Abstract
The predominant rotavirus electropherotypes (e-types) during 17 epidemic seasons (1980 through 1997) in Finland were established, and representative virus isolates were studied by nucleotide sequencing and phylogenetic analysis. The virus isolates were either P[8]G1 or P[8]G4 types. The G1 and G4 strains formed one G1 lineage (VP7-G1-1) and one G4 lineage, respectively. Otherwise, they belonged to two P[8] lineages (VP4-P[8]-1 and -2) unrelated to their G types. Phylogenetic analysis of partial sequences of all 11 RNA segments obtained from the strains also revealed genetic diversity among gene segments other than those defining P and G types. With the exception of segments 1, 3, and 10, the sequences of the other segments could be assigned to 2 to 4 different genetic clusters. The results of this study suggest that, in addition to the RNA segments encoding VP4 and VP7, the other RNA segments may segregate independently as well. In total, the 9 predominant e-types represented 7 different RNA segment combinations when the phylogenetic clusters of their 11 genes were determined. The extensive genetic diversity and number of e-types among rotaviruses are best explained by frequent genetic reassortment.
Human group A rotaviruses, members of the Reoviridae family, are enteric viruses causing gastroenteritis mainly in 1.5- to 3-year-old children (17). The first rotavirus vaccine was licensed in the United States in 1998, but its production has been discontinued because it was associated with the development of intussusception (6). Knowledge about the molecular epidemiology and genetics of rotaviruses is needed when vaccination strategies are planned and when the vaccine is improved.
The nonenveloped icosahedral rotavirus particle is composed of two protein layers: the inner layer (VP6) and the outer layer (VP7 and spike protein VP4) (reviewed in reference 17). Inside these layers there is the core (VP2), which contains the 11 double-stranded RNA gene segments, RNA polymerase (VP1), and guanylyltransferase/methylase (VP3). Cleavage of the spike protein into VP5* and VP8* by trypsin enhances the infectivity of the virus (reviewed in reference 17). In general, each RNA segment is monocistronic, with the exception of segment 11, which has an additional overlapping open reading frame (coding for NSP6). In the prototype simian rotavirus strain SA-11, the coding assignment of the genes is as follows: structural proteins VP1 to -4, VP6, and VP7 are encoded by segments 1 to 4, 6, and 9, respectively, and nonstructural proteins NSP1 to -6 are encoded by segments 5, 8, 7, 10, 11, and 11, respectively (reviewed in reference 17).
Proteins VP4, VP6, and VP7 are the primary antigens of rotaviruses. Although group A rotaviruses of humans and animals share common antigens on VP6, some subgroup-specific antigenic determinants are located on this protein as well. The cell attachment protein VP4 and the glycoprotein VP7, which is the major neutralization antigen, determine the P and G types. At least 11 P and 10 G types have been found in humans (17). In addition to direct antigen detection by serological means, P and G types can be defined based on partial nucleotide sequences of genes 4 and 9 (or 7 or 8, depending on the strain), respectively (19, 25). Sequence analysis has also revealed the existence of genetic lineages within the P and G types. For example, the strains of type P[8] form at least three lineages (23, 29, 36), and four lineages of type G1 have been found so far (15, 29, 30, 36, 55). Genetic clustering is also evident at least for segments 5 (16), 7 (44), 8 (41), and 10 (12, 17, 28). Phylogenetic analysis of human and animal segment-5 sequences has revealed distinct genetic groups according to species origin (16).
Another kind of genetic variation, gene rearrangement, also occurs among rotaviruses (reviewed in reference 13). The most common gene rearrangement found is that of segment 11, which exists in two forms, with the long form containing a partial duplication that the short form lacks (33, 38). Gene recombination other than rearrangement has been reported only once for human rotaviruses (47). By contrast, gene reassortment seems to be more important for their evolution, and it has been studied extensively in vitro (reviewed in reference 43). Although no restrictions for reassortment seem to exist, nonrandom reassortment with regard to progeny has been found in vitro (24) and some segment combinations occur only rarely in vivo. Rotavirus strains that are likely to have emerged through natural reassortment between human rotavirus isolates (see, e.g., references 29, 40, 51, 52, and 56) or between animal and human rotavirus isolates (37, 45) have been described in the literature.
Soon after rotaviruses were discovered by electron microscopy in 1973 (2), the separation of discrete RNA segments by polyacrylamide gel electrophoresis (PAGE) was proven to be a useful method for molecular epidemiological analysis. In gels the electropherotypes (e-types), or RNA profiles, expressing a high number of RNA segment migration differences among rotavirus isolates, could be distinguished. Changes in the migration of the 11 RNA segments do not reveal the nature of mutational events (7), but when the place and time scale are limited, an e-type seems to correspond to a virus strain (3, 8, 35). It is typical for a vast diversity of rotavirus strains with different P/G combinations to be cocirculating in a rotavirus epidemic season (3, 20). One of the strains may predominate, usually for one or two epidemic seasons (32). Dual infections, which offer opportunities for RNA segment reassortment, have been reported in rotavirus studies with frequencies of 0.9 to 23% (1, 5, 11, 50). In vitro experiments have shown that a slight asynchrony in the dual infection does not hinder the reassortment event (42) and may even favor it (49).
In Europe and the United States the P/G type combination P[8]G1 has been the most prevalent in recent years (see, e.g., references 5, 9, and 46). In the present study, nine predominant P[8]G1 and P[8]G4 e-types have been selected for genetic analysis. Information has been gathered on the one hand about the RNA segment patterns in gel electrophoresis and on the other hand about the nucleotide sequences of each of the 11 RNA segments for the viruses. Phylogenetic analysis revealed the clustering of the sequences for each RNA segment, and the segment cluster combinations of the rotaviruses were compared. One aim was to find out whether any of the combinations favor predominance. Possible mechanisms of generating these combinations are discussed.
MATERIALS AND METHODS
Samples.
