 |
| Tertile 2, medium VL (114) | 4.32 | 0.54 | <0.0001 | 20,892 |
| Tertile 3, high VL (114) | 5.00 | 0.47 | <0.0001 | 100,000 |
| Mean of the means (6–42 mo) | ||||
| Tertile 1, low VL (113) | 3.23 | 0.50 | Reference | 1,698 |
| Tertile 2, medium VL (114) | 4.34 | 0.20 | <0.0001 | 21,878 |
| Tertile 3, high VL (114) | 5.00 | 0.30 | <0.0001 | 100,000 |
The log10-transformed plasma HIV-1 RNA concentration.
GM, geometric mean.
TABLE A2.
Models summarizing effects of contributing CCR2-CCR5 haplotypes or genotypes on HIV-1 RNA concentrations, categorized by tertiles of viral load (VL) measured repeatedly in the 6- to 42-month interval after seroconversiona
| CCR genotype (n) | No. (%) in VL tertile
|
Uni- variate P for trendb | Multi- variate GLMcP | ||
|---|---|---|---|---|---|
| 1 (n = 113) | 2 (n = 114) | 3 (n = 114) | |||
| Model 1 | 0.0007 | ||||
| F*2, no G*2 (57) | 20 (17.7) | 18 (15.8) | 19 (16.7) | 0.9282 | 0.4311 |
| F*2/G*2 (9) | 6 (5.3) | 2 (1.8) | 1 (0.9) | 0.0880 | 0.0208 |
| G*2, no F*2 (57) | 29 (25.7) | 17 (14.9) | 11 (9.6) | 0.0044 | 0.0013 |
| E/E (39) | 6 (5.3) | 16 (14.0) | 17 (14.9) | 0.0427 | 0.1496 |
| Others (179) | 52 (46.0) | 61 (53.5) | 66 (57.9) | 0.1939 | Reference |
| Model 2 | 0.0005 | ||||
| A/G*2 (11) | 9 (8.0) | 1 (0.9) | 1 (0.9) | 0.0023d | 0.0029 |
| F*2/G*2 (9) | 6 (5.3) | 2 (1.8) | 1 (0.9) | 0.0880 | 0.0372 |
| E/E (39) | 6 (5.3) | 16 (14.0) | 17 (14.9) | 0.0427 | 0.0476 |
| Others (282) | 92 (81.4) | 95 (83.3) | 95 (83.3) | 0.9075 | Reference |
REFERENCES
- 1.Alexander, L., E. Weiskopf, T. C. Greenough, N. C. Gaddis, M. R. Auerbach, M. H. Malim, S. J. O’Brien, B. D. Walker, J. L. Sullivan, and R. C. Desrosiers. 2000. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 74:4361–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, P., M. P. Martin, G. W. Nelson, M. Carrington, M. W. Smith, K. Gong, D. Vlahov, S. J. O’Brien, and C. A. Winkler. 2000. Influence of CCR5 promoter haplotypes on AIDS progression in African-Americans. AIDS 14:2117–2122. [DOI] [PubMed] [Google Scholar]
- 3.Anzala, A. O., T. B. Ball, T. Rostron, S. J. O’Brien, F. A. Plummer, S. L. Rowland-Jones, and University of Nairobi Collaboration for HIV Research. 1998. CCR2–64I allele and genotype association with delayed AIDS progression in African women. Lancet 351:1632–1633. [DOI] [PubMed] [Google Scholar]
- 4.Barroga, C. F., C. Raskino, M. C. Fangon, P. E. Palumbo, C. J. Baker, J. A. Englund, and S. A. Spector. 2000. The CCR5Δ32 allele slows disease progression of human immunodeficiency virus-1-infected children receiving antiretroviral treatment. J. Infect. Dis. 182:413–419. [DOI] [PubMed] [Google Scholar]
- 5.Bratt, G., A. Karlsson, A. C. Leandersson, J. Albert, B. Wahren, and E. Sandstrom. 1998. Treatment history and baseline viral load, but not viral tropism or CCR-5 genotype, influence prolonged antiviral efficacy of highly active antiretroviral treatment. AIDS 12:2193–2202. [DOI] [PubMed] [Google Scholar]
- 6.Buseyne, P., G. Janvier, J. P. Teglas, S. Ivanoff, M. Burgard, E. Bui, M. J. Mayauk, S. Blanche, C. Rouzioux, and Y. Riviere. 1998. Impact of heterozygosity for the chemokine receptor CCR5 32-bp-deleted allele on plasma virus load and CD4 T lymphocytes in perinatally human immunodeficiency virus-infected children at 8 years of age. J. Infect. Dis. 178:1019–1023. [DOI] [PubMed] [Google Scholar]
