Abstract
Proteasome-dependent degradation of ubiquitinated proteins plays a key role in many important cellular processes. Ubiquitination requires the E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and frequently a substrate-specific ubiquitin protein ligase (E3). One class of E3 ubiquitin ligases has been shown to contain a common zinc-binding RING finger motif. We have previously shown that herpes simplex virus type 1 ICP0, itself a RING finger protein, induces the proteasome-dependent degradation of several cellular proteins and induces the accumulation of colocalizing conjugated ubiquitin in vivo. We now report that both full-length ICP0 and its isolated RING finger domain induce the accumulation of polyubiquitin chains in vitro in the presence of E1 and the E2 enzymes UbcH5a and UbcH6. Mutations within the RING finger region that abolish the in vitro ubiquitination activity also cause severe reductions in ICP0 activity in other assays. We conclude that ICP0 has the potential to act as an E3 ubiquitin ligase during viral infection and to target specific cellular proteins for destruction by the 26S proteasome.
Herpes simplex virus type 1 (HSV-1) is a significant human pathogen whose biological and clinical importance is emphasized by its ability to attain and reactivate from a latent state in sensory neurons (reviewed in reference 16). The mechanisms that control the balance between the lytic and latent states are of considerable interest yet are incompletely understood. Our past studies have concentrated on the functions and mechanisms of action of HSV-1 immediate-early regulatory protein ICP0, which is required for the efficient initiation of lytic cycle gene expression, reactivation of quiescent virus in cultured cells, and reactivation of latent virus in mouse models (for reviews, see references 5 and 15; see also references 18 and 19).
ICP0 is being actively studied in a number of laboratories, and a wide spectrum of possible functions, interactions, and mechanisms of action are being revealed. Early transfection studies showed that ICP0 is able to increase the expression of a wide variety of genes in cotransfected cells, and this effect does not depend on specific promoter sequences. Since ICP0 does not bind directly to DNA (13), it is likely that it functions via interactions with other proteins. Recent studies have proposed a number of possible interactions, including USP7 (a ubiquitin-specific protease) (11), cyclin D3 (25), elongation factor EF-1δ (23), the transcription factor BMAL1 (24), and the major HSV-1 transcriptional regulator ICP4 (42). ICP0 has also been suggested to activate cdk4 and to stabilize both cyclin D1 and cyclin D3 (40). Whatever the significance of these varied observations, it is now generally accepted that a major biological activity of ICP0 causes the disruption of specific nuclear structures known as ND10 or promyelocytic leukemia (PML) nuclear bodies in a process which correlates with the ability of ICP0 to stimulate viral infection and reactivation from quiescence (reviewed in reference 5).
We have investigated in some detail the mechanism by which ICP0 achieves the destruction of ND10. It has been shown that ICP0 induces the proteasome-dependent degradation of two major components of ND10, PML itself and Sp100, particularly their isoforms that are covalently modified by the ubiquitin-like protein SUMO-1 (1, 8, 31, 33). ICP0 alone is sufficient to abrogate the conjugation of SUMO-1 to PML nuclear bodies and then to induce its degradation (31, 33), and this activity accounts for the disruption of ND10. We have also detected other substrates for ICP0-induced degradation, including the catalytic subunit of DNA protein kinase (35) and the centromere proteins CENP-C (7) and CENP-A (27). Loss of the latter two proteins from the cell results in severe mitotic defects and must explain at least in part the cytotoxicity of ICP0 (7, 27). Since the proteasome inhibitor MG132 interferes with the ability of ICP0 to stimulate viral infection and reactivation from quiescence (14), it is likely that these effects on cellular proteins reflect a major biological function of ICP0. This has led to an investigation of the mechanisms by which ICP0 might target specific proteins for destruction.
Studies on the domains of ICP0 important for its function have consistently revealed that a zinc-binding RING finger domain located near the N terminus of the 775-residue protein plays a crucial role in its activities (reviewed in reference 5). RING finger domains are present in a wide variety of proteins, both viral and cellular, and in the recent past it has been found that several RING finger proteins take part in the ubiquitin-proteasome pathway by acting as E3 ubiquitin ligases (reviewed in references 17 and 21). Briefly, the ubiquitin cycle involves activation of ubiquitin by an enzyme known as E1 and then its transfer to a thiol ester linkage at the active site of an E2 ubiquitin conjugating enzyme. The activities and perhaps the specificities of E2 enzymes are controlled by one of many E3 ubiquitin ligases, which may comprise one or several components. Given the similarity between the ICP0 RING finger and a number of known cellular RING finger E3 ligase components and also the clear ability of ICP0 to induce proteasome-dependent degradation of selected target proteins, we have investigated whether ICP0 acts as an E3 ubiquitin ligase. We have recently shown that ICP0 induces the formation of colocalizing conjugated ubiquitin in transfected and infected cells (4). Here we demonstrate that full-length ICP0 and its isolated RING finger domain possess E3 ubiquitin ligase activity in vitro.
MATERIALS AND METHODS
Plasmids.
Plasmids of the pGEX series expressing wild-type or mutant RING finger domain regions of ICP0 were based on vector pGEX2T and derivatives. The initial parent, pGEX262, expresses glutathione S-transferase (GST) fused to the N-terminal 241 residues of ICP0 (encoded by gene IE1 exons 1 and 2) plus 21 residues encoded by sequences in intron 2. This plasmid was constructed using a cDNA fragment containing exon 1 and part of exon 2 from p110262 and a fragment of p110E11 containing the rest of exon 2 plus intron 2 sequences as far as the inserted EcoRI site in the intron of p110E11 (downstream of the stop codon used for the 262-residue peptide). Plasmid pGEX241 was derived from pGEX262 by PCR of the 3′ side of exon 2 to introduce a stop codon after residues 241, thus removing the intron-encoded residues from the expressed ICP0 peptide. Plasmid pGEX211 was constructed by deleting the coding sequences between the KpnI site in codon 212 and the stop codon of plasmid pGEX241. The mutant derivatives of pGEX262 and pGEX241 were constructed by inserting exon 2 fragments from previously characterized mutant plasmids in place of the wild-type fragment. The parents of these plasmids were originally described as follows: p110262 (9); p110K144E, p110Q148E, p110N151D, p110DR36c, and p110DR40 (12, 32); p110F1, p110R2, p110E8, p110E11, and p110E13, (2); and p110FXE, p110D1, and p110D22 (3).
Plasmid pEGFP-110wt expresses full-length ICP0 as a green fluorescent protein (GFP) fusion protein from vector pEGFP-C1 (26). Plasmid pCI-rtagUbcH5a contains the UbcH5a cDNA with an N-terminal UL30 tag (34) inserted into vector pCIneo, and pCMV2-FLAG-UbcH6 expresses Flag-tagged UbcH6 from a cytomegalovirus (CMV) vector.
Expression and purification of GST fusion proteins.