Rotavirus electron microscopy (EM)-positive stool samples from children, sent to the Department of Virology, University of Helsinki, during 1980 to 1997, were used in this study (Fig. 1). They were collected mainly from hospitalized children in the Helsinki metropolitan area, with a population of about 1 million inhabitants. To define the most predominant rotavirus strains, approximately 150 to 200 virus isolates per epidemic season from the 1981 to the 1986 season were run in gels, and all samples for the seasons from 1986 to 1997 were electrophoresed. The e-types during the years 1987 to 1990 had been determined previously (35). The most common e-type was considered predominant if its prevalence was higher than 30% during the season. The partial nucleotide sequences of segments 4 and 9 were determined from viruses of 26 e-types. From those, all of the nine predominant G1 and G4 viruses, as well as three nonpredominant e-types that shared RNA profiles with one of the predominant strains, were selected for more-detailed sequence and phylogenetic analysis.
FIG. 1.
Numbers of rotavirus isolates from epidemic seasons 1981 through 1997, used as the basic material in this study. The predominant e-type for each season is given above the bars. The G type of the most common but nonpredominant e-type is given in parentheses.
Gel electrophoresis.
RNA extraction, gel electrophoresis, and silver staining were performed as previously described (35). Briefly, the nucleic acid from a rotavirus-positive stool suspension was extracted by treatment with phenol and chloroform-isoamyl alcohol, followed by ethanol-acetate precipitation. RNA segments were resolved in a 7.5% polyacrylamide gel and visualized by silver staining.
RNA extraction and RT-PCR.
Viral RNA was extracted by using the phenol-containing reagent Tripure (Roche, Indianapolis, Ind.) as described previously (34). The primers used in reverse transcription (RT) and PCR are shown in Table 1. The RT was run separately for 1 h at 37°C, in a 20-μl reaction mixture containing 50 mM Tris-HCl (pH 8.8), 75 mM KCl, 10 mM dithiothreitol, 3 mM MgCl2, 0.5 mM deoxynucleoside triphosphates (Pharmacia, Uppsala, Sweden), 0.5 μM primer, 1 U of RNAsin inhibitor (Promega, Madison, Wis.)/μl, 10 U of Moloney murine leukemia virus reverse transcriptase (Bethesda Research Laboratories, Gaithersburg, Md.)/μl, and rotavirus RNA that had been pretreated by heating for 5 min in presence of 37.5% dimethyl sulfoxide and chilling on ice for 5 min. For PCR, 5 μl of the RT reaction mixture was added to a 100-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 0.1 mM deoxynucleoside triphosphates, 0.2 μM each primer, and 2.5 U of AmpliTaq DNA polymerase (Perkin Elmer, Branchburg, N.J.)/100 μl. The PCR program was as follows: 33 cycles of 94°C for 1.0 min, 40 to 42°C for 2.0 min, and 72°C for 2.0 min, followed by a final incubation for 7 min at 72°C.
TABLE 1.
Primers used to produce the partial sequences for the 11 rotavirus gene segments
| Segment | PCR and sequencing primersa (nt) | Sense | Sequenced region |
|---|---|---|---|
| 1 | beg, 5′-CAAAGAGCGTTCATGTCTTT-3′ (2371-2390) | + | 2500-2522 (153 nt) |
| end, 5′-GGTAGTGTTGGCATAAATTT-3′ (2710-2729) | − | ||
| seq, 5′-GTTAACAAATTATCTGACCA-3′ (2452-2471) | + | ||
| 2 | beg, 5′-CCTCAATGGCGTACAGGAA-3′ (13-20) | + | 67-342 (276 nt) |
| end, 5′-ATTCTCTACAGCCATATCTT-3′ (591-610) | − | ||
| seq, 5′-GCTAAACGTGAAAACTTACCA-3′ (38-59) | + | ||
| 3 | beg, 5′-CCTGGATGGAAATTAACATAT-3′ (407-427) | + | 509-850 (342 nt) |
| end, 5′-ATGGTGTGTCCAATGGATCC-3′ (970-989) | − | ||
| seq, 5′-GTCAAAACGCTGCAACAGATG-3′ (480-500) | + | ||
| 4 | beg, 5′-TGGCTTCGCTCATTTATAGACA-3′ (11-32) | + | 250-561 (312 nt) |
| end, 5′-ATTTCGGACCATTTATAACC-3′ (868-887)b | − | ||
| seq, 5′-GATGGTCCTTATCAGCC-3′ (205-221) | + | ||
| 5 | beg, 5′-CTAGATGTAGAAT(G/A)TTCACG-3′ (885-905) | + | 1153-1398 (246 nt) |
| end, 5′-TAGGCGCTACTCTAGTG-3′ (1524-1550) | − | ||
| seq, 5′-TCACAGGAAAAGATATATGA-3′ (1122-1141) | + | ||
| 6 | beg, 5′-ACTCTTAAAGATGCTAGGGA-3′ (51-70) | + | 207-512 (306 nt) |
| end, 5′-GCTGAATTAAT(T/A)ACTCTT-3′ (726-745) | − | ||
| seq, 5′-ACTATGAATGGAAATGA(A/T/C/G)TT-3′ (141-160) | + | ||
| 7 | beg, 5′-GA(T/C)ACTAT(T/A)GATTGGAAAT-3′ (521-539) | + | 705-968 (264 nt) |
| end, 5′-TTGACAGTGTTAGCTTTTAAC-3′ (1013-1033) | − | ||
| seq, 5′-ATGAACTCTCTTCAGAATGT-3′ (641-660) | + | ||
| 8 | beg, 5′-ATAGCTATTGGTCATTCAAA-3′ (457-476) | + | 739-1041 (303 nt) |
| end, 5′-CATAAGCGCTTTCTATTCTT-3′ (1034-1053) | − | ||
| seq, 5′-CATGGTAAAGGTCACTATAG-3′ (706-725) | + | ||
| 9 | beg, 5′-GGCTTTAAAAGAGAGAATTTCCGTCTGG-3′ (1-28)c | + | 100-498 (399 nt) |
| end, 5′-GGTCACATCATACAATTCTAATCTAAG-3′ (1036-1062)c | − | ||
| seq, 5′-GTATGGTATTGAATATACCAC-3′ (51-71) | + | ||
| 10 | beg, 5′-AGTTCTGTTCCGAGAGAGCG-3′ (11-30) | + | 105-506 (402 nt) |
| end, 5′-GTCACACTAAGACCATTCC-3′ (731-749) | − | ||
| seq, 5′-CACATTGAGTGTAATCACTT-3′ (68-87) | + | ||
| 11 | beg, 5′-TGGAAAATCTATTGGTAGGA-3′ (110-129) | + | 306-630 (325 nt) |
| end, 5′-AGTGGGGAGCTCCCTAGTG-3′ (631-649) | − | ||
| seq, 5′-CTGGCGTGTCTATGGATTCA-3′ (280-299) | + |
Sequencing.
