Abstract
Patients with chronic hepatitis C are more likely to have significant changes in their physical and mental well-being than patients with liver disease of other etiology, and hepatitis C virus (HCV) has been occasionally implicated in diseases of the central nervous system. We analyzed the presence of the HCV negative-strand RNA sequence, which is the viral replicative intermediary, in autopsy brain tissue samples from six HCV-infected patients. Negative-strand HCV RNA was searched for by a strand-specific Tth-based reverse transcriptase PCR, and viral sequences amplified from brain tissue and serum were compared by single-strand conformational polymorphism analysis and direct sequencing. HCV RNA negative strands were detected in brain tissue in three patients. In two of these patients, serum- and brain-derived viral sequences were different and classified as belonging to different genotypes. In one of the latter patients, HCV RNA negative strands were detected in lymph node and, while being different from serum-derived sequences, were identical to those present in the brain. The results of the present study suggest that HCV can replicate in the central nervous system, probably in cells of the macrophage/monocyte lineage.
Hepatitis C virus (HCV) is a common etiologic agent of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (1). Although HCV is a primary hepatotropic virus, there is mounting evidence that it can also replicate at extrahepatic sites, particularly under conditions of immunodeficiency associated with human immunodeficiency type 1 (HIV-1) infection (16, 17, 22).
Whether HCV can infect the central nervous system (CNS) remains unclear. HCV belongs to the Flaviviridae family, which includes several well-known neurotropic viruses (e.g., yellow fever, dengue, and tick-borne encephalitis viruses), and several reports have implicated HCV as an occasional cause of various CNS and peripheral nervous system pathologies (3, 6, 11, 13, 27). Moreover, HCV RNA has been detected in cerebrospinal fluid from both HIV-positive and HIV-negative patients (23, 25), and viral sequences have been amplified directly from brain tissue from a patient diagnosed with progressive encephalomyelitis (3). However, the presence of viral sequences in any particular compartment cannot be regarded as evidence for replication, and to prove the latter, the presence of replicative intermediates must be established. In the case of positive-strand viruses such as HCV, cells supporting replication should contain viral negative-strand RNA sequences.
In the present study we analyzed HCV RNA in autopsy brain tissue samples from six subjects, three of whom were HIV-1 positive. In addition to strand-specific detection of HCV RNA negative strands, we compared viral sequences amplified from various CNS structures and serum, assuming that in the presence of independent viral compartments they could be different, much like what has been described for HIV-1 (10). To our knowledge, this is the first attempt to detect HCV replicative intermediaries in brain tissue and to analyze viral sequences derived from various parts of the brain.
MATERIALS AND METHODS
Biological samples.
Serum and brain tissue samples were collected from six HCV-positive patients, five of whom had liver cirrhosis (Table 1). Brain tissue samples were obtained during routine autopsies conducted within 36 h after death and stored at −80°C until analysis. The following samples were collected: subcortical white matter and cerebral cortex from the frontal region, nucleus lentiformis, cerebellum, and medulla oblongata. However, in patient 1 only the last of these tissues was available for analysis. In addition, mediastinal lymph nodes were collected from four patients. After tissue homogenization, RNA was extracted by means of a modified guanidinium thiocyanate-phenol-chloroform technique using a commercially available kit (RNAzol; Gibco/BRL). Total RNA (1 and 5 μg as determined by spectrophotometry) was routinely used for reverse transcriptase PCR (RT-PCR). We found the latter amount of RNA to be the upper limit of the template, beyond which the amplification reactions would be commonly inhibited. In the case of serum, the amount of extracted RNA loaded into the reaction mixture corresponded to 100 μl. This study was approved by the respective ethical committees of the involved institutions.
TABLE 1.
Clinical and virologic data on six HCV-infected patients whose autopsy CNS tissue samples were analyzed for the presence of HCV replicationa
| Patient no. | Age (yr) | Gender | Diagnosis | Cause of death | HIV-1 status (CD4+ cell count) | Presence of HCV negative-strand RNA in:
|
HCV genotype
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Serum | CNS | Lymph node | Serum | CNS | Lymph node | ||||||
| 1 | 28 | M | i.v. drug abuse; decompensated cirrhosis; bacterial endocarditis | Sepsis | Pos (418) | Neg | Medulla oblongata | Neg | 1b | 3a | 1b |
| 2 | 70 | M | Decompensated cirrhosis; HCC | Liver failure | Neg | Neg | Cerebellum | Pos | 1b | 1a | 1ab |
| 3 | 45 | M | Alcoholism; decompensated cirrhosis; miliary tuberculosis | Acute pancreatitis | Neg | Neg | Subcortical white matter | NA | 1b | 1b | NA |
| 4 | 57 | M | Alcoholism, decompensated liver cirrhosis, HCC | Liver failure, hepatic coma | Neg | Neg | Neg | Neg | 1b | 1b | 1b |
| 5 | 34 | M | i.v. drug abuse | Drug overdose | Pos (440) | Neg | Neg | Neg | 1b | 1b | 1b |
| 6 | 29 | F | i.v. drug abuse; AIDS; alcoholism; cirrhosis | Drug overdose | Pos (120) | Neg | Neg | NA | 1b | 1b | NA |
Abbreviations: M, male; F, female; i.v., intravenous; HCC, hepatocellular carcinoma; Neg, negative; Pos, positive; NA, not available.