Plasmids of the pGEX series were transformed into BL21(DE3) pLysS bacteria and then fresh colonies were inoculated into 100 ml of yeast-tryptone broth and grown to mid-log phase. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.1 mM and then incubation was continued for 2 h. Bacteria were harvested by centrifugation at 3,000 rpm in a Sorvall RT 6000B tabletop centrifuge for 15 min, resuspended in 2 ml of phosphate-buffered saline (PBS), and lysed by probe sonication. NP-40 was added to 0.1% (vol/vol) and then cell debris were removed by centrifugation at 8,000 rpm in a Sorvall SS34 rotor for 20 min. The soluble protein extracts were stored at −20°C. GST fusion proteins were purified by incubating 300 μl of extract with 100 μl of glutathione-agarose beads (50% [vol/vol] in PBS) for 30 min, then washing the beads three times with PBS, and finally eluting bound proteins with 150 μl of 50 mM reduced glutathione in 250 mM Tris (pH 8.0), 0.1 M NaCl, 0.1% NP-40. Purified proteins were dialyzed against 50 mM Tris (pH 7.5), 50 mM NaCl, 2.5 mM β-mercaptoethanol where necessary and stored at −20°C.
Construction of recombinant baculoviruses and expression and purification of His-ICP0 and His-ICP0FXE.
Plasmid pUC9-myc110 (kindly provided by Wei-Li Hsu) contains the 2.2-kb ICP0 cDNA fragment modified at the 3′ end by the addition of EcoRV and HindIII sites just downstream of the ICP0 stop codon and at the 5′ end by an oligonucleotide containing an EcoRI site, an NdeI site containing an initiation codon, a myc tag sequence, and an NcoI site that links in frame with the NcoI site containing the normal initiation codon of ICP0. The ICP0 cDNA (without the myc tag) was excised from pUC9-myc110 as an NcoI-HindIII fragment and inserted between the NcoI and HindIII sites of pFastBac-HTa (Gibco BRL). The NcoI-KpnI fragment of pGEX262FXE (see above), containing the FXE RING finger deletion mutation, was isolated and inserted into pUC9-myc110 in place of the wild-type fragment and then the NcoI-HindIII fragment of the derivative was similarly inserted into pFastBac-HTa. The resultant plasmids, pFastBac-HTa-ICP0 and pFastBac-HTa-FXE, contain full-length ICP0 or ICP0-FXE open reading frames linked to N-terminal polyhistidine tags.
Recombinant baculoviruses were isolated using the Bac-To-Bac system (Gibco BRL) and grown according to the manufacturer’s guidelines. Sf21 insect cells were grown in suspension and infected for 72 h at a cell density of 106 cells/ml using a multiplicity of 2 PFU per cell. The cells were harvested by low-speed centrifugation, washed in cold PBS, and stored at −70°C until use. Cell pellets (equivalent to 75 ml of infected cell suspension) were resuspended in 5 ml of buffer A (100 mM Tris-HCl [pH 8.0], 500 mM NaCl, 10% glycerol, 20 mM β-mercaptoethanol, 1% NP-40) in the presence of a cocktail of protease inhibitors (Roche). The cell suspension was gently sonicated for 30 s in a Soni-bath to reduce viscosity, and the cells were incubated on ice for 30 min. For His-ICP0, extracts were diluted with an equal volume of 100 mM Tris-HCl (pH 8.0) before being clarified by centrifugation at 13,000 rpm in an MSE Micro Centaur microcentrifuge for 5 min at 4°C, while for His-ICP0FXE a similar dilution step was done after elution from the nickel column. The soluble supernatant fraction was mixed end-over-end with 200 μl of Ni-nitriloacetic acid agarose beads (Qiagen) for 45 min at 4°C. The beads were subsequently washed three times with 1 ml of buffer B (100 mM Tris-HCl [pH 8.0], 250 mM NaCl, 5% glycerol, 10 mM β-mercaptoethanol, 0.5% NP-40) and five times with 1 ml of buffer B containing 30 mM imidazole (pH 8.0) to remove nonspecifically bound proteins. Proteins were eluted from the beads in four aliquots of 300 μl of buffer B containing 250 mM imidazole (pH 8.0) and then stored at −70°C.
In vitro ubiquitin E3 ligase assays.
E1 ubiquitin activating enzyme was purified from HeLa cell S100 extracts by ubiquitin affinity chromatography, or it was expressed with an N-terminal polyhistidine tag by a recombinant baculovirus and purified from extracts of infected cells by nickel affinity chromatography. The accession numbers of the E2 ubiquitin conjugating enzyme cDNAs that were used in these experiments are as follows: Ubch2b, P23567; Ubch5a, P51668; Ubch6, P51965; Ubch7, P51966; Ubch10, O00762; Ubch13, Q16763; Ubch16, AF161499; Ubch18, P56554; and Cdc34a, P49427. The E2 enzymes were expressed in recombinant Escherichia coli as polyhistidine or GST fusion proteins and were purified from soluble extracts by nickel or glutathione affinity chromatography. The GST moiety was subsequently removed by thrombin cleavage. All E2 preparations were competent in forming covalent thiol ester linkages with ubiquitin and, with the exception of Ubch16 and Ubch18, had been demonstrated to have activity with cognate ubiquitin E3 ligases in other assays (data not shown). Ubiquitin was purchased from Sigma, and His-tagged ubiquitin was from Affiniti.
In vitro ubiquitin conjugation assays were carried out in a buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM ATP. The precise concentrations of the proteins and the reaction conditions used varied between assays over the following ranges: 5 to 50 nM E1, 50 to 250 nM E2, 25 to 50 μM ubiquitin, and approximately 50 ng of ICP0 full-length protein or 5 to 50 ng of GST-ICP0 fusion proteins. The mixtures were incubated at 37 or 30°C for 30 to 60 min and then stopped with 3× gel-loading buffer and subjected to electrophoresis on 7.5% sodium dodecyl sulfate (SDS)-polyacrylamide or 4-to-12% NuPAGE gels (Invitrogen). The proteins were transferred to nitrocellulose filters by Western blotting and then probed sequentially using one or more of the following antibodies at the indicated dilutions: anti-conjugated ubiquitin monoclonal antibody FK2 (Affiniti; 1/5,000); antiubiquitin monoclonal P4D1 (Santa Cruz Biotechnology; 1/1,000); anti-ICP0 monoclonal antibody 11060 (1/2,000); antipolyhistine monoclonal directly conjugated to horseradish peroxidase (Sigma; 1/1,000). After thorough washing, the membranes were incubated with horseradish peroxidase-linked sheep anti-mouse immunoglobulin G (IgG), and bound antibody was detected using an enhanced chemiluminescence reagent (NEN) and exposure to film.
Cells, transfection, immunofluorescence, and confocal microscopy.