Sequencing was carried out as described earlier (36). Biotinylated PCR products were bound to avidin-coated beads (IDEXX, Westbrook, Maine), and the separated DNA strands were sequenced by the dideoxynucleotide chain termination method (48). The Sequenase DNA sequencing kit, version 2.0 (U.S. Biochemicals, Cleveland, Ohio), was used with [35S]ATP for manual sequencing. Reaction products were resolved on thin 6% polyacrylamide-7 M urea gels in taurine buffer (89 mM Tris, 29 mM taurine, 0.5 mM EDTA [pH 8.8]). The sequenced region of each segment and the sequencing primers are shown in Table 1. Regions expressing much nucleotide variation were selected for sequencing after comparison of the rotavirus nucleotide sequences in the GenBank database. Random parts were selected for segments 1 and 3, since insufficient sequence data to allow for accurate sequence comparison were available in the database at that time.
Sequence analysis.
The CLUSTAL V software package (27) was used to perform multiple alignments, and a neighbor-joining phylogenetic tree was built for each RNA segment. These methods were applied for nucleotide sequences with 1,000 bootstrap replicates. Programs of the Genetics Computer Group, especially the BLAST program, which searches for sequences similar to a query sequence, were also used (14).
Nucleotide sequence accession numbers.
Nucleotide sequences of rotavirus RNA segments 1 to 11 were deposited in GenBank/EMBL under accession numbers AJ272451 to -8, AJ287448 to -55, AJ287456 to -64, AJ276278 to -91, AJ287465 to -71, AJ275894 to -901, AJ275902 to -9 and AJ292380, AJ275942 to -9, AJ288000 to -17, AJ275950 to -7, and AJ272531 to -6, respectively. Partial nucleotide sequences of segments 1 and 3 of strain Wa are available under accession numbers AJ292378 and AJ292379. The accession numbers of segment-4 and -9 sequences for the previously determined rotavirus isolates are as follows: G1-87-(101:1), Z80234 and Z80271; G4-89-(305:8) and G4-90-(417:4), Z80317 (segment 4) and Z80330 (segment 9), respectively.
RESULTS
P/G combinations among rotavirus e-types and their locations in the genetic lineages defined by genes encoding VP4 and VP7.
The material for this study included rotavirus-positive stool samples from the Helsinki metropolitan area spanning a period of 17 years, 1981 to 1997, with epidemics occurring regularly during the winter months (Fig. 1). The predominant rotaviruses were identified by PAGE screening. In 10 seasons a single e-type was predominant (accounting for more than 30% of all isolates [range, 31 to 74%]), while in the other 7 seasons no such prevalence was seen. The nine predominant e-types (for which the temporal distribution is shown in Fig. 1) with long electrophoresis patterns (Fig. 2) were selected for further analysis.
FIG. 2.
Polyacrylamide gel showing the RNA gel patterns of the nine predominant rotavirus e-types during 1980 to 1997 (lanes 1 to 9). Simian rotavirus SA-11 was used as a marker.
The genes coding for the antigenically important proteins VP4 and VP7 were partially sequenced. From these sequences the P and G types of the 9 predominant rotavirus strains and 17 other rotavirus strains throughout the period were deduced. All virus isolates sequenced showed a P/G combination of either P[8]G1 or P[8]G4. The particular VP4 and VP7 genetic lineages to which the rotaviruses belonged were also defined (amino acid signature motifs), as described previously (36). As Table 2 shows, the P[8]G1 lineage combination P[8]-1 G1-1 was the most common among the G1 rotavirus strains. It was remarkable that all predominant G1 strains showed this combination, whereas among other G1 rotavirus strains the combinations P[8]-1 G1-2, P[8]-1 G1-4, and P[8]-2 G1-3 were also found. The type G4 rotavirus sequences belonged to the same VP7 genetic lineage. Unlike the situation with G1 strains, both VP4 genetic lineages, P[8]-1 and P[8]-2, were found among predominant G4 strains.
TABLE 2.
P/G lineage combinations among the 9 predominant and 17 other rotavirus e-types in 1981 through 1997
| P/G lineage combinationa | No. of e-types with the indicated combination
|
|
|---|---|---|
| Predominant e-types | Other e-typesb | |
| P[8]-1 G1-1 | 4/9 | 7/17 |
| P[8]-1 G1-2 | 2 | |
| P[8]-1 G1-4 | 2 | |
| P[8]-2 G1-3 | 2 | |
| P[8]-1 G4 | 3 | 3 |
| P[8]-2 G4 | 2 | 1 |
Lineages are as presented in reference 36. Amino acid signature motifs: VP4 positions, amino acids 121, 125, 131, and 135; VP7 positions, amino acids 29, 37, 41, 49, 55, 57, 65, and 66.
Other rotavirus strains were isolated in 1981 through 1983, 1985, 1988 and 1990 through 1996.
Phylogenetic analysis of the 11 RNA segments of the predominant rotaviruses.