- 7.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O’Brien. 1999. HLA and HIV-1: heterozygosity advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control. 1987. Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. Morb. Mortal. Wkly. Rep. 36:3S–15S. [Google Scholar]
- 9.Childs, E. A., R. H. Lyles, O. A. Selnes, B. Chen, E. N. Miller, B. A. Cohen, J. T. Becker, J. Mellors, and J. C. McArthur. 1999. Plasma viral load and CD4 lymphocytes predict HIV-associated dementia and sensory neuropathy. Neurology 52:607–613. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, A. L., S. Li, J. Juarez, G. Lynch, M. Alali, and H. Naif. 2000. The level of HIV infection of macrophages is determined by interaction of viral and host cell genotypes. J. Leukoc. Biol. 68:311–317. [PubMed] [Google Scholar]
- 11.D’Aquila, R. T., L. Sutton, A. Savara, M. D. Hughes, V. A. Johnson, and NIAID AIDS Clinical Trials Group Protocol 241 Virology Team. 1998. CCR5/Δ(ccr5) heterozygosity: a selective pressure for the syncytium-inducing human immunodeficiency virus type 1 phenotype. J. Infect. Dis. 177:1549–1553. [DOI] [PubMed] [Google Scholar]
- 12.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Franscisco Cohort Study, ALIVE Study, and S. J. O’Brien. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a common deletion allele of the chemokine receptor 5 structural gene. Science 273:1856–1862. [DOI] [PubMed] [Google Scholar]
- 13.Dyer, W. B., A. F. Geczy, S. J. Kent, L. B. McIntyre, S. A. Blasdall, J. C. Learmont, and J. S. Sullivan. 1997. Lymphoproliferative immune function in the Sydney Blood Bank Cohort, infected with natural nef/long terminal repeat mutants, and in other long-term survivors of transfusion-acquired HIV-1 infection. AIDS 11:1565–1574. [DOI] [PubMed] [Google Scholar]
- 14.Easterbrook, P. J., T. Rostron, N. Ives, M. Troop, B. G. Gazzard, and S. L. Rowland-Jones. 1999. Chemokine receptor polymorphisms and human immunodeficiency virus disease progression. J. Infect. Dis. 180:1096–1105. [DOI] [PubMed] [Google Scholar]
- 15.Eugen-Olsen, J., A. K. Iversen, T. L. Benfield, U. Koppelhus, and P. Garred. 1998. Chemokine receptor CCR2b 64I polymorphism and its relation to CD4 T-cell counts and disease progression in a Danish cohort of HIV-infected individuals. Copenhagen AIDS cohort. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18:110–116. [DOI] [PubMed] [Google Scholar]
- 16.Faure, S., L. Meyer, D. Costagliola, C. Vaneensberghe, E. Genin, B. Autran, J. F. Delfraissy, D. H. McDermott, P. M. Murphy, P. Debre, I. Theodorou, and C. Combadiere. 2000. Rapid progression to AIDS in HIV+ individuals with a structural variant of the chemokine receptor CX3CR1. Science 287:2274–2277. [DOI] [PubMed] [Google Scholar]
- 17.Fideli, U. S., S. Allen, R. Musunda, S. Trask, B. Hahn, J. Mulenga, F. C. Kasolo, S. H. Vermund, and G. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 (HIV-1) in Africa. AIDS Res. Hum. Retroviruses 17:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, P. M., L. A. Kalish, J. Pitt, H. Minkoff, T. C. Quinn, S. K. Burchett, J. Kornegay, B. Jackson, J. Moye, C. Hanson, C. Zorrilla, and J. F. Lew. 1999. Maternal levels of plasma human immunodeficiency virus type 1 RNA and the risk of perinatal transmission. Women and Infants Transmission Study Group. N. Engl. J. Med. 341:394–402. [DOI] [PubMed] [Google Scholar]