HEp-2 cells were grown in Dulbecco’s modified Eagles’ medium supplemented with 10% fetal calf serum and seeded onto 12-mm coverslips in 24-well dishes at a density of 105 cells per well. The cells were transfected using Lipofectamine Plus reagent (Gibco BRL) according to the manufacturer’s instructions and then stained for immunofluorescence about 24 h later. The cells were washed with PBS, fixed with formaldehyde (5% [vol/vol] in PBS containing 2% sucrose), and then permeabilized with 0.5% NP-40 in PBS with 10% sucrose. The primary antibodies were diluted in PBS containing 1% newborn calf serum. Anti-ICP0 monoclonal antibody 11060 was used at a dilution of 1/1,000, anti-UL30 tag rabbit serum r113 (34) was used at 1/5,000, and anti-FLAG tag monoclonal M2 (Sigma) was used at 1/200. After incubation at room temperature for 1 h, the coverslips were washed at least six times and then treated with secondary antibodies at the following dilutions: fluorescein isothiocyanate-conjugated goat anti-mouse IgM (Sigma; 1/100); Cy5-conjugated goat anti-rabbit IgG (Amersham; 1/500). After a further 60-min incubation, the coverslips were again washed at least six times and mounted using Citifluor AF1.
Samples were examined using a Zeiss LSM 510 confocal microscope with three lasers giving excitation lines at 633, 543, and 488 nm. The data from the channels were collected sequentially using the appropriate band-pass filters built into the instrument. Data were collected with fourfold averaging at a resolution of 1,024 by 1,024 pixels using optical slices of between 0.5 and 1 μm. The microscope was a Zeiss Axioplan utilizing a 63× oil immersion objective lens, numerical aperture 1.4. Data sets were processed using the LSM 510 software and then exported for preparation for printing using PhotoShop. Scanning conditions were adjusted to ensure that signal overlap between channels was eliminated.
RESULTS
ICP0 stimulates ubiquitin conjugating enzymes UbcH5a and UbcH6.
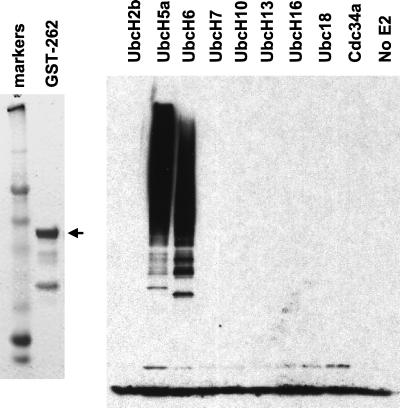
We have adopted a standard method for the detection of the E3 ubiquitin ligase activity of RING finger proteins in vitro. Previous studies have shown that while some RING finger E3 ligases (for example, c-Cbl [22]) direct the ubiquitination of specific, biologically relevant substrates in the absence of factors other than a member of the E2 family of ubiquitin conjugating enzymes, many RING finger proteins stimulate substrate-independent assembly of multiubiquitin chains in simple reaction mixes containing ubiquitin, E1 enzyme, the appropriate E2 enzyme, and the RING finger protein itself (29). A truncated ICP0 protein comprising the N-terminal 241 residues (including the RING domain) plus 21 residues derived from intron sequences (Fig. 1) was expressed as a GST fusion protein (GST-262) and purified. The 262-residue ICP0 truncation protein (ICP0-262, also known as ICP0R [36]) is expressed in limited amounts during a normal viral infection as a result of failure to splice out the second intron from the primary transcript (6), and in transfection assays it acts as an inhibitor of gene expression (36). Since both full-length ICP0 and ICP0-262 induce colocalizing conjugated ubiquitin in transfected cells (4), we suspected that these proteins might have E3 ubiquitin ligase activity in vitro. Incubation of GST-262 with all the components of a standard in vitro ubiquitination assay and a range of human enzymes revealed a strong stimulation of the activity of UbcH5a and UbcH6. The other E2 enzymes tested were not stimulated by GST-262 (Fig. 2).
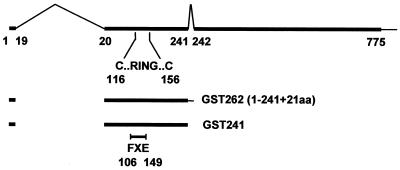
FIG. 1.
Map of the ICP0 transcript and coding region. The upper line shows the ICP0 coding region (thick line), the introns (thin inverted v-lines), and the 3′ noncoding region (thin line). The numbers refer to the codon positions at the intron junctions and the last coding triplet. The position of the RING finger in exon 2 is indicated with the locations of the first and last zinc-coordinating cysteine residues. Below the map are the ICP0 residues expressed as a GST fusion protein. GST-262 includes residues 1 to 241 encoded in exons 1 and 2 (thick line) plus 21 residues derived from intron 2 sequences. The ICP0 portion of the GST fusion protein is equivalent to ICP0-262 (ICP0R). GST-241 includes only ICP0 residues 1 to 241. The position of the RING finger deletion mutation in clones derived from mutant FXE is indicated.
FIG. 2.
Typical expression and purification of a GST-ICP0 RING finger domain fusion protein and its activity in an in vitro ubiquitin conjugation assay. GST-262 was expressed in bacteria and purified using glutathione-agarose beads as described in Materials and Methods. The left panel shows a Coomassie-stained gel of the fusion protein that migrates at approximately 60 kDa (arrow). The right panel shows the results of an in vitro ubiquitin conjugation assay, probed using antiubiquitin antibody P4D1, as described in the text. A panel of nine different E2 enzymes were screened for activity in the presence of GST-262. The related proteins UbcH5a and UbcH6 were stimulated by GST-262, but the others were not.
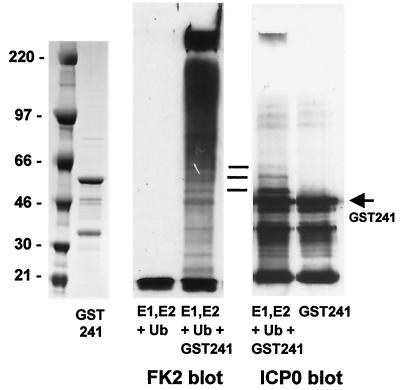
Previous work had shown that ICP0-262 could be isolated as a monoubiquitinated form from transfected cells, due to conjugation of a single ubiquitin molecule to a lysine residue encoded by the intron sequences (41). To investigate whether sequences in ICP0-262 that are not part of authentic ICP0 contribute to the E3 ligase activity, a plasmid was constructed that expresses GST-linked ICP0-241, a protein containing only residues derived from exons 1 and 2 of full-length ICP0. This protein (GST-241) was also active in the ubiquitination assay (Fig. 3), and under the conditions used in this assay much of the product is retained in the stacking gel. Reprobing of the Western blot for ICP0 demonstrated that some of the ubiquitination activity is directed against the GST-241 fusion protein itself (Fig. 3), a common finding in similar RING finger-dependent assays (29). However, comparison of the intensities of the conjugated ubiquitin and apparently ubiquitinated GST-241 signals suggests that the vast majority of the conjugated ubiquitin signal is not linked to GST-241. Similar analysis of the other components in the mixture (E1 and UbcH5a) showed that both components formed monoubiquitinated thiol ester derivatives as expected but that neither was significantly modified into the very-low-mobility ubiquitin conjugates detected by FK2 (data not shown). Therefore, it is likely that much of the conjugated ubiquitin product comprises unanchored polyubiquitin chains whose molecular weight or branched structure precludes efficient gel migration.
FIG. 3.