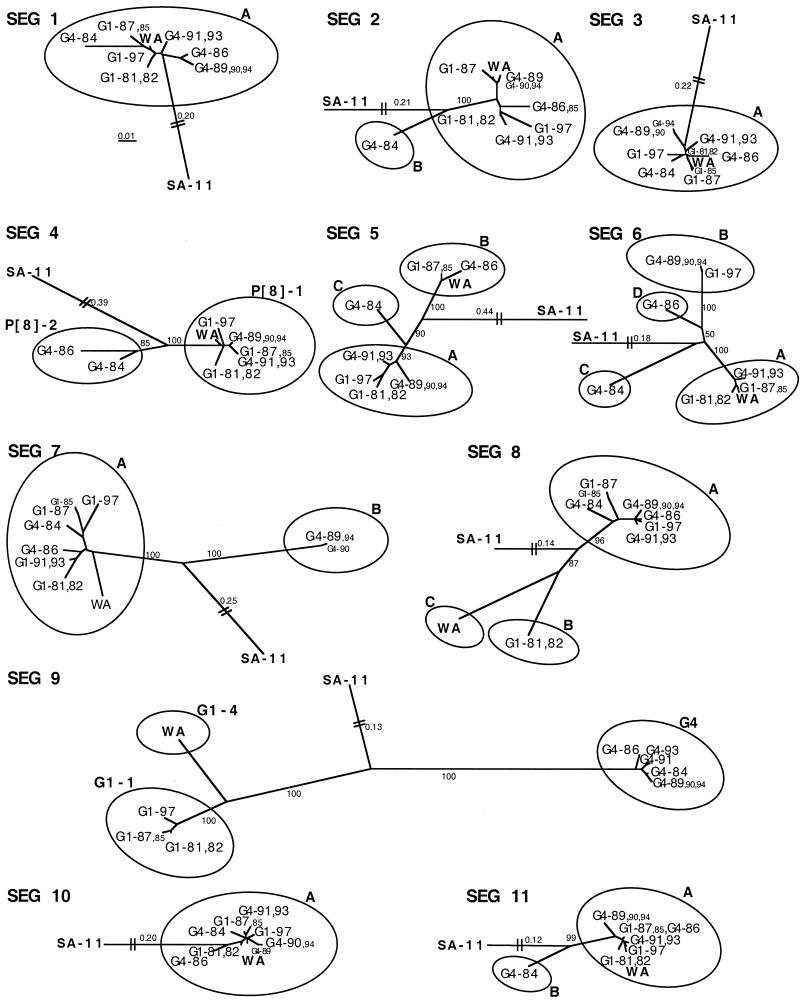
Partial nucleotide sequences of all RNA segments were determined for the nine predominant e-types. The 11 phylogenetic trees, in which the G1 prototype strain Wa and a genetically distant simian rotavirus, SA-11, were included, revealed clustering of the sequences (Fig. 3). For most segments, the genetic clusters (circled in Fig. 3) were supported by high bootstrap values. In the phylogenetic trees of segments 4 and 9, the G1 and P[8] lineages or clusters of Table 2 are visualized. As mentioned above, in the segment-9 tree, the predominant viruses grouped to G1-1 while strain Wa belonged to type G1-4; all the G4 viruses formed one cluster. The clusters in segment 9 defining types G1 and G4 were genetically farther apart from each other (nucleotide difference, about 30%) than clusters in any other segment (maximum nucleotide difference, 16%). In all trees the nucleic acid sequence variation between the clusters was greater than 5%. Only one strain, with e-type G4-84 (Fig. 2, lane 3), contained an insert: segment 2 contained 12 extra nucleotides and segment 9 contained 3 extra nucleotides.
FIG. 3.
Phylogenetic trees derived from partial RNA gene segments 1 to 11 of the nine predominant rotaviruses (normal font) and three rotaviruses with nonpredominant e-types sharing an identical RNA profile with one of the predominant rotaviruses (small font). Trees were constructed by the neighbor-joining method using the CLUSTAL V software package. The sequenced regions are shown in Table 1. Phylogenetic clusters (circled) are marked by letters A through D (cluster A contains the largest number of strains, and so forth) or numbers (P[8]-1 or -2, G1-1 or -4). Percent bootstrap support values obtained from 1,000 replicates are given between the clusters. All trees were plotted to the scale indicated by the bar (segment-1 tree). The branch lengths are related to the degree of divergence between the sequences. The branches of SA-11 are shortened, and the distances are indicated by numbers.
The lowest degree of heterogeneity was found in segments 1, 3, and 10, where all our sequences and strain Wa clustered together; only minor genetic variation existed within the clusters. In segments 2, 7, and 11, most sequences grouped to one main cluster, A, with only one diverging sequence (cluster B [Fig. 3]). In segments 2 and 11, the diverging sequence originated from a strain with the e-type G4-84. In segment 7, another strain, with the e-type G4-89, had a unique sequence unrelated to any of the sequences deposited in databanks so far, while other sequences resembled strain Wa.
The sequences of segments 5 and 8 grouped into three clusters. In segment 5 the main cluster, A, included six sequences which were found to resemble that of the prototype strain K8 of type P[9]G1 when a BLAST search was performed in GenBank (identity, 97 to 98%; not included in Fig. 3). The strain with type G4-84 (cluster C) again diverged from the main cluster, as did the two e-types from 1986 and 1987 (different G types), which clustered with strain Wa (cluster B). In segment 8 all our sequences diverged from strain Wa (cluster C). The e-types G1-81 and G1-82 with identical sequences (cluster B) also diverged from the main cluster, A, which included seven predominant strains. In the GenBank comparison, the cluster A sequences showed a closer relationship to porcine strain OSU than to strain Wa (identity, 94 versus 87%).
The segment-6 sequences showed the most genetic variation at the nucleic acid level and formed four clusters. The main cluster, A, included the prototype strain Wa. In the GenBank comparison the cluster B sequences most closely resembled the prototype strain E210 of type P[4]G2 (subgroup II) (40), with an identity of 98%. The G4-84 sequence (cluster C) showed 96% identity with strain RV-3 of type P[6]G3.
In most segments the same genetic clusters were defined both at the nucleic acid and amino acid levels, since roughly the same percentages of nucleic acid and amino acid differences between the clusters were calculated (5 to 16 versus 4 to 16%; in segment 9, 30 versus 24% [data not shown]). In segments 6 and 8, however, the clusters formed by nucleic acid analysis (genetic difference, 6 to 11%) merged at the amino acid level (differences, 0 to 4%).
RNA segment cluster combinations of the predominant rotaviruses.