The positive HCV RNA strand was type 1b, while the negative strand was type 1a.
Strand-specific RT-PCR.
Strand specificity of our RT-PCR for the detection of HCV negative-strand RNA was ascertained by conducting cDNA synthesis at high temperature using the thermostable enzyme Tth. The sensitivity and strand specificity of this reaction were established using synthetic RNAs as templates. A detailed description of our strand-specific assay and sequence of employed primers was published previously (17, 20). In brief, the cDNA was generated in 20 μl of a reaction mixture containing 50 pM sense primer, 1× RT buffer (Perkin-Elmer), 1 mM MnCl2, 200 μM concentrations of each deoxynucleoside triphosphate, and 5 U of Tth (Perkin-Elmer). After 20 min at 65°C, Mn2+ was chelated with 8 μl of 10× EGTA chelating buffer (Perkin-Elmer), 50 pM antisense primer was added, the volume was adjusted to 100 μl, and the MgCl2 concentration was adjusted to 2.2 mM. The amplification was performed in a Perkin-Elmer GenAmp PCR System 9600 thermocycler as follows: initial denaturing for 1 min at 94°C; 50 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s; followed by a final extension at 72°C for 7 min. Twenty microliters of the final product was analyzed by agarose gel electrophoresis and Southern hybridization with a 32P-labeled internal oligoprobe. For the detection of positive-strand RNA, the primers were added in reverse order.
The strand-specific assay was capable of detecting approximately 100 genomic eq molecules of the correct strand while unspecifically detecting ≥108 genomic eq of the incorrect strand. The addition of 1 to 5 μg of total cellular RNA extracted from human tissues would lower the sensitivity of the reaction by no more than 1 log, while the specificity of the assay was not affected. Thus, the strand-specific assay was capable of detecting between 102 and 103 viral genomic eq in 1 μg of RNA. In serum the approximate detection limit was 103 eq/ml. The sensitivity and specificity of our assay for the detection of the positive strand were identical to those for the detection of the negative strand.
Standard RT-PCR.
Moloney murine leukemia virus RT-based detection of HCV has been described in detail previously (17). This assay was capable of detecting approximately 10 genomic eq of the correct synthetic template but was not strand specific. Similarly to Tth-based assay, the addition of cellular RNA would slightly lower the sensitivity by up to 1 log. The established detection limit was approximately 10 to 100 genomic eq per 1 μg of total RNA. In serum the approximate detection limit was 100 genomic eq per 1 ml.
The NS5 region was amplified by RT-PCR using primers described previously (21). Appropriate measures, described elsewhere (17, 20), were employed to prevent and detect contamination. Nested protocols, which are prone to carryover contamination, were not used for detection purposes. All RT-PCR runs included positive controls consisting of end point dilutions of respective RNA strands, and negative controls included brain tissue samples from uninfected subjects and normal sera.
Analysis of HCV quasispecies.
The analysis was conducted on the stable 5′ untranslated region (5′UTR) because a small number of expected viral variants within quasispecies allows for reliable comparison and we have previously found that variations in this region may correlate with extrahepatic replication (16, 17). In addition, comparison of highly variable E2 regions may be unreliable due to selective adsorption by human cells of viral quasispecies differing in the E2 region (18). For the purpose of sequence comparison, nested protocols were used to maximize the yield of PCR product. Amplification of the 5′UTR was conducted by using the RT-PCR assay as previously described (17).
HCV quasispecies were compared by the single-strand conformation polymorphism (SSCP) assay as described elsewhere (17), with minor modifications. In brief, PCR products were purified with a DNA binding resin system (Wizard PCR; Promega, Madison, Wis.) and resuspended in 50 μl of water. Next, 2 to 4 μl of the purified product was diluted in15 μl of low-ionic-strength solution (10% saccharose, 0.5% bromophenol blue, 0.5% xylene cyanol), denatured by heating at 97°C for 3 min, immediately cooled on ice, and subjected to nondenaturing 8% polyacrylamide gel electrophoresis in 1× Tris-borate-EDTA buffer with 400 V applied for 5 to 6 h at a constant temperature of 25°C. The bands were visualized with silver staining (Silver Stain; Promega). This assay enables detection of minor variants representing ≥3% of the whole population (17).
All analyzed products were sequenced directly in both directions using a Perkin-Elmer ABI 377 automatic sequencer. To rule out incorporation errors by Taq polymerase, direct sequencing was repeated from a new amplification reaction. HCV genotypes were determined by direct sequencing of the NS5 region (33).