Activity and autoubiquitination of a GST fusion protein including the first 241 residues of ICP0. The left panel shows a Coomassie-stained gel of purified GST-241. The middle panel shows the in vitro ubiquitination activity of the purified GST-241 protein (right) next to a control sample without added GST-241 (left), as detected with antibody FK2 that detects conjugated ubiquitin. The right panel shows a portion of the same filter reprobed to detect GST-241 after incubation with E1, UbcH5a, and ubiquitin (left), next to a control untreated sample (right). The horizontal lines indicate modified forms of GST-241 that comigrate with minor conjugated ubiquitin bands and that appear only after incubation in the in vitro conjugation reaction mixture.
Titration of the components of the standard reaction mixtures revealed that E1 and E2 concentrations of 2 and 50 nM (respectively) were sufficient, while GST-241 activity could be detected with as little as 5 ng of protein (10 nM). The reactions analyzed in Fig. 3 had been incubated for 60 min, but a time course experiment produced detectable product after only a 5-min incubation with 50 nM GST-241 (data not shown). Therefore, the RING finger domain of ICP0 appears to have a very strong E3 ligase activity in vitro.
ICP0 E3 ligase activity requires the RING finger and distal C-terminal sequences.
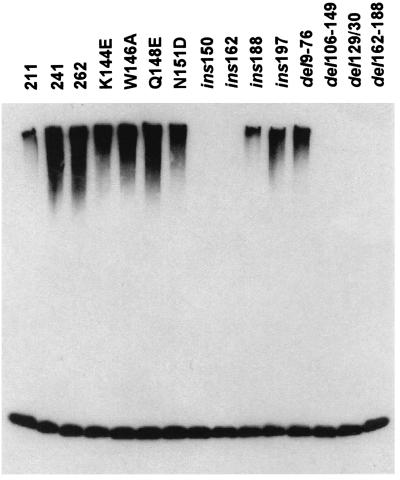
A number of well-characterized mutations in the ICP0 RING finger and surrounding regions (Table 1) were transferred to the GST-262 or GST-241 backgrounds and tested for ubiquitin E3 ligase activity with UbcH5a. The results (Fig. 4 and Table 1) indicated that sequences both within and on the C-terminal side of the RING finger are important for the activity. Neither truncation of the C-terminal end at residue 211 nor removal of residues 9 to 76 eliminated the activity, which isolates the active moiety to the 135 residues between 77 to 211. Deletion of most of the RING finger (FXE; del106-149), two residues in loop 1 in the RING structure (del129/130), and insertion of four residues in the helix region (ins150) all eliminated the activity, while some insertion and deletion mutations on the C-terminal side of the RING (ins162, del162-188) also abrogated the activity. All of the mutations that abolish the in vitro E3 ligase activity of ICP0 have previously been shown to interfere with the ability of ICP0 to stimulate gene expression, induce the degradation of cellular proteins, or induce the formation of colocalizing conjugated ubiquitin (Table 1).
TABLE 1.
Details of the GST fusion protein constructs used in this studyf
| Plasmida | Derivationb | Activationc | FK2d | E3 ligasee |
|---|---|---|---|---|
| pGEX262 | p110262 | (+++) | ++ | + |
| pGEX262del106-149 | p110FXE | − | − | − |
| pGEX241 | This study | (+++) | ND | + |
| pGEX211 | This study | (+++) | ND | + |
| pGEX241K144E | p110K144E | − | − | + |
| pGEX241W146A | p110DR36c | +++ | +++ | + |
| pGEX241Q148E | p110Q148E | ++ | + | + |
| pGEX241N151D | p110N151D | + | − | + |
| pGEX241ins150 | p110F1 | − | − | − |
| pGEX241ins162 | p110R2 | − | − | − |
| pGEX241ins188 | p110E8 | − | − | + |
| pGEX241ins197 | p110E13 | − | − | + |
| pGEX241del9-76 | p110D1 | ++ | ND | + |
| pGEX241del129/130 | p110DR40 | − | − | − |
| pGEX241del162-188 | p110D22 | − | − | − |
The names of the various GST fusion proteins expressed from pGEX vector plasmids indicate the background ICP0 segment (262 is residues 241 plus 21 residues encoded in intron 2; 241 is residues 1 to 241; 211 is residues 1 to 211) plus the various deletion, insertion, or substitution mutations.
The names of the plasmids (as referenced in Materials and Methods) that were used as sources of the mutations present in the pGEX series plasmids.
Activation refers to the ability of otherwise full-length versions of ICP0 carrying these lesions to activate gene expression from either the ICP6 promoter (mutants FXE, K144E, W146A, Q148E, and N151D [12]), the tk promoter (mutant DR40 [32]), or the gD promoter (all other mutants [2, 3]). −, less than 10% of wild-type ICP0 activity; +, 10 to 40%; ++, 40 to 70%; +++, >70%. The entries shown in parentheses indicate that these proteins contain segments of wild-type ICP0.
The effects of these mutations on the ability of ICP0 to induce colocalizing conjugated ubiquitin in transfected and (where known) infected cells, as described in reference 4. The scoring system is a qualitative comparison of the intensity of induced signal compared to a wild-type, full-length ICP0 control. −, undetectable activity. ND, not done.
Ability of the protein expressed by the relevant pGEX plasmid to stimulate the production of high-molecular-weight ubiquitin chains in vitro.
The effects of the mutations on activation of gene expression and production of colocalizing conjugated ubiquitin induced by full-length ICP0 in vivo are compared with the results of in vitro ubiquitin E3 ligase assays using GST fusion proteins containing the same mutations.
FIG. 4.
Comparison of the effects of mutations in the first and second exons of ICP0 in the in vitro conjugation reaction. GST fusion proteins based on either GST-262 or GST-241 were purified after bacterial expression. The locations and phenotypes of the mutations are shown in Table 1. All the proteins had similar expression, solubility, and stability properties (data not shown). The products of the in vitro ubiquitin conjugation reaction using E1, ubiquitin, UbcH5a, and added GST fusion protein were detected by probing a Western blot with the antiubiquitin antibody P4D1.
Interestingly, point mutations within the RING finger helix (K144E, W146A, Q148E, and N151D) and some insertion mutations in the C-terminal portion of the fusion protein (ins188 and ins197) had little or no significant effect on the in vitro activity. Mutant W146A had no detectable phenotype in other assays (32), but the others showed a spectrum of deleterious effects on the ability of ICP0 to stimulate gene expression and disrupt ND10 in transfection assays (2, 12). Of particular interest is the insertion ins197, which lies within a region implicated in binding to cyclin D3 (39). Although a single amino acid substitution at position 198 was reported not to affect the ability of ICP0 to disrupt ND10 (40), we found in transfection assays that the more invasive ins197 mutation removed the ability of ICP0 to disrupt ND10 and centromeres (R. D. Everett, unpublished data), and the mutant protein failed to induce the formation of colocalizing conjugated ubiquitin (4). Given that deletion of sequences downstream of residue 211 does not completely remove the in vitro E3 ligase activity, it is likely that ins197 lies towards the C-terminal side of the active E3 ligase domain.