In Table 3 all the different gene cluster combinations of the nine predominant rotaviruses and strain Wa are aligned according to the phylogenetic analysis results shown in Fig. 3. The results revealed that for segments other than 4 and 9, the division of the G1 and G4 viruses into different clusters also varied, suggesting frequent segment reassortment between these G types. As many as seven different RNA segment cluster combinations, none of which was identical with that for strain Wa, were found among the virus strains studied. The predominance of these virus strains was obviously not related to a particular RNA segment cluster combination.
TABLE 3.
Genetic cluster combinations for all 11 RNA segments for the nine predominant rotavirus e-types and strain Wa
| e-type | Clustera for the following RNA segment:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| G1-81 | A | A | A | P[8]-1 | A | A | A | B | G1-1 | A | A |
| G1-82 | A | A | A | P[8]-1 | A | A | A | B | G1-1 | A | A |
| G4-84 | A | B | A | P[8]-2 | C | C | A | A | G4 | A | B |
| G4-86 | A | A | A | P[8]-2 | B | D | A | A | G4 | A | A |
| G1-87 | A | A | A | P[8]-1 | B | A | A | A | G1-1 | A | A |
| G4-89 | A | A | A | P[8]-1 | A | B | B | A | G4 | A | A |
| G4-91 | A | A | A | P[8]-1 | A | A | A | A | G4 | A | A |
| G4-93 | A | A | A | P[8]-1 | A | A | A | A | G4 | A | A |
| G1-97 | A | A | A | P[8]-1 | A | B | A | A | G1-1 | A | A |
| Wa | A | A | A | P[8]-1 | B | A | A | C | G1-4 | A | A |
Cluster designations refer to the genetic clusters in Fig. 3.
Three (G1-81, G1-87, and G1-97) out of the four G1 e-types had different RNA segment cluster combinations; thus, e-types G1-81 and G1-82, which had identical nucleotide sequences and identical RNA profiles in PAGE (Fig. 2, lanes 1 and 2), were isolates of the same virus strain that predominated during two successive seasons. The cluster combinations of the three different G1 strains clearly indicate that the virus strains have not evolved from each other by accumulation of point mutations. A change of one or several RNA segments from one cluster to another during a period of 6 or 16 years is more likely to have happened by reassortment. Very similar results were found for the five G4 rotavirus strains. Four different RNA segment cluster combinations by e-types G4-84, G4-86, G4-89, and G4-91 appeared; e-types G4-91 and G4-93 had identical RNA profiles (Fig. 2, lanes 7 and 8) and identical nucleotide sequences with the exception of one silent nucleotide difference in segment 9. Among the different type G4 strains several genomic reassortments probably had occurred as well.
Comparison of the gene cluster combinations of G1 and G4 strains reveals that these strains shared the same genetic clusters and could exchange several of their RNA segments with each other. Furthermore, comparisons of the sequences with those in GenBank (see above) showed that some of these genetic pools were probably also shared with other P and G types (P[4], P[6], P[9], G2, and G3) that were not investigated in this study. Based on the results of Table 3, a directed selection of segments into predominant strains seems unlikely.
Stability of the rotavirus e-types.
In addition to the predominant rotaviruses, three nonpredominant e-types that were found to have RNA profiles identical with those of some predominant e-types were included in the detailed sequence analysis (Fig. 3, e-types in small font). All 11 sequences of e-type G1-85, which had an RNA profile identical to that of the predominant e-type G1-87, were almost identical with the G1-87 sequences. In total only four nucleotide differences in the partial sequences of segments 3, 7, and 8 were found. The predominant e-type G4-89 had an RNA profile identical with that of nonpredominant e-types G4-90 and G4-94. Their sequences also shared a high level of identity, as seen in the phylogenetic trees of Fig. 3. The results implied that these particular rotavirus strains remained relatively stable over the observation period, although they predominated only during one epidemic season.
DISCUSSION
In this study the nine predominant rotavirus strains identified over a 17-year period (1981 to 1997) in Finland were analyzed. Predominant G4 rotavirus strains were detected more frequently than G1 strains (five versus four seasons). In 1992, Woods et al. (54) reported that in combined materials from many countries, type G1 was the most common global G type in circulation. In several recent studies, P[8]G4 strains have been found to be the second most prevalent strains after P[8]G1 strains (5, 9). Type G9, a newly emerging type that has been found in many countries (see, e.g., references 31 and 53), was not found among the predominant strains in this study.
In the present study the P components of the predominant G1 and G4 types were always of type P[8]. The P sequences belonged to the genetic lineages P[8]-1 and P[8]-2, i.e., they were Wa- and F45-like (23), respectively. The third established lineage, P[8]-3 (29, 36), was not found in this study. The finding that the G1 and G4 strains shared the same P[8] lineages agrees well with previous reports (29, 36).
The G1-VP7 sequences have been analyzed perhaps in greater detail than those of any other G type, and different nomenclatures for lineages have been proposed. Xin et al. (55) divided VP7 sequences into three subtypes (A, B, and intermediate). Jin et al. (30) found four G1 lineages (I through IV) among vaccine failure strains. The existence of at least four lineages has been confirmed (29, 36). In our study all four predominant G1 strains had VP7 sequences belonging to lineage G1-1 (or lineage II [30]), although strains representing the other three lineages (G1-2 to G1-4) were found circulating during the 17-year period. Lineage G1-1 (lineage II) isolates have been classified as monotype G1a (15), and predominant rotavirus strains of this monotype have also been reported as circulating in Australia (39). The P components of our predominant G1 strains belonged to lineage P[8]-1. For reasons not known, this P/G-lineage combination, P[8]-1 G1-1, has been most favored. Among the predominant G4 strains, P components of P[8]-1 and -2 lineages were observed, but the G component always belonged to the same VP7 lineage. These strains probably are of subtype 4A, since strain ST3 (P[6]G4), representing subtype 4A (21), also belongs to this lineage.