The presence of CD2 (T cells), CD19 (B cells), and CD14 (monocyte/macrophage) phenotypes was determined by RT-PCR as described by others (34). The following primers were used: for CD2, 5′-AGACCGATGATCAGGATAT-3′ and 5′-TGGGAAGTTGCTGGATTCTG-3′ (expected product size, 547 bp); for CD14, 5′-ATGCATGTGGTCCAGCGCCC-3′ and 5′-CCACCGACAGGGTCGAACG-3′ (expected product size, 266 bp); for CD19, 5′-GACCTCACCATGGCCCCTGG-3′ and 5′-CTGGCCGAGCAGTGATCTCC-3′ (expected product size, 277 bp). To prevent contamination with genomic DNA, extracted RNA was digested with DNase I (1 U/μg of RNA), extracted with phenol-chloroform, and ethanol precipitated. In addition, to check the integrity of isolated RNA and to detect contaminating genomic DNA, β-actin primers specific for two different exons separated by an intron were used (5′-TCATGTTTGAGACCTTCAA-3′ and 5′-GTCTTTGCGGATGTCCACG-3′). Amplification of genomic DNA would result in a 607-bp product, while amplification of cDNA would result in a 513-bp product.
RESULTS
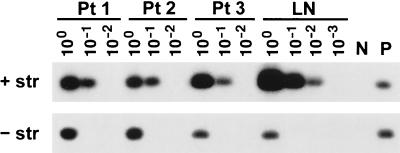
Tests for the presence of HCV RNA were positive in serum and in every sample of brain tissue analyzed from all six patients, the only exception being patient 3, from whom viral sequences could not be amplified from medulla oblongata. HCV RNA titers, determined by analyzing 10-fold serial dilutions of the RNA template, ranged from 105 to 107 genomic eq/ml and 101 to 103 genomic eq/μg of RNA in serum and brain tissue, respectively (Table 2). In patients 4 to 6, viral negative strand was not detected in any of the analyzed brain tissue samples in two independent sets of experiments using RNA from two separate extractions. However, medulla oblongata from patient 1 and cerebellum and subcortical white matter from patients 2 and 3, respectively, were repeatedly positive for the presence of negative-strand HCV RNA. HCV negative-strand RNA was also detected in one out of the four analyzed lymph nodes (patient 2), while none of the six analyzed sera tested positive. These results are summarized in Tables 1 and 2, and Fig. 1 illustrates detection of HCV negative-strand RNA in patients 1 to 3. These reactions were unlikely to represent false-positive results, because nonspecific detection of the incorrect strand might be expected when the latter is present at high number, at least 108 genomic eq/reaction. However, the approximate concentration of HCV RNA in brain samples containing the viral negative strand was only 103 genomic eq/μg of total RNA while the concentration of HCV RNA in lymph node tissue in patient 2 was 104 genomic eq/μg of total RNA (Table 2).
TABLE 2.
The detection and titers of positive and negative strands of HCV RNA in serum, lymph nodes, and different parts of brain in six subjectsa
| Patient no. | Serum titer (eq/ml) in serum
|
Titer (eq/μg of RNA) in:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cerebral cortex
|
Subcortical white matter
|
Medulla oblongata
|
Nucleus lentiformis
|
Cerebellum
|
Lymph node
|
|||||||||
| +str | −str | +str | −str | +st | −str | +str | −str | +str | −str | +str | −str | +str | −str | |
| 1 | 107 | Nc | NDd | ND | ND | ND | 103 | 102 | ND | ND | ND | ND | 104 | N |
| 2 | 106 | N | 101b | N | 102 | N | 102 | N | 102 | N | 103 | 102 | 104 | 102 |
| 3 | 106 | N | 102 | N | 103 | 102 | N | N | 101 | N | 101 | N | ND | ND |
| 4 | 107 | N | 102 | N | 102 | N | 102 | N | 103 | N | 101 | N | 103 | N |
| 5 | 105 | N | 102 | N | 102 | N | 101 | N | 102 | N | 102 | N | 103 | N |
| 6 | 106 | N | 101 | N | 102 | N | 102 | N | 101 | N | 101 | N | ND | ND |
The titers of the negative strand (−str) were determined by Tth-based strand-specific assays, while the titers of the positive strand (+str) were determined by a Tth-based assay or, when negative, by the more sensitive MMLV-based assay.
A titer of 101 represents a sample which was negative by the Tth-based assay but was positive by the MMLV-based assay in 5 μg of RNA template only.
N, negative.
ND, not done.
FIG. 1.
Detection of negative-strand HCV RNA in various brain tissue samples and lymph nodes (LN) in patients 1 to 3 (Pt 1 to Pt 3). The presence of viral negative strands was determined using strand-specific Tth-based RT-PCR. Twenty microliters (20%) of the reaction mixture was fractionated on agarose, transferred to a nylon membrane by Southern blotting, and subsequently hybridized to a 32P-labeled probe. The amount of RNA loaded into each reaction mixture was 5 μg; in the case of serum (S), it corresponded to 100 μl. The examined brain tissue samples included cerebral cortex (CC), subcortical white matter (WM), nucleus lentiformis (NL), cerebellum (C), and medulla oblongata (MO). In patient 1 only medulla oblongata was available for study. Positive sensitivity controls (lanes P) consisted of 103 genomic eq of the correct synthetic strand mixed with 5 μg of RNA extracted from brain tissue from an HCV-negative subject. Negative controls (lanes N) consisted of 5 μg of RNA extracted from brain tissue from uninfected patients.
To determine the proportion of positive-strand HCV RNA to negative-strand HCV RNA at the sites of putative replication, serial dilutions of extracted RNA were tested for the presence of positive- and negative-strand HCV RNA using Tth-based strand-specific RT-PCR. As can be seen in Fig. 2, negative-strand HCV RNA titers in brain tissue were 1 log lower than titers of the positive strand. In lymph node from patient 2, this difference was 2 logs.