It is possible that the in vitro ubiquitin E3 ligase assay is not sufficiently flexible to detect any reduction in activity caused by the point mutations in the RING finger helix region, because these generally have less-pronounced phenotypes than the insertions and deletions in transfected and infected cells. However, it is likely that the mutations that compromise in vivo but not in vitro function affect residues that are involved in the assembly of a substrate- or conformation-specific activity in the authentic active complex in vivo. For example, some RING finger E3 ligase proteins require several additional components for substrate-specific activity in vivo (reviewed in reference 21; see also reference 30). Thus, the mutations in the RING finger helix region and towards the C-terminal side of the exon 2 residues of ICP0 that affect its in vivo but not in vitro activities are likely to be compromising interactions with other cellular components that are required for specific, regulated in vivo activity.
Full-length ICP0 has E3 ligase activity.
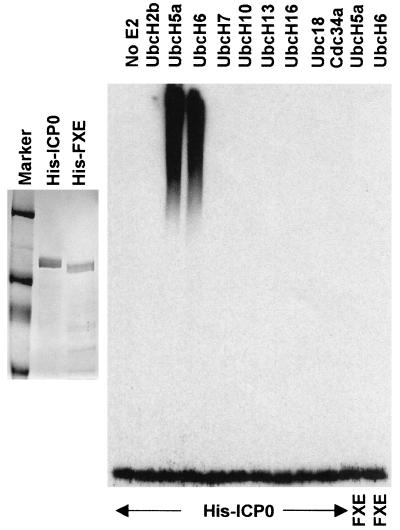
To determine whether full-length ICP0 also has E3 ubiquitin ligase activity in vitro, His-tagged versions of wild-type (His-ICP0) and FXE RING finger deletion ICP0 (His-FXE) were expressed from recombinant baculoviruses and partially purified by nickel-chelate affinity chromatography (Fig. 5A). As with the GST-262 and GST-241 bacterially expressed proteins, 100 nM His-ICP0 gave a substantial stimulation of UbcH5a and UbcH6 ubiquitination activity in vitro but did not stimulate the other E2 enzymes tested (notably cdc34; see below). Comparable purified preparations of His-FXE were inactive in this assay (Fig. 5B, rightmost two tracks). Therefore, the activity of the His-ICP0 fractions can be attributed to ICP0 itself and specifically its RING finger domain rather than any contaminating insect cell E3 ligase activities.
FIG. 5.
Full-length ICP0 stimulates the activity of UbcH5a and UbcH6 in vitro. Wild-type and RING finger mutant full-length ICP0 proteins with N-terminal His-tags were expressed in insect cells using a recombinant baculovirus and purified by nickel affinity chromatography. The left panel shows typical samples of purified His-ICP0 and His-FXE, stained by Coomassie blue. The right panel shows the results of a screen for stimulation of the activity of a panel of E2 ubiquitin conjugation enzymes, probed for high-molecular-weight ubiquitin conjugates using antibody P4D1. Like the GST fusion constructs containing ICP0 RING domains, His-ICP0 stimulates the activity of UbcH5a and UbcH6. The rightmost two tracks show a similar assay using purified RING finger deletion mutant ICP0 (FXE) with UbcH5a and UbcH6.
Lack of a stable interaction between ICP0 and UbcH5a.
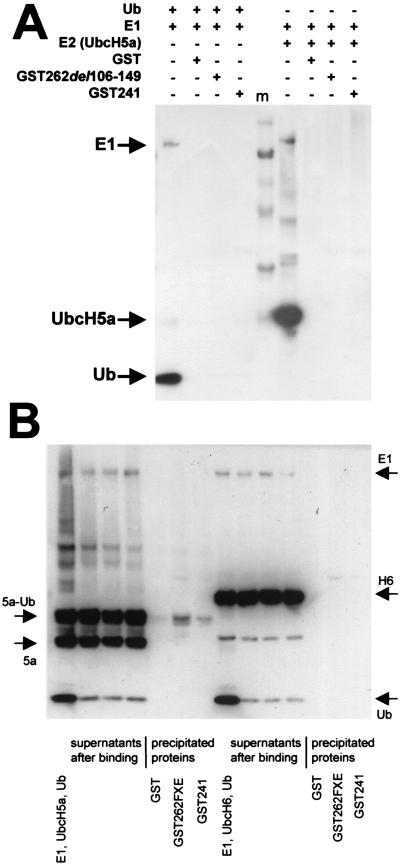
In many cases, the ubiquitin E3 ligase activity of a RING finger protein can be correlated with an interaction with the cognate E2 enzyme in vitro and/or in yeast two-hybrid assays. Accordingly, we attempted to detect an interaction between ICP0 and UbcH5a by a number of approaches including yeast two-hybrid assays, interaction of full-length His-ICP0 with GST-linked UbcH5a, and interaction of in vitro-translated or bacterially expressed His-tagged UbcH5a with GST-linked ICP0 RING finger proteins. Despite the clear functional data showing that ICP0 stimulates UbcH5a activity, none of these approaches detected a stable interaction between ICP0 and UbcH5a. Figure 6A shows the data from a GST pull-down experiment using GST-241 incubated with E1 plus ubiquitin or E1 and UbcH5a (all including polyhistidine tags). It is clear that GST-241 does not interact stably with E1, ubiquitin, or UbcH5a. To test whether GST-241 interacts with the ubiquitin-charged thiol ester version of UbcH5a, a standard reaction mixture of E1, ubiquitin, and UbcH5a was preincubated at 37°C to produce the UbcH5a-Ub thiol ester and then the mixture was incubated on ice with GST-241 beads. Under these conditions the thiol ester intermediate was stable, but the amount of UbcH5a-Ub coprecipitating with the beads was very minor compared to the unbound fraction, and in any case it was not significantly greater in the GST-241 sample compared to the GST control (Fig. 6B). A similar experiment using UbcH6 showed that GST-241 did not stably interact with this protein either (Fig. 6B). The lack of a stable interaction between GST-241 and any of the components of active E2 (ubiquitin, UbcH5a, and the thiol ester form) implies that GST-241 is stimulating UbcH5a activity in vitro via transient interactions in a catalytic manner, so that the E2 activity is repeatedly stimulated to produce very long polyubiquitin chains.
FIG. 6.