- 19.Goedert, J. J., R. J. Biggar, D. M. Winn, D. L. Mann, D. P. Byar, D. M. Strong, R. A. DiGioia, R. J. Grossman, W. C. Sanchez, R. G. Kase et al. 1985. Decreased helper T lymphocytes in homosexual men. II. Sexual practices. Am. J. Epidemiol. 121:637–644. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez, E., M. Bamshad, N. Sato, S. Mummidi, G. Dhanda, G. Catano, S. Cabrera, M. McBride, X.-H. Cao, G. Merrill, P. O’Connell, D. W. Bowden, B. I. Fredman, S. A. Anderson, E. A. Walters, J. S. Evans, K. T. Stephan, R. A. Clark, S. Tyagi, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 1999. Race-specific HIV-1 disease modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. USA 96:12004–12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, E., R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, S. A. Anderson, E. A. Walter, K. T. Stephan, M. F. Hammer, A. Mangano, L. Sen, R. A. Clark, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1α: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerin, S., L. Meyer, I. Theodorou, F. Boufassa, M. Magierowska, C. Goujard, C. Rouzioux, P. Debre, and J. F. Delfraissy. 2000. CCR5 Δ32 deletion and response to highly active antiretroviral therapy in HIV-1-infected patients. AIDS 14:2788–2790. [DOI] [PubMed] [Google Scholar]
- 23.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O’Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 162:6942–6946. [PubMed] [Google Scholar]
- 24.Hendel, H., N. Henon, H. Lebuanec, A. Lachgar, H. Poncelet, S. Caillat-Zucman, C. A. Winkler, M. W. Smith, L. Kenefic, S. O’Brien, W. Lu, J. M. Andrieu, D. Zagury, F. Schachter, J. Rappaport, and J. F. Zagury. 1998. Distinctive effects of CCR5, CCR2, and SDF1 genetic polymorphisms in AIDS progression. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:381–386. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman, T. L., R. R. MacGregor, H. Burger, R. Mick, R. W. Doms, and R. G. Collman. 1997. CCR5 genotypes in sexually active couples discordant for human immunodeficiency virus type 1 infection status. J. Infect. Dis. 176:1093–1096. [DOI] [PubMed] [Google Scholar]
- 26.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240–1243. [DOI] [PubMed] [Google Scholar]
- 27.Ioannidis, J. P., P. S. Rosenberg, J. J. Goedert et al. 2001. Effects of CCR5-Δ32, CCR2–64I and SDF-1 3′A polymorphisms on HIV disease progression: an international meta-analysis of individual patient data. Ann. Intern. Med. 135:782–795. [DOI] [PubMed] [Google Scholar]
- 28.Ioannidis, J. P. A., J. J. Goedert, P. G. McQueen, C. Enger, and R. A. Kaslow. 1999. Comparison of viral load and human leukocyte antigen statistical and neural network predictive models for the rate of HIV-1 disease progression across two cohorts of homosexual men. J. Acquir. Immune Defic. Syndr. 20:129–136. [DOI] [PubMed] [Google Scholar]
- 29.Kaslow, R. A., and J. M. McNicholl. 1999. Genetic determinants of HIV-1 infection and its manifestations. Proc. Assoc. Am. Physicians 111:299–307. [DOI] [PubMed] [Google Scholar]
- 30.Kaslow, R. A., D. G. Ostrow, R. Detels, J. P. Phair, B. F. Polk, and C. R. Rinaldo, Jr. 1987. The Multicenter AIDS Cohort Study (MACS). I. Rationale, organization and selected characteristics of the participants. Am. J. Epidemiol. 126:310–318. [DOI] [PubMed] [Google Scholar]