FIG. 2.
The detection of HCV negative-strand (−str) and positive-strand (+str) RNA in brain tissue samples from patients 1 to 3 (Pt 1 to Pt 3). The analyzed tissues were medulla oblongata, cerebellum, and subcortical white matter in patients 1, 2, and 3, respectively. In addition, lymph node (LN) from patient 2 was studied. Tenfold serial dilutions of extracted RNA were tested for the presence of positive- and negative-strand HCV RNA by Tth-based RT-PCR. The amount of RNA loaded into the reaction mixture at dilution 100 corresponds to 5 μg. Negative controls (lanes N) consisted of RNA extracted from brain tissue from uninfected subjects, and positive/sensitivity controls (lanes P) consisted of 103 genomic eq of the correct synthetic strand mixed with 5 μg of RNA from an uninfected subject.
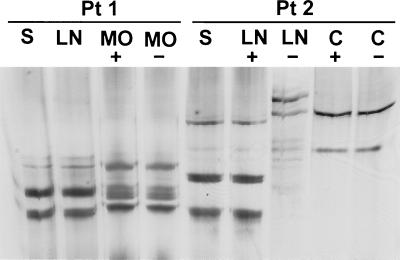
In the next step, viral sequences amplified from different parts of brain were compared by the SSCP assay with one another and with respective viral sequences amplified from serum. In patients 4 to 6, in whom viral negative strands were not detected in brain tissue, all the band patterns were identical. Similarly, no differences were observed for patient 3. However, in patients 1 and 2, negative- and positive-strand viral sequences recovered from medulla oblongata and cerebellum, respectively, were different from those derived from serum (Fig. 3). Importantly, in the latter patient cerebellum-derived sequences were identical to the viral negative strand amplified from the lymph node. The discrepancy between lymph node-derived positive and negative HCV RNA strands can be explained by the fact that the former represents in large part serum-derived contamination and the latter represents indigenous replicating virus. To lower the risk that these results are artifactual, all SSCP analysis was duplicated in an independent experiment using new RNA template.
FIG. 3.
(A) Analysis by SSCP of 5′UTR HCV sequences amplified from serum (S) and various autopsy brain tissue samples from patients 1 to 3 (Pt 1 to Pt 3). The following brain tissue samples were examined: cerebral cortex (CC), subcortical white matter (WM), nucleus lentiformis (NL), cerebellum (C), and medulla oblongata (MO). In patient 1 only medulla oblongata was available for study, while in patient 3 HCV RNA from medulla oblongata could not be amplified. In patients 1 and 2, mediastinal lymph nodes (LN) were also available for analysis. HCV RNA negative strands were detected in medulla oblongata in patient 1, in lymph node and cerebellum in patient 2, and in subcortical white matter in patient 3. The presence of identical and dissimilar viral sequences in the analyzed samples was verified by direct sequencing. Symbols: +, positive strand; −, negative strand. (B) Analysis by SSCP of 5′UTR HCV sequences amplified from serum (S) and various autopsy brain tissue samples from patients 4 to 6. Abbreviations denoting various analyzed tissues are the same as those for panel A. HCV RNA negative strands were not detected in any of the samples. The presence of identical viral sequences in the analyzed samples was verified by direct sequencing.
Direct sequencing confirmed the presence of identical master sequences in serum and brain tissue in patients 3 to 6. However, both positive- and negative-strand “master” sequences recovered from medulla and cerebellum of patients 1 and 2, respectively, differed by several nucleotide substitutions from the serum consensus sequences (Fig. 4). Thus, in two out of three patients in whom HCV negative-strand RNA was found in the CNS, sequences amplified from brain tissue were different from those circulating in serum.
FIG. 4.
Nucleotide sequence alignment of 5′UTR fragments of HCV recovered from serum (S) and brain tissue samples from patients 1 to 3 (Pt1 to Pt3). Sequences are compared with the prototype sequence published by Choo et al. (7) shown on the top line. Symbols and abbreviations: -, sequence identity; +, positive strand; −, negative strand; MO, medulla oblongata; WM, subcortical white matter; LN, lymph node; C, cerebellum.
To determine the genotype of infecting strains, the NS5 region of HCV was amplified from all analyzed samples. When serum samples were analyzed, all patients were found to be infected with genotype 1b strains. In patients 4 to 6, the genotypes of strains amplified from the CNS were concordant with those of strains circulating in serum. However, in patient 1 the viral strain found in medulla oblongata was classified as type 3a, while in patient 2 the strain found in cerebellum was classified as type 1a.
To further clarify the issue of different HCV genotypes in the CNS and circulation in patients 1 and 2, we conducted SSCP analysis of NS5 region sequences amplified from serum, lymph nodes, and pertinent brain structures. Positive and negative strands were amplified using the Tth-based assay. As can be seen in Fig. 5, similar to the results of 5′UTR analysis, positive and negative strands amplified from medulla oblongata and cerebellum in patients 1 and 2, respectively, were different than those amplified from serum. In addition, in patient 2 the pattern for the positive strand from the lymph node was identical to the serum pattern, while the negative strand’s pattern resembled the pattern found in the brain (some additional bands present in neither the serum- nor brain-derived viral sequences were also present). Direct sequencing of the viral negative strand amplified from this lymph node allowed its classification as type 1a, while the positive strand was classified as type 1b.