ICP0 does not form a stable complex in vitro with E1, ubiquitin, UbcH5a, UbcH6, or the UbcH5a-ubiquitin thiol ester. (A) Purified His-E1 and His-ubiquitin (lefthand set of tracks) or His-E1 and His-UbcH5a (righthand set of tracks) were preincubated at 37°C for 15 min in E3 ligase reaction buffer and then precleared by incubation with glutathione-agarose beads charged with GST for 10 min on ice. Glutathione-agarose beads charged with GST, GST-241, and the RING finger mutant GST-262(del106-149) (FXE) were prepared and incubated on ice for 20 min with aliquots of the respective mixtures. The beads were washed five times with 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5% glycerol, and 0.05% NP-40 and then pelleted, and coprecipitating proteins were released by boiling in SDS gel loading buffer without reducing agent, so as to preserve any ubiquitin-thiol ester linkages. The samples were analyzed by SDS-PAGE followed by Western blotting with an antipolyhistidine antibody. Each set of four tracks shows, from left to right, a sample of the target reaction mixture and proteins in the pellets of GST, GST-262FXE, and GST-241 beads, respectively. The sets are separated by molecular weight markers (m), and the ingredients in each incubation mix are indicated above the gel lanes. (B) The lefthand set of seven tracks shows the results from a similar experiment in which the UbcH5a-ubiquitin thiol ester intermediate was prepared by incubating E1, ubiquitin, and UbcH5a in E3 ligase buffer at 37°C for 15 min. All subsequent operations were performed on ice to preserve the thiol ester linkage. After preclearing the initial mixture as above, aliquots were incubated with beads charged with GST, GST-241, and the RING finger deletion derivative and then the samples were washed and analyzed as above. From left to right the tracks show the initial reaction mixture, the supernatants, and the pellets after binding. Equivalent proportions of the total bead and supernatant fractions were loaded onto the gel to illustrate the true relative amounts of bound and unbound protein. Although some UbcH5a-Ub was present in the pellets, this was minor compared to the unbound fraction and was also present in the negative control. The righthand part of the panel shows a similar experiment with UbcH6. In this example, the formation of the UbcH6-ubiquitin thiol ester linkage was considerably less efficient than that with UbcH5a.
Some other RING finger E3 ligases do form stable complexes with their cognate E2 enzymes in the absence of other factors, and indeed the structure of the c-Cbl RING domain complexed to a substrate peptide and UbcH7 has been solved (44). In contrast, as noted above, some other RING domain E3 ligases require additional components for formation of the complete active complex. Because the RING finger helix region of ICP0 is related to that of c-Cbl by virtue of a tryptophan residue that is conserved in a subset of RING finger domains, we examined the structures of the c-Cbl-UbcH7 complex in comparison with the structure of the RING domain of Eg63 (EICP0), the EHV-1 equivalent of ICP0 (the structure of the RING domain of ICP0 itself has not been solved). A portion of UbcH7 docks into a groove in the c-Cbl RING domain, but a similar interaction with the Eg63 RING (and thus probably the ICP0 RING) appears unlikely because this groove is not present in the viral protein (P. N. Barlow, personal communication). We also failed to detect any interaction between GST-241 or full-length ICP0 with UbcH1, UbcH7, and UbcH8 in a variety of in vitro assays.
ICP0 partially sequesters UbcH5a and UbcH6 in transfected cells.
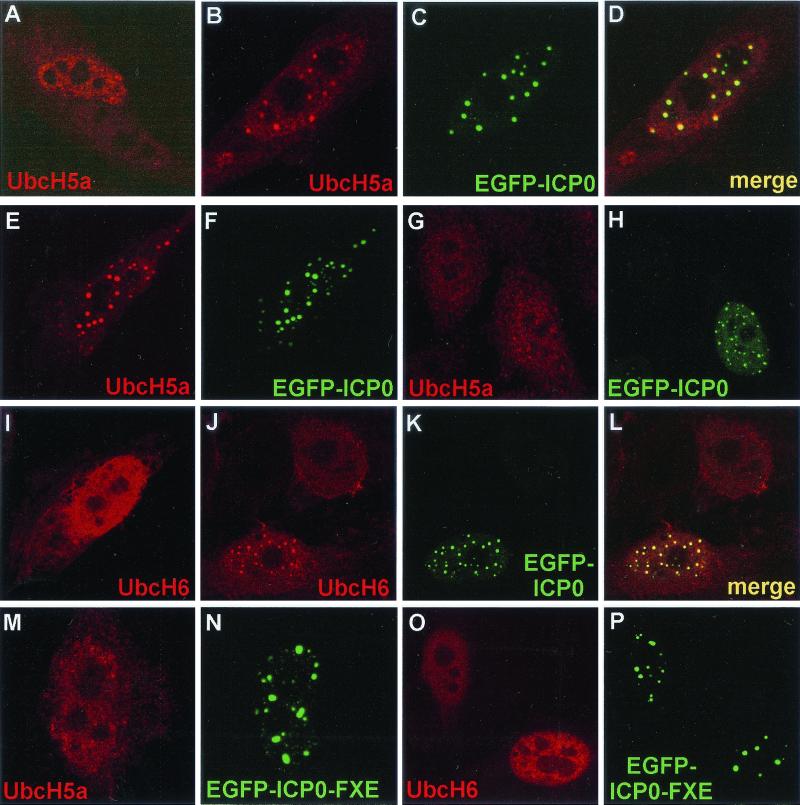
As an alternative approach to assess the biological significance of the in vitro stimulation of UbcH5a and UbcH6 by ICP0, we investigated the distribution of these E2 enzymes in transfected cells expressing ICP0. Because no antibodies that recognize endogenous UbcH5a or UbcH6 are available, we cotransfected plasmids expressing ICP0 and tagged derivatives of the E2 enzymes. The distribution of the tagged E2 proteins in singly transfected cells was diffuse throughout the cell, but it was generally more intense in the nucleus. The proportion of cytoplasmic staining shown in the presented images is to some extent dependent on the position of the image capture plane in the z axis. Coexpression of enhanced GFP (EGFP)-linked ICP0 led to local accumulations of UbcH5a in association with the ICP0 foci in many cells. This occurred in both the nucleus (Fig. 7B to D) and in the cytoplasm when ICP0 formed such local cytoplasmic concentrations (Fig. 7E and F). The extent of the effect varied from cell to cell and, while in many cells it was dramatic (panels B to F), in some cases it was less obvious (panels G and H). Similar results were obtained with UbcH6 (I-L). The RING finger FXE mutant (del106-149) derivative of EFGP-linked ICP0 had no significant effect on the localizations of UbcH5a (panels K and L) and UbcH6 (panels M and N). These results were obtained using EGFP-linked ICP0 so that complications due to possible antibody specificity overlaps were eliminated, and the use of Cy5-conjugated secondary antibody to detect the E2 enzymes eliminated optical overlap effects. However, similar results were obtained when both ICP0 and the E2 proteins were detected using a double antibody labeling approach (data not shown).
FIG. 7.
ICP0 sequesters UbcH5a and UbcH6 in transfected cells in a RING finger-dependent manner. HEp-2 cells were transfected with plasmids expressing tagged UbcH5a or UbcH6 and EGFP-linked or native ICP0 and ICP0 RING finger mutant derivatives. The E2 enzymes were detected by the UL30 tag (r113) (UbcH5a) or the FLAG-tag (UbcH6) and the appropriate Cy5-conjugated secondary antibodies. (A) The Cy5-labeled UbcH5a single transfection control. (B to D) The single and merged channels for a cell coexpressing UbcH5a and EGFP-ICP0. (E, F, G, and H) The separated channels of further examples of cotransfected wild-type EGFP-ICP0-expressing cells. (I) A typical cell expressing UbcH6 alone. (J to L) The separated and merged channels of an image of a singly UbcH6-transfected cell (top) and one doubly transfected with EGFP-ICP0. (M and N) A cell expressing UbcH5a and ICP0 RING finger mutant FXE (del 106–149) linked to EGFP. (O and P) A similar experiment using UbcH6.