- 31.Kasten, S., A. Goldwich, M. Schmitt, A. Rascu, M. Grunke, C. Dechant, J. R. Kalden, and T. Harrer. 2000. Positive influence of the Δ32CCR5 allele on response to highly active antiretroviral therapy (HAART) in HIV-1 infected patients. Eur. J. Med. Res. 5:323–328. [PubMed] [Google Scholar]
- 32.Katzenstein, T. L., J. Eugen-Olsen, B. Hofmann, T. Benfield, C. Pedersen, A. K. Iversen, A. M. Sorensen, P. Garred, U. Koppelhus, A. Svejgaard, and J. Gerstoft. 1997. HIV-infected individuals with the CCR Δ32/CCR5 genotype have lower HIV RNA levels and higher CD4 cell counts in the early years of the infection than do patients with the wild type. Copenhagen AIDS Cohort Study Group. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 16:10–14. [DOI] [PubMed] [Google Scholar]
- 33.Keet, I. P., J. Tang, M. R. Klein, S. LeBlanc, C. Enger, C. Rivers, R. J. Apple, D. Mann, J. J. Goedert, F. Miedema, and R. A. Kaslow. 1999. Consistent associations of HLA class I and class II and transporter gene products with progression of human immunodeficiency virus-1 infection in homosexual men. J. Infect. Dis. 180:299–309. [DOI] [PubMed] [Google Scholar]
- 34.Kim, A., M. Pettoello-Mantovani, and H. Goldstein. 1998. Decreased susceptibility of peripheral blood mononuclear cells from individuals heterozygous for a mutant CCR5 allele to HIV infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:145–149. [DOI] [PubMed] [Google Scholar]
- 35.Kostrikis, L. G., Y. Huang, J. P. Moore, S. M. Wolinsky, L. Zhang, Y. Guo, L. Deutsche, J. Phair, A. U. Neumans, and D. D. Ho. 1998. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat. Med. 4:350–353. [DOI] [PubMed] [Google Scholar]
- 36.Kostrikis, L. G., A. U. Neumann, B. Thomson, B. T. Korber, P. McHardy, R. Karanicolas, L. Deutsch, Y. Huang, J. F. Lew, K. McIntosh, H. Pollack, W. Borkowsky, H. M. L. Spiegel, P. Palumbo, J. Oleske, A. Bardeguez, K. Luzuriaga, J. Sullivan, S. M. Wolinsky, R. A. Koup, D. D. Ho, and J. P. Moore. 1999. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J. Virol. 73:10264–10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, H. L., D. Chao, E. E. Nakayama, H. Taguchi, M. Goto, X. M. Xin, J. Takamatsu, H. Saito, Y. Ishikawa, T. Akaza, T. Juji, Y. Takebe, T. Ohishi, K. Fukutake, Y. Maruyama, S. J. Yashiki, S. Sonoda, T. Nakamura, Y. Nagai, A. Iwamoto, and T. Shioda. 1999. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc. Natl. Acad. Sci. USA 96:4581–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lockett, S. F., A. Alonso, R. Wyld, M. P. Martin, J. R. Robertson, S. M. Gore, C. L. S. Leen, R. P. Brettle, D. L. Yirrell, M. Carrington, and A. J. L. Brown. 1999. Effect of chemokine receptor mutations on heterosexual human immunodeficiency virus transmission. J. Infect. Dis. 180:614–620. [DOI] [PubMed] [Google Scholar]
- 39.Lyles, R. H., A. Munoz, T. E. Yamashita, H. Bazmi, R. Detels, C. R. Rinaldo, J. B. Margolick, J. P. Phair, and J. W. Mellors. 2000. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J. Infect. Dis. 181:872–880. [DOI] [PubMed] [Google Scholar]
- 40.Magierowska, M., I. Theodorou, P. Debre, F. Sanson, B. Autran, Y. Riviere, D. Charron, and D. Costagliola. 1999. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood 93:936–941. [PubMed] [Google Scholar]
- 41.Mangano, A., E. Gonzalez, R. Dhanda, G. Catano, M. Bamshad, A. Bock, R. Duggirala, K. Williams, S. Mummidi, R. A. Clark, S. S. Ahuja, M. J. Dolan, R. Bologna, L. Sen, S. K. Ahuja, and L. Sen. 2001. Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus. J. Infect. Dis. 183:1574–1585. [DOI] [PubMed] [Google Scholar]