FIG. 5.
Analysis by SSCP of NS5 region sequences amplified from serum (S), lymph node (LN), and brain tissue samples from patients 1 and 2. Symbols and abbreviations: MO, medulla oblongata; C, cerebellum; +, positive strand; −, negative strand.
In patients 1 and 2, HCV strains replicating in the CNS and those circulating in serum were different and classified as belonging to different genotypes. However, the other strain could have been present below the sensitivity level of direct sequencing (20 to 25%) and even that of SSCP (∼3%). A commonly used alternative, sequencing of cloned PCR products, would require a large number of clones to be processed and sequenced, making it laborious and thereby impractical.
We decided to use sequence-specific primers that would allow specific amplification of one sequence from the background of other sequences. This strategy takes advantage of the observation that mismatches localized at the 3′ terminus of the primer can dramatically decrease amplification efficiency (14, 26). However, as relatively few differences were present between the brain-derived and serum-derived strains in the 5′UTR, the analysis was conducted on the NS5 region. We previously used this approach to analyze virological outcome in cases of infection with multiple HCV strains (21).
In both cases, strain-specific primers were designed to match either the serum- or brain-derived sequence but to provide a 3′ end mismatch with respect to the other strain (Fig. 6). Thus, these primers should preferentially amplify only one of the strains present. To provide the control template necessary to check the specificity of the reactions, the first-round PCR product representing either the serum- or brain-derived viral sequence was end point diluted so that no more than one in five reactions was positive when amplified with the second round of primers. In this case, the second-round PCR product would be for the most part derived from single template copies, thus ensuring that it is homogenous (32). The sequence of these control templates was ascertained by direct sequencing.
FIG. 6.
Nucleotide sequence alignment of the NS5 region fragments of HCV recovered from patients 1 and 2 (Pt 1 and Pt 2) from serum (S) and autopsy brain tissue. In patient 1 viral fragments recovered from medulla oblongata (MO) and in patient 2 sequences recovered from cerebellum (C) were different from those found in respective serum. Sequence differences between the serum- and brain-derived strains were exploited to design strain-specific primers which would allow specific amplification of one strain from the background of the other strain. The underlined sequence segments show the location of the strain-specific primers. The nucleotide numbering system follows that of the type 1a wild-type strain described by Choo et al. (8).
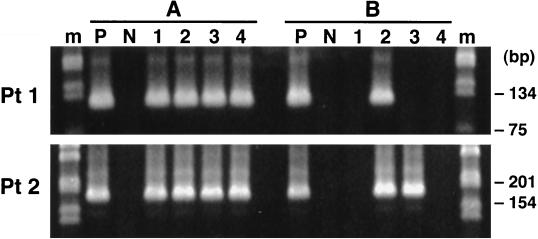
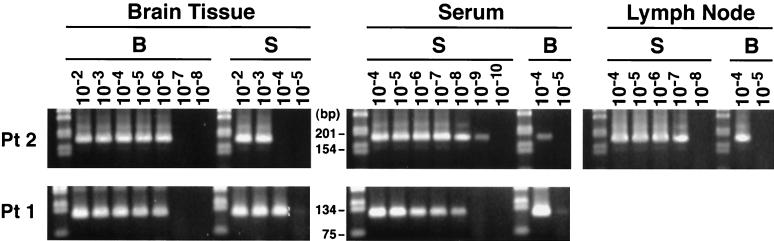
As seen in Fig. 7, in both patients serum-specific sequences were detected in the CNS and brain-specific sequences were detected in serum. However, the latter detection was not uniform from experiment to experiment, most likely due to stochastic phenomena related to low-copy template number. All positive reactions were sequenced directly, and sequence analysis confirmed that indeed the strain-specific PCR detected the proper strain in each case. By analysis of serial dilutions of the PCR products, it was determined that the titers of the minor strains were several logs lower than the titers of the major strains (Fig. 8).
FIG. 7.
Specific detection of serum-derived HCV sequences in brain tissue (A) and of brain-derived sequences in serum (B) in two patients with evidence of viral replication in the CNS. One microgram of RNA extracted from medulla oblongata (patient 1) (Pt1) and cerebellum (patient 2) (Pt2) or RNA corresponding to 100 μl of serum was subjected to 35 cycles of RT-PCR, after which the product was diluted 1:10 and 1 μl was amplified for another 35 cycles with strain-specific primers. Each reaction was repeated in 4 independent experiments (lanes 1 to 4). As can be seen, serum-derived sequences were detected from the background of brain-derived sequences in all four independent experiments, while the brain-derived sequences were detected from the background of serum-derived sequences less uniformly. The RT-PCR products were sequenced and it was determined that they matched the sequences they were designed to amplify. The positive controls (P) contain approximately 104 to 105 template copies (as determined by optical density readings) of the correct template, while negative controls (N) contain approximately 1010 template copies of the incorrect template. Samples were analyzed by agarose gel electrophoresis (3% NuSieve). Lane m, 1-kb molecular ladder (Gibco/BRL).