These data were extended by examining a panel of ICP0 proteins with insertion and deletion mutations in the RING finger region. As shown in Fig. 7, the RING finger mutant FXE (del106-149) did not recruit either E2 enzyme. Similarly, none of the (otherwise full-length) ICP0 proteins containing the lesions that inactivate the in vitro E3 ligase activity of ICP0 (Fig. 4) recruited UbcH5a into discrete foci (data not shown). However, this was also the case for the insertion mutants ins188, ins197, and ins212 (data not shown), which retained in vitro E3 ligase activity (Fig. 4, by implication for ins212). Since these mutations all eliminated the ability of ICP0 to induce formation of colocalizing conjugated ubiquitin in transfected cells (4), these results are consistent with the proposal that these distal sequences are required for the formation of the complete active complex in vivo.
DISCUSSION
Evidence that ICP0 functions via the ubiquitin-proteasome pathway has been accumulating for several years. The initial indication that this might be the case was the finding that ICP0 binds very strongly to USP7, a member of the ubiquitin-specific protease family of enzymes (11). Subsequently, ICP0 expressed during viral infection was found to induce, in a RING finger-dependent manner, the proteasome-dependent degradation of PML (8), the catalytic subunit of DNA-dependent protein kinase (35), Sp100 (33), CENP-C (7), and CENP-A (27). ICP0 function was also connected to the ubiquitin pathway by the finding that it partially protects cyclin D3 from the rapid proteasome-dependent degradation that occurs during infection by ICP0 mutant viruses (25). Proteasome inhibitors block the ability of ICP0 to stimulate viral infection and reactivation from quiescence (14), and they change the localization of ICP0 both at the initiation of infection (14) and more dramatically at late stages of infection (28). The above studies using virus-infected cells have been complemented by transfection studies using ICP0 expressed in the absence of other viral proteins. The effects of ICP0 on the stability of PML and Sp100 (and/or their SUMO-1-modified forms) and CENP-A have been directly reproduced in transfection assays (27, 31, 33), and the degradation of DNA-dependent protein kinase and CENP-C during virus infection is paralleled by a loss of fluorescence signal from these proteins in transfected cells expressing ICP0 (7, 35). More recently it was shown that ICP0 induces the formation of colocalizing conjugated ubiquitin in both infected and transfected cells (4) and that cytoplasmic ICP0 structures formed after high-level expression become associated with proteasomes after treatment of cells with MG132 (28). This report firmly establishes that ICP0 stimulates the synthesis of conjugated ubiquitin in vitro and that this activity correlates very well with its observed effects in transfected and infected cells.
In recent years there have been numerous reports that identify cellular RING finger proteins as essential components of ubiquitin E3 ligase complexes. ICP0 appears to be acting in an analogous manner by stimulating the activity of UbcH5a and UbcH6. The UbcH5 family comprises three closely related proteins that have been implicated in the function of several E3 ligase complexes, including examples of both the RING finger and SCF classes (29, 37). UbcH5b and UbcH5c are of very similar length to UbcH5a, and the three proteins are almost 90% identical. UbcH6 is extensively related to members of the UbcH5 family (75% identity over the length of the UbcH5a sequence), but it has an additional N-terminal extension. Other members of the UbcH series of E2 enzymes share less than 50% identity with UbcH5a. Therefore, ICP0 may be interacting with sequence elements conserved within the UbcH5/6 group of related E2 proteins. However, despite extensive attempts, we were unable to demonstrate a stable interaction between ICP0 and either UbcH5a or UbcH6. That some kind of interaction is occurring can be inferred from the very strong stimulation of the activity of these E2 proteins induced by ICP0, in a manner controlled by the concentrations of both the E2 and the ICP0 RING finger. The data suggest that the interaction may be transient, perhaps occurring only during a particular step in the catalysis of the formation of polyubiquitin chains.
Of direct relevance to our results, a recent report also suggests ICP0 has the properties of an ubiquitin E3 ligase (38). However, there are substantial differences between the two studies. Because these other investigators found that a modified form of the E2 enzyme cdc34 (UbcH3) could be immunoprecipitated with proteasomes from HSV-1-infected cells, they questioned whether the ICP0 RING domain could stimulate cdc34 activity in vitro. Consistent with our data, their results using this combination of reagents were negative. However, they found that cdc34 was precipitated with the ICP0 RING domain GST fusion protein if the reaction mixture was subsequently incubated with glutathione beads. Surprisingly, it was reported that the activity of cdc34 could be stimulated by a GST fusion protein containing ICP0 residues 543 to 768, although this C-terminal domain did not interact with cdc34. Based on these data, it was suggested that ICP0 has a novel E3 ligase activity in which the RING domain interacts with the E2 (cdc34), but that the actual E3 ligase domain lies elsewhere. We note that the in vitro E3 ligase assays of the previous study used 5 μg of the ICP0 543-768 GST fusion protein and did not achieve high levels of activation, whereas as little as 5 ng of GST-241 can produce very high levels of E3 ligase activity with UbcH5a in our assays. Their proposal that the E3 ligase activity of ICP0 utilizes a C-terminal domain and cdc34 is not consistent with the lack of activity of full-length ICP0 with cdc34 in vitro (Fig. 5) and is difficult to reconcile with the ability of an ICP0 mutant lacking residues 594 to 775 to induce degradation of cellular proteins and accumulation of conjugated ubiquitin in vivo (4, 33). Furthermore, an ICP0 truncation mutant containing residues 1 to 547 gave a positive result similar to that of the truncation containing residues 1 to 593 in terms of conjugated ubiquitin detection in transfected cells (data not shown). Finally, ICP0 did not recruit cdc34 into colocalizing foci in cotransfected cells (data not shown). On the other hand, the role of the ICP0 RING finger in the degradation of cellular proteins and induction of colocalizing conjugated ubiquitin in vivo has been extensively documented, and the mutational analyses of these previous studies are consistent with the in vitro assay data presented here.
An obvious question concerns the relevance of our findings on the in vitro stimulation of UbcH5a and UbcH6 by ICP0 to virus infection in vivo. The partial association of UbcH5a and UbcH6 with ICP0 in cotransfected cells supports the notion that either of these two related proteins (or other members of the UbcH5 family) may form part of the active ICP0 E3 ligase complex that targets PML and other proteins for degradation in vivo. We have attempted to induce ICP0-mediated ubiquitination of PML in our in vitro assays, so far unsuccessfully. This could be due to several reasons. Firstly, a technical problem is that full-length PML is not easy to purify or express adequately in a soluble form for use in vitro. Secondly, although full-length PML can be expressed by in vitro translation, this material may not be properly folded for use in these assays. Thirdly, modification of PML by phosphorylation (or even SUMO-1 conjugation) may be necessary for it to be targeted by ICP0. Fourthly, other components may be required for assembly of the active, substrate-specific, in vivo complex. Last, it remains possible that PML is not directly ubiquitinated in vivo by the E3 ligase activity of ICP0, but rather it is degraded as a consequence of the ubiquitination activity of ICP0 on some other ND10 component. While some RING finger E3 ligases appear to interact directly with both substrate and cognate E2, others require additional components and interact with substrate only indirectly. It may be relevant that a number of mutations in the region of the RING finger of ICP0 did not reduce its substrate-independent in vitro activity yet did reduce the ability of ICP0 to degrade target proteins and induce colocalizing conjugated ubiquitin in vivo. It is possible that these mutations affect surfaces of ICP0 that interact with other components of the complete, substrate-specific E3 ligase complex in vivo.