Partial sequences of all 11 RNA segments from the nine predominant strains were determined. The sequenced areas covered the most variable regions of the genes, as determined from the sequence data in the GenBank database. In segments 1, 3, and 10 no clustering was observed, but there were only minor genetic variations between the sequences. In segment 3 most of the few nucleic acid differences also resulted in amino acid shifts. Although the segment-1, -3, and -10 sequences of the Finnish strains occurred in one cluster, genetic variation in these segments has been found. For example, corresponding sequences of the human rotavirus strain KU (GenBank accession numbers, AB022765 and AB022767) have nucleotide identities of only 90 to 91% with the sequences of the Finnish strains and would cluster separately. The segment-10 (NSP4) sequences especially showed an unexpected lack of heterogeneity, since as many as four human NSP4 genotypes have been described (reviewed in reference 17).
In gene segments other than 1, 3, and 10, several genetic clusters were formed. Differences in the nucleic acid sequences were the main reason for clustering; insertions were found only in the G4-84 rotavirus strain. As expected, the genetically most distant clusters were found in the VP7-encoding segments of the G1 versus G4 rotaviruses, reflecting the antigenic importance of VP7. The prototype strain Wa, type P[8]G1, usually coclustered with some of the sequences found in this study in the phylogenetic trees, with the exception of the 8th segment of strain Wa. In segment 8 the closest sequence in GenBank to our sequences was of porcine origin, strain OSU (P[7]G5). The very few human rotavirus segment-8 sequences in the database may explain this finding. Two other porcine rotavirus sequences in GenBank were found to be closely related to human rotavirus sequences. The segment-7 sequence of strain Price had a nucleotide identity of 98% with G4-84 and G1-87 (cluster A), and the segment-11 sequence of strain YM had an identity of 96% with G4-84 (cluster B). In fact, the G4-84 strain that resembled some porcine rotavirus strains showed the greatest sequence heterogeneity in the majority of segments in this study. Brazilian piglets have been found to be infected with P[8]G1 strains, traditionally regarded as human viruses (45). Furthermore, a phylogenetic analysis of segment-5 sequences has revealed a close relationship between some human and porcine rotaviruses (16).
The present data indicate frequent reassortment between most gene segments of G1 and G4 rotavirus strains. Segment exchange is not likely to be limited to P[8]G1 and P[8]G4 rotaviruses, analyzed in this study. The GenBank comparisons suggested that exchange of some RNA segments between these rotaviruses and the P[4], P[6], and P[9] G2 and G3 rotaviruses is not ruled out. Several recent reports have also described possible rotavirus reassortants (e.g., references 29, 45, 50, 52, and 56). Gouvea et al. (23) found that G components of G5 and G9 rotaviruses shared the same genetic P[8] lineages as G1 and G4 strains. In the United Kingdom the P components of P[8]G9 strains were found to be closely related to those of cocirculating G1, G3, and G4 strains (29). Interaction by reassortment of other segments may be freer of restrictions than that of segments 4 and 9, in which certain G/P combinations seem to be preferred even at the cluster level (Table 2) (29). The expanding rotavirus diversity (recently reviewed in reference 10) seems to have developed mainly on the basis of genomic reassortment.
The cluster combinations of the rotaviruses for all 11 RNA segments were compared with each other. Our aim was to look for some common combinations that would lead to predominance of these particular strains. No such combination was found, but all predominant e-types either were totally identical, e.g., the same strain, or their segment cluster combinations differed randomly. No directed selection of RNA segments into predominant strains was found.
Until now electropherotyping and hybridization have been the methods most commonly used to characterize the entire genomes of different rotavirus strains. Clearly, even partial sequencing of the 11 RNA segments gives better and more specific information. Genetic clusters of rotavirus strains can be defined on the basis of the sequences of relatively short gene fragments, since recombination within genes is very unlikely to occur frequently. Such fragments have been identified in gene segments in this and a previous study (36). These results were obtained with virus isolates from Finland, a relatively sparsely populated, developed country. The probability of reassortments taking place must be considered much higher in developing countries.
The overall mutation rate of segment 11 has been estimated as <5 × 10−5 per replicated base for porcine rotaviruses, which would result in 1 mutation per replication round for the entire genome (4). In spite of our relatively long study period of 17 years, it was not possible to find evidence for straightforward evolutionary changes by sequential point mutations (genetic drift) for rotaviruses similar to those reported for hemagglutinin genes of influenza A viruses (18). One rotavirus strain in the present study could be found unchanged during a period of 5 years. A few nucleotide differences were detected in the isolates of different epidemic seasons, but in total, no more changes were found during a longer period than among isolates of one season (36).
Like rotaviruses, influenza viruses show reassortment of cocirculating viruses in humans but especially in animals (H5N1, in geese, chickens, and ducks [26]). Rotaviruses seem to circulate in humans, like influenza A virus in bird populations, without any pronounced exclusion of strains. This offers ample opportunities for reassortment.
Acknowledgments
We thank Hannu Fritze for the electrophoretic analysis of 92 rotavirus samples in 1981 and 1982 and Alexander Plyusnin and Antti Vaheri for critical reading of the manuscript.