FIG. 8.
The determination of the proportion of brain- and serum-derived sequences in serum and brain tissue from patient 1 and 2 (Pt 1 and Pt 2). In addition, in patient 2 lymph node was also studied. One microgram of RNA extracted from medulla oblongata (patient 1) and cerebellum (patient 2) or RNA corresponding to 100 μl of serum was subjected to 20 cycles of RT-PCR, after which the product was serially diluted 1:10 and 1 μl was amplified for another 35 cycles with primers specific for the serum (S)- or brain (B)-derived strain. As seen, in brain tissue the approximate ratio of serum- to brain-derived strains was 1:102 to 1:103, while in serum the brain-derived virus was present at a level 3 to 5 logs lower than that of the major serum-derived virus. In the lymph node in patient 2 the ratio between the two strands was approximately 3 logs. Leftmost lane on each gel, 1-kb molecular ladder (Gibco/BRL).
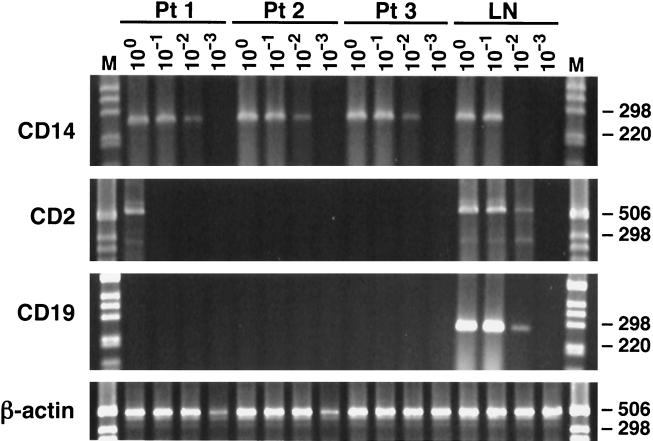
There remains a theoretical possibility that the detectable HCV RNA negative strands were originating in peripheral blood mononuclear cells (PBMC) present in contaminating blood. To exclude the PBMC as the significant source of HCV negative-strand RNA, we conducted mRNA phenotyping by RT-PCR of cells present in the implicated brain tissue from patients 1 to 3. Tested phenotypes were CD2 (T cells), CD19 (B cells), and CD14 (monocytes/macrophages). As can be seen in Fig. 9, expression of CD14 was found in all three brain samples, CD19 was not detected, and CD2 expression was found only in the brain tissue sample from patient 1, but its titer was low. Importantly, in the latter patient viral negative strands were not detected in the lymph node, which makes it even less likely that lymphoid cells in blood were the direct source of viral replicative forms detectable in his brain.
FIG. 9.
mRNA phenotyping by RT-PCR of cells present in brain tissue from patients 1 to 3 (Pt 1 to Pt 3). Tested phenotypes were CD2 (T cells), CD14 (moncytes/macrophages), and CD19 (B cells). The studied tissues were medulla oblongata, cerebellum, and subcortical white matter in patients 1, 2, and 3, respectively. In addition, as a positive control, lymph node (LN) from patient 2 was analyzed. Tenfold serial dilutions of extracted RNA were tested; the amount of RNA loaded into the reaction mixture at dilution 100 corresponds to 1 μg. The expected product sizes were 266 bp for CD14, 547 bp for CD2, and 277 bp for CD19. As can be seen, expression of CD14 was found in all three brain samples, CD19 was not detected, and CD2 expression was found in the brain tissue sample from patient 1 (but the titer was low). To check the integrity of isolated RNA and to detect contaminating genomic DNA, β-actin primers specific for two different exons separated by an intron were used. Amplification of genomic DNA results in a 607-bp product (not seen), while amplification of cDNA results in a 513-bp product.
DISCUSSION
The findings of a recently published study demonstrated the common presence of HCV replication in blood macrophages/monocytes in HIV-infected subjects (16). As brain microglia cells are essentially tissue-resident macrophages of blood monocytic origin (8), we hypothesized that they could support HCV replication as well. The present study supports this conjecture, as HCV replicative forms were detected in the CNS of three out of six studied patients, and in two of these subjects viral sequences in serum and the CNS differed enough to be classified as belonging to different genotypes. These two independent lines of evidence, viral sequence differences and the presence of negative-strand RNA, argue for the genuine presence of HCV replication in the CNS, most likely in cells of the macrophage/monocyte lineage. Moreover, negative-strand HCV RNA titers were 1 log lower than titers of the positive strand, which is the same proportion as that found commonly in the liver (15, 19). The assumption about lymphoid origin of infected cells is supported by the observation that in patient 2 the strain replicating in the CNS, while being different from that circulating in serum, was the same genotype as the one replicating in the lymph node.