- 42.Mangano, A., F. Prada, A. Roldan, G. Picchio, R. Bologna, and L. Sen. 1998. Distribution of CCR-5 Δ32 allele in Argentinean children at risk of HIV-1 infection: its role in vertical transmission. AIDS 12:109–110. [PubMed] [Google Scholar]
- 43.Marmor, M., H. W. Sheppard, D. Donnell, S. Bozeman, C. Celum, S. Buchbind, B. A. Koblin, G. R. Seage, and H. I. V. Network for Prevention Trials Vaccine Preparedeness Protocol Team. 2001. Homozygous and heterozygous CCR5-Δ32 genotypes are associated with resistance to HIV infection. J. Acquir. Immune Defic. Syndr. 27:472–481. [DOI] [PubMed] [Google Scholar]
- 44.Martin, M. P., M. Dean, M. W. Smith, C. Winkler, B. Gerrard, N. L. Michael, B. Lee, R. W. Doms, J. Margolick, S. Buchbinder, J. J. Goedert, T. S. O’Brien, M. W. Hilgartner, D. Vlahov, S. O’Brien, and M. Carrington. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907–1911. [DOI] [PubMed] [Google Scholar]
- 45.McDermott, D. H., J. S. Colla, C. A. Kleeberger, M. Plankey, P. S. Rosenberg, E. D. Smith, P. A. Zimmerman, C. Combadiere, S. F. Leitman, R. A. Kaslow, J. J. Goedert, E. A. Berger, T. R. O’Brien, and P. M. Murphy. 2000. Genetic polymorphism in CX3CR1 and risk of HIV disease. Science 290:2031. [DOI] [PubMed] [Google Scholar]
- 46.McDermott, D. H., P. A. Zimmerman, F. Guignard, C. A. Kleeberger, S. F. Leitman, The Multicenter AIDS Cohort Study, and P. M. Murphy. 1998. CCR5 promoter polymorphism and HIV-1 disease progression. Lancet 352:866–870. [DOI] [PubMed] [Google Scholar]
- 47.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167–1170. [DOI] [PubMed] [Google Scholar]
- 48.Meyer, L., M. Magierowska, J. B. Hubert, C. Rouzioux, C. Deveau, F. Sanson, P. Debre, J. F. Delfraissy, I. Theodorou, and the SEROCO Study Group. 1997. Early protective effect of CCR-5 Δ32 heterozygosity on HIV-1 disease progression: relationship with viral load. AIDS 11:F73–F78. [DOI] [PubMed] [Google Scholar]
- 49.Michael, N. L. 1999. Host genetic influences on HIV-1 pathogenesis. Curr. Opin. Immunol. 11:466–474. [DOI] [PubMed] [Google Scholar]
- 50.Michael, N. L., G. Chang, L. G. Louie, J. R. Mascola, D. Dondero, D. L. Birx, and H. Sheppard. 1997. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat. Med. 3:338–340. [DOI] [PubMed] [Google Scholar]
- 51.Michael, N. L., L. Louie, A. L. Rohrbaugh, K. A. Schultz, D. E. Dayhoff, C.-E. Wang, and H. W. Sheppard. 1997. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat. Med. 3:1160–1162. [DOI] [PubMed] [Google Scholar]
- 52.Mummidi, S., M. Bamshad, S. S. Ahuja, E. Gonzalez, P. M. Feuillet, K. Begum, M. C. Galvis, V. Kostecki, A. J. Valente, K. K. Murthy, L. Haro, M. J. Dolan, J. S. Allan, and S. K. Ahuja. 2000. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J. Biol. Chem. 275:18946–18961. [DOI] [PubMed] [Google Scholar]
- 53.O’Brien, S. J., G. W. Nelson, C. A. Winkler, and M. W. Smith. 2000. Polygenic and multifactorial disease gene association in man: lessons from AIDS. Annu. Rev. Genet. 34:563–591. [DOI] [PubMed] [Google Scholar]
- 54.O’Brien, T. R., D. H. McDermott, J. P. Ioannidis, M. Carrington, P. M. Murphy, D. V. Havlir, and D. D. Richman. 2000. Effect of chemokine receptor gene polymorphisms on the response to potent antiretroviral therapy. AIDS 14:821–826. [DOI] [PubMed] [Google Scholar]