REFERENCES
- 1.Ahmed, M. U., S. Urasawa, K. Taniguchi, T. Urasawa, N. Kobayashi, F. Wakasugi, A. I. M. M. Islam, and H. A. Sakikh. 1991. Analysis of human rotavirus strains prevailing in Bangladesh in relation to nationwide floods brought by the 1988 monsoon. J. Clin. Microbiol. 29:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, R. F., G. P. Davidson, I. H. Holmes, and B. J. Ruck. 1973. Virus particles in epithelial cells of duodenal mucosa from children with viral gastroenteritis. Lancet ii:1281-1283. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, R. F., L. E. Unicomb, and G. L. Barnes. 1991. Epidemiology of rotavirus serotypes in Melbourne, Australia, from 1973 to 1989. J. Clin. Microbiol. 29:862-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackhall, J., A. Fuentes, and G. Magnusson. 1996. Genetic stability of a porcine rotavirus RNA segment during repeated plaque isolation. Virology 225:181-190. [DOI] [PubMed] [Google Scholar]
- 5.Bon, F., C. Fromantin, S. Aho, P. Pothier, and E. Kohli. 2000. G and P genotyping of rotavirus strains circulating in France over a three-year period: Detection of G9 and P[6] strains at low frequencies. J. Clin. Microbiol. 38:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Intussusception among recipients of rotavirus vaccine—United States, 1998-1999. Morb. Mortal. Wkly. Rep. 48:577-581, 1007. [PubMed] [Google Scholar]
- 7.Chanock, S. J., E. A. Wenske, and B. N. Fields. 1983. Human rotaviruses and genome RNA. J. Infect. Dis. 148:45-50. [DOI] [PubMed] [Google Scholar]
- 8.Coulson, B. 1987. Variation in neutralization epitopes of human rotaviruses in relation to genomic RNA polymorphism. Virology 159:209-216. [DOI] [PubMed] [Google Scholar]
- 9.Cubitt, W. D., A. D. Steele, and M. Iturriza. 2000. Characterisation of rotaviruses from children treated at a London hospital during 1996: emergence of strains G9PA[6] and G3P2A[6]. J. Med. Virol. 61:150-154. [DOI] [PubMed] [Google Scholar]
- 10.Cunliffe, N. A., J. S. Bresee, J. R. Gentsch, R. I. Glass, and C. A. Hart. 2002. The expanding diversity of rotaviruses. Lancet 359:640-642. [DOI] [PubMed] [Google Scholar]
- 11.Cunliffe, N. A., J. S. Gondwe, R. L. Broadhead, M. E. Molyneux, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and C. A. Hart. 1999. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J. Med. Virol. 57:308-312. [PubMed] [Google Scholar]
- 12.Cunliffe, N. A., P. A. Woods, J. P. G. Leite, B. K. Das, M. Ramachandran, M. K. Bhan, C. A. Hart, R. I. Glass, and J. R. Gentsch. 1997. Sequence analysis of NSP4 gene of human rotavirus allows classification into two main genetic groups. J. Med. Virol. 53:41-50. [PubMed] [Google Scholar]
- 13.Desselberger, U. 1996. Genome rearrangements of rotaviruses. Adv. Virol. 46:69-95. [DOI] [PubMed] [Google Scholar]
- 14.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diwakarla, C. S., and E. A. Palombo. 1999. Genetic and antigenic variation of capsid protein VP7 of serotype G1 human rotavirus isolates. J. Gen. Virol. 80:341-344. [DOI] [PubMed] [Google Scholar]
- 16.Dunn, S. J., T. L. Cross, and H. B. Greenberg. 1994. Comparison of the rotavirus nonstructural protein NSP1 (NS53) from different species by sequence analysis and Northern blot hybridization. Virology 203:178-183. [DOI] [PubMed] [Google Scholar]
- 17.Estes, M. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 18.Fitch, W. M., J. M. E. Leiter, X. Li, and P. Palese. 1991. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. USA 88:4270-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerna, G., S. Arista, N. Passarani, A. Sarasini, and M. Battaglia. 1987. Electropherotype heterogeneity within serotypes of human rotavirus strains circulating in Italy. Arch. Virol. 95:129-135. [DOI] [PubMed] [Google Scholar]
- 21.Gerna, G., A. Sarasini, A. di Matteo, M. Parea, P. Orsolini, and M. Battaglia. 1988. Identification of two subtypes of serotype 4 human rotavirus by using VP7-specific neutralizing monoclonal antibodies. J. Clin. Microbiol. 26:1388-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z.-Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouvea, V., R. C. C. Lima, R. E. Linhares, H. F. Clark, C. M. Nosawa, and N. Santos. 1999. Identification of two lineages (WA-like and F45-like) within the major rotavirus genotype P[8]. Virus Res. 59:141-147. [DOI] [PubMed] [Google Scholar]
- 24.Graham, A., G. Kudesia, A. M. Allen, and U. Desselberger. 1987. Reassortment of human rotavirus possessing genome rearrangements with bovine rotavirus: evidence for host cell selection. J. Gen. Virol. 68:115-122. [DOI] [PubMed] [Google Scholar]
- 25.Green, K., J. Sears, K. Taniguchi, K. Midthun, Y. Hoshino, M. Gorziglia, K. Nishikawa, S. Urasawa, A. Kapikian, R. Chanock, and J. Flores. 1988. Prediction of human rotavirus serotype by nucleotide sequence analysis of the VP7 protein gene. J. Virol. 62:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan, Y., M. Peiris, K. F. Kong, K. C. Dyrting, T. M. Ellis, T. Sit, L. J. Zhang, and K. F. Shortridge. 2002. H5N1 influenza viruses isolated from geese in Southeastern China: evidence for genetic reassortment and interspecies transmission to ducks. Virology 292:16-23. [DOI] [PubMed] [Google Scholar]
- 27.Higgins, D. G., A. J. Bleasby, and R. Fuchs. 1991. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biol. Sci. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 28.Horie, Y., O. Masamune, and O. Nakagomi. 1997. Three major alleles of rotavirus NSP4 proteins identified by sequence analysis. J. Gen. Virol. 78:2341-2346. [DOI] [PubMed] [Google Scholar]
- 29.Iturriza-Gomara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin, Q., R. L. Ward, D. R. Knowlton, Y. B. Gabbay, A. C. Linhares, R. Rappaport, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Divergence of VP7 genes of G1 rotaviruses isolated from infants vaccinated with reassortant rhesus rotaviruses. Arch. Virol. 141:2057-2076. [DOI] [PubMed] [Google Scholar]