An important question is that of how the HCV got into the CNS. In theory, HCV could gain access to the brain by way of cerebrospinal fluid as occurs in visna virus infection (12). Alternatively, and more likely, neuroinvasion is related to trafficking of infected cells of monocyte/macrophage lineage through the blood-brain barrier, in a process similar to that postulated for HIV-1 infection (28, 35). Subsequently, there could be a secondary spread of HCV to permissive resident microglial cells within the brain. Replication in the CNS could be facilitated by immunosuppression, some degree of which was likely to be present in all of our patients. This possibility is supported by the observations that while HCV negative-strand RNA is rarely detected in PBMC from normal subjects (15, 24), it is commonly found in HIV-coinfected patients or liver transplant recipients (16, 30). Moreover, HCV replication was demonstrated in hematopoietic cells inoculated into severe combined immunodeficiency mice (5). However, HCV replication in bone marrow was also found in some obviously immunocompetent subjects (29, 31).
The observed concomitant infection of the same host by two different HCV strains, each replicating in a different compartment, is probably the consequence of coinfection or superinfection with strains manifesting different tropisms for different cells. For example, it has been demonstrated for lymphocytic choriomeningitis virus that strains differing by a single amino acid substitution, when inoculated together into a mouse, are competitively selected either by the liver and spleen or by neurons (9). We have recently reported that infection with multiple HCV strains results in rapid predominance of a single strain that presumably replicates in the liver and that all other strains are either eliminated or constitute a tiny fraction of circulating virions (21). It is thus possible that HCV adaptation to an extrahepatic niche may be a strategy to elude competitive exclusion by the dominant strain that replicates in the liver.
In the present study the HCV negative-strand RNA was detected in one site of the brain in each of the three patients. However, as the titer of the replicating virus seemed to be low, replication may have been present at other brain sites but below the level of detection. This could be compounded by the fact that the studied biological material constituted autopsy tissues which were obtained within 36 h after death and therefore some RNA might have been degraded. Interestingly, HIV-1, which undoubtedly infects brain microglia cells, is also not uniformly detected throughout the brain, even in the same subjects (2, 4).
In summary, we found evidence of HCV replication in CNS autopsy samples from three out of six studied patients. However, the consequences of this are presently unclear.
Acknowledgments
This work was supported in part by National Institutes of Health grant DA13760.
REFERENCES
- 1.Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, R. E. Sampliner, E. L. Meeks, and M. J. Beach. 1992. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N. Engl. J. Med. 327:1899–1905. [DOI] [PubMed] [Google Scholar]
- 2.Bagasra, O., E. Lavi, L. Bobroski, K. Khalili, J. P. Pestaner, R. Tawadros, and R. J. Pomerantz. 1996. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS 10:573–585. [DOI] [PubMed] [Google Scholar]
- 3.Bolay, H., F. Soylemezoglu, G. Nurlu, S. Tuncer, and K. Varli. 1996. PCR detected hepatitis C virus genome in the brain of a case with progressive encephalomyelitis with rigidity. Clin. Neurol. Neurosurg. 98:305–308. [DOI] [PubMed] [Google Scholar]
- 4.Brew, B. J., M. Rosenblum, K. Cronin, and R. W. Price. 1995. AIDS dementia complex and HIV-1 brain infection: clinical-virological correlations. Ann. Neurol. 38:563–570. [DOI] [PubMed] [Google Scholar]
- 5.Bronowicki, J. P., M. A. Loriot, V. Thiers, Y. Grignon, A. L. Zignego, and C. Brechot. 1998. Hepatitis C virus persistence in human hematopoietic cells injected into SCID mice. Hepatology 28:211–218. [DOI] [PubMed] [Google Scholar]
- 6.Caudai, C., D. Maimone, P. Almi, P. Annunziata, I. Bastianoni, C. A. Boggiano, G. C. Guazzi, M. Padula, and P. E. Valensin. 1997. The potential role of hepatitis C virus in the pathogenesis of the neurological syndrome in chronic hepatitis C. Gut 41:411–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, E. J., T. D. Foster, and W. E. Thomas. 1994. Cellular forms and functions of brain microglia. Brain Res. Bull. 34:73–78. [DOI] [PubMed] [Google Scholar]
- 9.Dockter, J., C. F. Evans, A. Tishon, and M. B. Oldstone. 1996. Competitive selection in vivo by a cell for one variant over another: implications for RNA virus quasispecies in vivo. J. Virol. 70:1799–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein, L. G., C. Kuiken, B. M. Blumberg, S. Hartman, L. R. Sharer, M. Clement, and J. Goudsmit. 1991. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology 180:583–590. [DOI] [PubMed] [Google Scholar]
- 11.Fujita, H., Y. Chuganji, M. Yaginuma, M. Momoi, and T. Tanaka. 1999. Case report: acute encephalitis immediately prior to acute onset of hepatitis C virus infection. J. Gastroenterol. Hepatol. 14:1129–1131. [DOI] [PubMed] [Google Scholar]