It is intriguing to consider the relevance of the present study to earlier observations of the strong and specific interaction between sequences in the C-terminal region of ICP0 and ubiquitin-specific protease USP7 (11). While the in vitro evidence demonstrates that the N-terminal RING finger region of ICP0 is involved in ubiquitin conjugation, the C-terminally bound USP7 would be expected to cleave polyubiquitin chains. The association of ICP0 with apparently opposing functions could be explained in a number of ways. Firstly, if USP7 counteracts ICP0-mediated ubiquitination, ICP0 might bind USP7 in order to inhibit the latter’s activity. Secondly, ICP0 might recruit an opposing USP enzyme as part of a feedback regulatory mechanism. Thirdly, by recruiting USP7, ICP0 may be stimulating the recycling of conjugated ubiquitin in situ, thereby increasing the availability of ubiquitin monomers for the conjugation activity associated with the RING finger. Fourthly, USP7 may be protecting ICP0 from autoubiquitination. Each of these suggestions would be consistent with the observation that the ability to bind USP7 contributes to but is not essential for ICP0 activity (10). Whatever the explanation, other examples of an association between a RING finger protein and an enzyme that cleaves ubiquitin conjugates have been reported. The proto-oncogene brca1 binds to the type I ubiquitin protease BAP1 via sequences in the former’s RING finger (20), while USP7 itself has recently been shown to bind to TRAF-6, a RING finger protein involved in tumor necrosis factor signaling (43).
After many years of research, the understanding of ICP0 function has now extended to the ability to study an important aspect of its activity in a simple in vitro assay. Since this is an enzymatic activity requiring specific cellular components, it should in principle be possible to design or screen for inhibitors of ICP0 function that are more specific and less toxic than the proteasome inhibitors already identified. Such a reagent would be of potential therapeutic benefit in the treatment of recurrent HSV-1 infections.
Acknowledgments
This work was supported by the Medical Research Council (R.D.E.), Millennium Pharmaceuticals (S.S.), and the Human Frontiers Science Programme (a fellowship awarded to C.B. to work in the laboratory of R.D.E.).
We are very grateful for the supply of materials from a number of people: Delia O’Rourke and Peter O’Hare for certain mutants in the ICP0 RING domain, Phil Robinson and Cecile Pickard for clones of selected E2 enzymes, and Kazuhiro Iwai for a baculovirus expressing His-E1. We are also grateful for the comparative structural analysis of the Eg63 and c-Cbl RING domains conducted by Paul Barlow. The constructive comments of Duncan McGeoch were much appreciated.
REFERENCES
- 1.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941. [DOI] [PubMed] [Google Scholar]
- 2.Everett, R. D. 1987. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 6:2069–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Everett, R. D. 1988. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J. Mol. Biol. 202:87–96. [DOI] [PubMed] [Google Scholar]
- 4.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994–10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761–770. [DOI] [PubMed] [Google Scholar]
- 6.Everett, R. D., A. Cross, and A. Orr. 1993. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology 197:751–756. [DOI] [PubMed] [Google Scholar]
- 7.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, R. D., M. Meredith, and A. Orr. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., P. O’Hare, D. O’Rourke, P. Barlow, and A. Orr. 1995. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J. Virol. 69:7339–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., A. Orr, and M. Elliott. 1991. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 19:6155–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., C. M. Preston, and N. D. Stow. 1991. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication, p.49–76. In E. K. Wagner (ed.), Herpesvirus transcription and its regulation. CRC Press, Inc., Boca Raton, Fla.
- 16.Fields, B. N., D. M. Knipe, and P. M. Howley (ed.). 1996. Virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Freemont, P. S. 2000. RING for destruction? Curr. Biol. 10:R84–R87. [DOI] [PubMed] [Google Scholar]
- 18.Halford, W. P., C. D. Kemp, J. A. Isler, D. J. Davido, and P. A. Schaffer. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 75:6143–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, D. E., M. Proctor, S. T. Marquis, H. P. Gardner, S. I. Ha, L. A. Chodosh, A. M. Ishov, N. Tommerup, H. Vissing, Y. Sekido, J. Minna, A. Borodovsky, D. C. Schultz, K. D. Wilkinson, G. G. Maul, N. Barlev, S. L. Berger, G. C. Prendergast, and F. J. Rauscher III. 1998. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16:1097–1112. [DOI] [PubMed] [Google Scholar]
- 21.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552. [DOI] [PubMed] [Google Scholar]
- 22.Joazeiro, C. A., S. S. Wing, H. Huang, J. D. Leverson, T. Hunter, and Y. C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286:309–312. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71:1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829–5835. [DOI] [PubMed] [Google Scholar]
- 28.Lopez, P., C. Van Sant, and B. Roizman. 2001. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J. Virol. 75:3832–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuzawa, S., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7:915–926. [DOI] [PubMed] [Google Scholar]
- 31.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Rourke, D., G. Elliott, M. Papworth, R. Everett, and P. O’Hare. 1998. Examination of determinants for intranuclear localization and transactivation within the RING finger of herpes simplex virus type 1 IE110k protein. J. Gen. Virol. 79:537–548. [DOI] [PubMed] [Google Scholar]
- 33.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, J., and R. D. Everett. 2001. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 induce the formation of colocalizing, conjugated ubiquitin. J. Virol. 75:5357–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spatz, S. J., E. C. Nordby, and P. C. Weber. 1996. Mutational analysis of ICP0R, a transrepressor protein created by alternative splicing of the ICP0 gene of herpes simplex virus type 1. J. Virol. 70:7360–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer, E., J. Jiang, and Z. J. Chen. 1999. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 13:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Sant, C., Y. Kawaguchi, and B. Roizman. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc. Natl. Acad. Sci. USA 96:8184–8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Sant, C., P. Lopez, S. J. Advani, and B. Roizman. 2001. Role of cyclin D3 in the biology of herpes simplex virus 1 ICP0. J. Virol. 75:1888–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber, P. C., S. J. Spatz, and E. C. Nordby. 1999. Stable ubiquitination of the ICP0R protein of herpes simplex virus type 1 during productive infection. Virology 253:288–298. [DOI] [PubMed] [Google Scholar]
- 42.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zapata, J. M., K. Pawlowski, E. Haas, C. F. Ware, A. Godzik, and J. C. Reed. 2001. A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J. Biol. Chem. 276:24242–24252. [DOI] [PubMed] [Google Scholar]
- 44.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102:533–539. [DOI] [PubMed] [Google Scholar]