- 31.Kirkwood, C. D., J. R. Gentsch, Y. Hoshino, H. F. Clark, and R. I. Glass. 1999. Genetic and antigenic characterization of a serotype P[6]G9 human rotavirus strain isolated in the United States. Virology 256:45-53. [DOI] [PubMed] [Google Scholar]
- 32.Konno, T., T. Sato, H. Suzuki, S. Kitaoka, N. Katsushima, M. Sakamoto, N. Yazaki, and N. Ishida. 1984. Changing RNA patterns in rotaviruses of human origin: demonstration of a single dominant pattern at the start of an epidemic and various patterns thereafter. J. Infect. Dis. 149:683-687. [DOI] [PubMed] [Google Scholar]
- 33.Matsui, S. M., E. R. Mackow, S. Matsuno, P. S. Paul, and H. B. Greenberg. 1990. Sequence analysis of gene 11 equivalents from “short” and “supershort” strains of rotavirus. J. Virol. 64:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maunula, L., H. Piiparinen, and C.-H. von Bonsdorff. 1999. Confirmation of Norwalk-like virus amplicons after RT-PCR by microplate hybridization and direct sequencing. J. Virol. Methods 83:125-134. [DOI] [PubMed] [Google Scholar]
- 35.Maunula, L., and C.-H. von Bonsdorff. 1995. Rotavirus serotypes and electropherotypes in Finland from 1986 to 1990. Arch. Virol. 140:877-890. [DOI] [PubMed] [Google Scholar]
- 36.Maunula, L., and C.-H. von Bonsdorff. 1998. Short sequences define lineages: phylogenetic analysis of group A rotaviruses based on partial sequences of genome segments 4 and 9. J. Gen. Virol. 79:321-332. [DOI] [PubMed] [Google Scholar]
- 37.Nakagomi, O., A. Ohshima, Y. Aboudy, I. Shif, M. Mochizuki, T. Nakagomi, and S. T. Gotlieb. 1990. Molecular identification by RNA-RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J. Clin. Microbiol. 28:1198-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuttall, S. D., C. P. Hum, I. H. Holmes, and M. L. Dyall-Smith. 1989. Sequences of VP9 genes from short and supershort rotavirus strains. Virology 171:453-457. [DOI] [PubMed] [Google Scholar]
- 39.Palombo, E. A., R. F. Bishop, and R. G. H. Cotton. 1993. Intra- and inter-season genetic variability in the VP7 gene of serotype 1 (monotype 1a) rotavirus clinical isolates. Arch. Virol. 130:57-69. [DOI] [PubMed] [Google Scholar]
- 40.Palombo, E. A., H. C. Bugg, P. J. Masendycz, B. S. Coulson, G. L. Barnes, and R. F. Bishop. 1996. Multiple-gene rotavirus reassortants responsible for an outbreak of gastroenteritis in central and northern Australia. J. Gen. Virol. 77:1223-1227. [DOI] [PubMed] [Google Scholar]
- 41.Patton, J. T., L. Salter-Cid, A. Kalbach, E. A. Mansell, and M. Kattoura. 1993. Nucleotide and amino acid sequence analysis of the rotavirus nonstructural RNA-binding protein NS35. Virology 192:438-446. [DOI] [PubMed] [Google Scholar]
- 42.Ramig, R. F. 1990. Superinfecting rotaviruses are not excluded from genetic interactions during asynchronous mixed infections in vitro. Virology 176:308-310. [DOI] [PubMed] [Google Scholar]
- 43.Ramig, R. F., and R. L. Ward. 1991. Genomic segment reassortment in rotaviruses and other reoviridae. Adv. Virus Res. 39:163-207. [DOI] [PubMed] [Google Scholar]
- 44.Rao, C., M. Das, P. Ilango, R. Lalwani, B. Rao, and K. Gowda. 1995. Comparative nucleotide and amino acid sequence analysis of the sequence-specific RNA-binding rotavirus nonstructural protein NSP3. Virology 207:327-333. [DOI] [PubMed] [Google Scholar]
- 45.Santos, N., C. C. Lima, C. M. Nozawa, R. E. Linhares, and V. Gouvea. 1999. Detection of porcine rotavirus type G9 and a mixture of types G1 and G5 associated with Wa-like VP4 specificity: evidence of natural human-porcine genetic reassortment. J. Clin. Microbiol. 37:2734-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos, N., M. Riepenhoff-Talty, H. F. Clark, P. Offit, and V. Gouvea. 1994. VP4 genotyping of human rotavirus in the United States. J. Clin. Microbiol. 32:205-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, Y., T. Gojobori, and O. Nakagomi. 1998. Intragenic recombinations in rotaviruses. FEBS Lett. 427:183-187. [DOI] [PubMed] [Google Scholar]
- 48.Tabor, S., and C. C. Richardson. 1990. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. J. Biol. Chem. 265:8322-8328. [PubMed] [Google Scholar]
- 49.Tauscher, G. I., and U. Desselberger. 1997. Viral determinants of rotavirus pathogenicity in pigs: production of reassortants by asynchronous coinfection. J. Virol. 71:853-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward, R. L., O. Nakagomi, D. R. Knowlton, M. M. McNeal, T. Nakagomi, J. D. Clemens, D. A. Sack, and G. M. Schiff. 1990. Evidence for natural reassortants of human rotaviruses belonging to different genogroups. J. Virol. 64:3219-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe, M., T. Nakagomi, Y. Koshimura, and O. Nakagomi. 2001. Direct evidence for genome segment reassortment between concurrently-circulating human rotavirus strains. Arch. Virol. 146:557-570. [DOI] [PubMed] [Google Scholar]
- 53.Widdowson, M.-A., G. J. J. Doornum, W. H. M. van der Poel, A. S. de Boer, U. Mahdi, and M. Koopmans. 2000. Emerging group-A rotavirus and a nosocomial outbreak of diarrhoea. Lancet 356:1161. [DOI] [PubMed] [Google Scholar]
- 54.Woods, P. A., J. Gentsch, V. Gouvea, L. Mata, A. Simhon, M. Santosham, Z. Bai, S. Urasawa, and R. I. Glass. 1992. Distribution of serotypes of human rotavirus in different populations. J. Clin. Microbiol. 30:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xin, K.-Q., S. Morikawa, Z.-Y. Fang, A. Mukoyama, K. Okuda, and H. Ushijima. 1993. Genetic variation in VP7 gene of human rotavirus serotype 1 (G1 type) isolated in Japan and China. Virology 197:813-816. [DOI] [PubMed] [Google Scholar]
- 56.Zao, C.-L., W.-N. Yu, C.-L. Kao, K. Taniguchi, C.-Y. Lee, and C.-N. Lee. 1999. Sequence analysis of VP1 and VP7 genes suggests occurrence of a reassortant of G2 rotavirus responsible for an epidemic of gastroenteritis. J. Gen. Virol. 80:1407-1415. [DOI] [PubMed] [Google Scholar]