- 12.Haase, A. T. 1986. Pathogenesis of lentivirus infections. Nature 322:130–136. [DOI] [PubMed] [Google Scholar]
- 13.Heckmann, J. G., C. Kayser, D. Heuss, B. Manger, H. E. Blum, and B. Neundorfer. 1999. Neurological manifestations of chronic hepatitis C. J. Neurol. 246:486–491. [DOI] [PubMed] [Google Scholar]
- 14.Kwok, S., D. E. Kellogg, N. McKinney, D. Spasic, L. Goda, C. Levenson, and J. J. Sninsky. 1990. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 18:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanford, R. E., D. Chavez, F. V. Chisari, and C. Sureau. 1995. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J. Virol. 69:8079–8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskus, T., M. Radkowski, A. Piasek, M. Nowicki, A. Horban, J. Cianciara, and J. Rakela. 2000. Hepatitis C virus in lymphoid cells of patients coinfected with human immunodeficiency virus type 1: evidence of active replication in monocytes/macrophages and lymphocytes. J. Infect. Dis. 181:442–448. [DOI] [PubMed] [Google Scholar]
- 17.Laskus, T., M. Radkowski, L. F. Wang, S. J. Jang, H. Vargas, and J. Rakela. 1998. Hepatitis C virus quasispecies in patients infected with HIV-1: correlation with extrahepatic viral replication. Virology 248:164–171. [DOI] [PubMed] [Google Scholar]
- 18.Laskus, T., M. Radkowski, L. F. Wang, M. Nowicki, and J. Rakela. 2000. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J. Virol. 74:1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskus, T., M. Radkowski, L. F. Wang, H. Vargas, and J. Rakela. 1998. Detection of hepatitis G virus replication sites by using highly strand-specific Tth-based reverse transcriptase PCR. J. Virol. 72:3072–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskus, T., M. Radkowski, L. F. Wang, H. Vargas, and J. Rakela. 1997. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J. Virol. 71:7804–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskus, T., L. F. Wang, M. Radkowski, H. Vargas, M. Nowicki, J. Wilkinson, and J. Rakela. 2001. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J. Virol. 75:2059–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerat, H., S. Rumin, F. Habersetzer, F. Berby, M. A. Trabaud, C. Trepo, and G. Inchauspe. 1998. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood 91:3841–3849. [PubMed] [Google Scholar]
- 23.Maggi, F., M. Giorgi, C. Fornai, A. Morrica, M. L. Vatteroni, M. Pistello, G. Siciliano, A. Nuccorini, and M. Bendinelli. 1999. Detection and quasispecies analysis of hepatitis C virus in the cerebrospinal fluid of infected patients. J. Neurovirol. 5:319–323. [DOI] [PubMed] [Google Scholar]
- 24.Mellor, J., G. Haydon, C. Blair, W. Livingstone, and P. Simmonds. 1998. Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J. Gen. Virol. 79:705–714. [DOI] [PubMed] [Google Scholar]
- 25.Morsica, G., M. T. Bernardi, R. Novati, C. Uberti Foppa, A. Castagna, and A. Lazzarin. 1997. Detection of hepatitis C virus genomic sequences in the cerebrospinal fluid of HIV-infected patients. J. Med. Virol. 53:252–254. [DOI] [PubMed] [Google Scholar]
- 26.Nassal, M., and A. Rieger. 1990. PCR-based site-directed mutagenesis using primers with mismatched 3′-ends. Nucleic Acids Res. 18:3077–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Origgi, L., M. Vanoli, A. Carbone, M. Grasso, and R. Scorza. 1998. Central nervous system involvement in patients with HCV-related cryoglobulinemia. Am. J. Med. Sci. 315:208–210. [DOI] [PubMed] [Google Scholar]
- 28.Price, R. W., J. Sidtis, and M. Rosenblum. 1988. The AIDS dementia complex: some current questions. Ann. Neurol. 23:S27–S33. [DOI] [PubMed] [Google Scholar]
- 29.Radkowski, M., J. Kubicka, E. Kisiel, J. Cianciara, M. Nowicki, J. Rakela, and T. Laskus. 2000. Detection of active hepatitis C virus and hepatitis G virus/GB virus C replication in bone marrow in human subjects. Blood 95:3986–3989. [PubMed] [Google Scholar]
- 30.Radkowski, M., L. F. Wang, H. E. Vargas, J. Rakela, and T. Laskus. 1998. Detection of hepatitis C virus replication in peripheral blood mononuclear cells after orthotopic liver transplantation. Transplantation 66:664–666. [DOI] [PubMed] [Google Scholar]
- 31.Sansonno, D., C. Lotesoriere, V. Cornacchiulo, M. Fanelli, P. Gatti, G. Iodice, V. Racanelli, and F. Dammacco. 1998. Hepatitis C virus infection involves CD34+ hematopoietic progenitor cells in hepatitis C virus chronic carriers. Blood 92:3328–3337. [PubMed] [Google Scholar]
- 32.Simmonds, P., P. Balfe, J. F. Peutherer, C. A. Ludlam, J. O. Bishop, and A. J. Brown. 1990. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J. Virol. 64:864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmonds, P., E. C. Holmes, T. A. Cha, S. W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391–2399. [DOI] [PubMed] [Google Scholar]
- 34.Thoma, S. J., C. P. Lamping, and B. L. Ziegler. 1994. Phenotype analysis of hematopoietic CD34+ cell populations derived from human umbilical cord blood using flow cytometry and cDNA-polymerase chain reaction. Blood 83:2103–2114. [PubMed] [Google Scholar]
- 35.Zheng, J., and H. E. Gendelman. 1997. The HIV-1 associated dementia complex: a metabolic encephalopathy fueled by viral replication in mononuclear phagocytes. Curr. Opin. Neurol. 10:319–325. [PubMed] [Google Scholar]