- 55.Ometto, L., M. Zanchetta, A. Cabrelle, G. Esposito, M. Mainardi, L. Chieco-Bianchi, and A. De Rossi. 1999. Restriction of HIV type 1 infection in macrophages heterozygous for a deletion in the CC-chemokine receptor 5 gene. AIDS Res. Hum. Retroviruses 15:1441–1452. [DOI] [PubMed] [Google Scholar]
- 56.Operskalski, E. A., D. O. Stram, M. P. Busch, W. Huang, M. Harris, S. L. Dietrich, E. R. Schiff, E. Donegan, J. W. Mosley, and Transfusion Safety Study Group. 1997. Role of viral load in heterosexual transmission of human immunodeficiency virus type 1 by blood transfusion recipients. Am. J. Epidemiol. 146:655–661. [DOI] [PubMed] [Google Scholar]
- 57.O’Shea, S., T. Rostron, A. S. Hamblin, S. J. Palmer, and J. E. Banatvala. 1991. Quantitation of HIV: correlation with clinical, virological, and immunological status. J. Med. Virol. 35:65–69. [DOI] [PubMed] [Google Scholar]
- 58.Pedraza, M. A., J. del Romero, F. Roldan, S. Garcia, M. C. Ayerbe, A. R. Noriega, and J. Alcami. 1999. Heterosexual transmission of HIV-1 is associated with high plasma viral load levels and a positive viral isolation in the infected partner. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 21:120–125. [PubMed] [Google Scholar]
- 59.Quillent, C., E. Oberlin, J. Braun, D. Rousset, G. Gonzalez-Canali, P. Metais, L. Montagnier, J.-L. Virelizier, F. Arenzana-Seisdedos, and A. Beretta. 1998. HIV-1 resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet 351:14–18. [DOI] [PubMed] [Google Scholar]
- 60.Quinn, T. C., M. J. Wawer, N. Sewankambo, D. Serwadda, C. Li, F. Wabwire-Mangen, M. O. Meehan, T. Lutalo, and R. H. Gray. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 342:921–929. [DOI] [PubMed] [Google Scholar]
- 61.Rabkin, C. S., Q. Yang, J. J. Goedert, G. Nguyen, H. Mitsuya, and S. Sei. 1999. Chemokine and chemokine receptor gene variants and risk of non-Hodgkin’s lymphoma in human immunodeficiency virus-1-infected individuals. Blood 93:1838–1842. [PubMed] [Google Scholar]
- 62.Rizzardi, G. P., R. A. Morawetz, E. Vicenzi, S. Ghezzi, G. Poli, A. Lazzarin, G. Pantaleo, and The Swiss HIV Cohort. 1998. CCR2 polymorphism and HIV disease. Nat. Med. 4:252–253. [DOI] [PubMed] [Google Scholar]
- 63.Saah, A. J., D. R. Hoover, S. Weng, M. Carrington, J. Mellors, C. R. J. Rinaldo, D. Mann, R. Apple, J. P. Phair, R. Detels, S. O’Brien, C. Enger, P. Johnson, and R. A. Kaslow. 1998. Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. Multicenter AIDS Cohort Study. AIDS 12:2107–2113. [DOI] [PubMed] [Google Scholar]
- 64.Schinkel, J., M. W. Langendam, R. A. Coutinho, A. Krol, M. Brouwer, and H. Schuitemaker. 1999. No evidence for an effect of the CCR5 Δ32/+ and CCR2b 64I/+ mutations on human immunodeficiency virus (HIV)-1 disease progression among HIV-1-infected injecting drug users. J. Infect. Dis. 179:825–831. [DOI] [PubMed] [Google Scholar]
- 65.Semba, R. D., N. Kumwenda, D. R. Hoover, T. E. Taha, T. C. Quinn, L. Mtimavalye, R. J. Biggar, R. Broadhead, P. G. Miotti, L. J. Sokoll, L. van der Hoeven, and J. D. Chiphangwi. 1999. Human immunodeficiency virus load in breast milk, mastitis, and mother-to-child transmission of human immunodeficiency virus type 1. J. Infect. Dis. 180:93–98. [DOI] [PubMed] [Google Scholar]
- 66.Shearer, W. T., T. C. Quinn, P. LaRussa, J. F. Lew, L. Mofenson, S. Almy, K. Rich, E. Handelsman, C. Diaz, M. Pagano, V. Smeriglio, and L. A. Kalish. 1997. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. Women and Infants Transmission Study Group. N. Engl. J. Med. 336:1337–1342. [DOI] [PubMed] [Google Scholar]
- 67.Shin, H., C. Winkler, J. C. Stevens, J. Bream, H. Young, J. J. Goedert, T. R. O’Brien, D. Vlahov, S. Buchbinder, J. Giorgi, C. Rinaldo, S. Donfield, A. Willoughby, S. J. O’Brien, and M. W. Smith. 2000. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc. Natl. Acad. Sci. USA 97:14467–14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O’Brien, L. P. Jacobson, R. A. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, W. Hilgartner, HGDS, MHCS, MACS, SFCC, ALIVE-Study, and S. J. O’Brien. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science 277:959–965. [DOI] [PubMed] [Google Scholar]
- 69.Tang, J., C. Costello, I. P. M. Keet, C. Rivers, S. LeBlanc, E. Karita, S. Allen, and R. A. Kaslow. 1999. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses 15:317–324. [DOI] [PubMed] [Google Scholar]
- 70.Tang, J., C. Rivers, E. Karita, C. Costello, S. Allen, P. N. Fultz, E. E. Schoenbaum, and R. A. Kaslow. 1999. Allelic variants of human beta-chemokine receptor 5 (CCR5) promoter: evolutionary relationships and predictable association with HIV-1 disease progression. Genes Immun. 1:20–27. [DOI] [PubMed] [Google Scholar]
- 71.Taylor, J. M., Y. Wang, L. Ahdieh, J. S. Chmiel, R. Detels, J. V. Giorgi, R. Kaslow, L. Kingsley, and J. Margolick. 2000. Causal pathways for CCR5 genotype and HIV progression. J. Acquir. Immune Defic. Syndr. 23:160–171. [DOI] [PubMed] [Google Scholar]
- 72.Valdez, H., S. F. Purvis, M. M. Lederman, M. Fillingame, and P. A. Zimmerman. 1999. Association of the CCR5Δ32 mutation with improved response to antiretroviral therapy. JAMA 282:734. [DOI] [PubMed] [Google Scholar]
- 73.van Rij, R. P., S. Proersen, J. Goudsmit, R. A. Coutinho, and H. Schuitemaker. 1998. The role of a stromal cell-derived factor-1 chemokine gene variant in the clinical course of HIV-1 infection. AIDS 12:F85–F90. [PubMed] [Google Scholar]
- 74.Visco-Comandini, U., C. Hultgren, C. Broström, M. Birk, S. Kim, and M. Sällberg. 1998. Human immunodeficiency virus type 1 disease progression, CCR5 genotype, and specific immune responses. Clin. Diagn. Lab. Immunol. 5:463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winkelstein, W., Jr., D. M. Lyman, N. Padian, R. Grant, M. Samuel, J. A. Wiley, R. E. Anderson, W. Lang, J. Riggs, and J. A. Levy. 1987. Sexual practices and risk of infection by the human immunodeficiency virus. The San Francisco Men’s Health Study. JAMA 257:321–325. [PubMed] [Google Scholar]
- 76.Winkler, C., W. Modi, M. W. Smith, G. W. Nelson, X. Wu, M. Carrington, M. Dean, T. Honjo, K. Tashiro, D. Yabe, S. Buchbinder, E. Vittinghoff, J. J. Goedert, T. R. O’Brien, L. P. Jacobson, R. Detels, S. Donfield, A. Willoughby, E. Gomperts, D. Vlahov, J. Phair, and S. J. O’Brien. 1998. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science 279:389–393. [DOI] [PubMed] [Google Scholar]
- 77.Zimmerman, P. A., A. Buckler-White, G. Alkhatib, T. Spalding, J. Kubofcik, C. Combadiere, D. Weissman, O. Cohen, A. Rubbert, G. Lam, M. Vaccarezza, P. E. Kennedy, V. Kumaraswami, J. V. Giorgi, R. Detels, J. Hunter, M. Chopek, E. A. Berger, A. S. Fauci, T. B. Nutman, and P. M. Murphy. 1997. Inherited resistance of HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol. Med. 3:23–36. [PMC free article] [PubMed] [Google Scholar]