Abstract
Retroviral integration is mediated by a preintegration complex (PIC) which contains the viral DNA made by reverse transcription together with associated protein factors. Prior to association with target DNA, the PIC must avoid suicidal intramolecular integration of its viral DNA (autointegration). We have demonstrated that barrier-to-autointegration factor (BAF) blocks the autointegration of Moloney murine leukemia virus (MoMLV) PICs in vitro. In this study, we show that BAF is an authentic component of MoMLV. Analysis of the sedimentation properties of initial, salt-stripped, and BAF-reconstituted PICs reveals that the viral DNA within the PIC is reversibly compacted by BAF, consistent with the functional role of BAF in protecting the viral DNA from autointegration. Furthermore, we find that BAF can promote the association of PICs with target DNA. Thus, our data suggest that BAF plays critical roles in promoting preferential intermolecular integration by both blocking autointegration and stimulating the capture of target DNA.
Retroviral virions contain both genomic RNA and many proteins that are required for completing the early phase of infection in the host cell. After the entry of a virion into the target cell, reverse transcription of viral RNA occurs to produce a copy of viral DNA within a higher-order nucleoprotein complex, the preintegration complex (PIC), which is derived from the core components of the infecting virion. Subsequently, this viral DNA is integrated into chromosomal DNA to produce a provirus in the infected cell. This integration step is essential in the retroviral replication cycle (1, 3, 8, 18, 19, 20) and comprises 3′-end processing, strand transfer, and gap repair measures in vivo (5, 10, 16, 30). Although purified integrase protein alone can carry out the steps of 3′-end processing and joining of viral DNA with simple DNA substrates in vitro (9, 11, 21, 22, 26), the reaction lacks the full fidelity of DNA integration mediated by PICs; most products result from the integration of only a single viral DNA end into one strand of target DNA (6). In contrast, PICs isolated from infected cells efficiently insert both viral DNA ends into target DNA in a pairwise manner in vitro, with all the hallmarks of the reaction in the cell (2, 5, 13, 16). Therefore, PICs may include an additional factor(s) that is required for the authentic retroviral DNA integration reaction and that is absent in the simplified systems using integrase protein alone.
A striking feature of PICs is their strong preference for intermolecular integration into a target DNA and their avoidance of intramolecular integration into the viral DNA, a reaction termed autointegration. The avoidance of autointegration of the viral DNA can be disrupted by incubating PICs with a high concentration of salt (salt stripping) (23). The strong preference for intermolecular integration can be restored by incubation with a cytoplasmic extract from uninfected cells (23). By using this reconstitution reaction as an assay, a protein termed barrier-to-autointegration factor (BAF) that could substitute for cytoplasmic extract in the reconstitution assay was identified (24). BAF is a cellular protein that blocks autointegration and promotes intermolecular integration of salt-stripped Moloney murine leukemia virus (MoMLV) and human immunodeficiency virus type 1 (HIV-1) PICs (7, 24). BAF is a homodimeric protein with 89 amino acid residues and is highly conserved among species (24, 28, 31). BAF interacts with cellular components, including lamin-associated polypeptide 2, which is associated with the inner nuclear membrane (17, 27, 29). In vitro studies have revealed that BAF bridges double-stranded DNA (dsDNA) with no detectable sequence specificity (24, 31). DNA bridging results in intramolecular compaction at low DNA concentrations and intermolecular aggregation at high DNA concentrations. We have proposed that the DNA-bridging activity of BAF may block the autointegration of retroviral DNA in PICs by compacting the DNA into a rigid structure, making it inaccessible as a target for autointegration (24). Here, we have investigated the role of BAF in the nucleoprotein organization of MoMLV PICs and the mechanism of its regulatory role in preventing autointegration and stimulating the intermolecular integration of viral DNA.
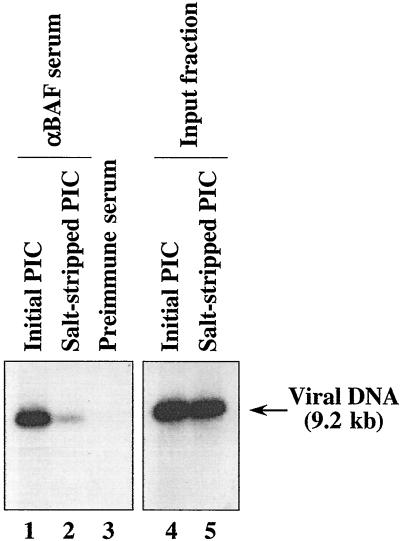
The intermolecular integration activities of MoMLV PICs are abolished by treatment with high salt concentrations and restored by reconstitution with either cell extract or purified BAF (24). On the basis of this functional reconstitution activity, it was concluded that BAF was initially associated with the PIC. However, we could not exclude the formal possibility that a different factor was initially responsible for the intermolecular integration preference, even though reconstitution with purified BAF restored this preference. To unambiguously resolve this issue, we tested whether BAF is associated with PICs by immunoprecipitation using anti-BAF serum from a rabbit immunized with a previously immunogenic BAF peptide (17). MoMLV PICs were prepared by coculture of NIH 3T3 cells and clone 4 cells, a MoMLV-producing cell line (16, 23). Cytoplasmic extract of MoMLV-infected cells was isolated by using buffer A (20 mM HEPES-NaOH [pH 7.5], 5 mM MgCl2, 150 mM KCl, 10 mM dithiothreitol [DTT], 20 μg of aprotinin [Sigma] per ml) containing 0.025% digitonin (Sigma). The isolated extract was then concentrated with a Centriprep 100 concentrator (Amicon) and stored until use at −70°C in buffer B (20 mM HEPES-NaOH [pH 7.5], 5 mM MgCl2, 1 mM DTT, 6 mM EDTA, 6% sucrose) containing 150 mM KCl (fraction I). To remove free BAF, fraction I was gel filtrated through a Sephacryl S-1000 Superfine spin column (Amersham Pharmacia) equilibrated with buffer B containing 150 mM KCl, thus producing fraction II. Fraction II was incubated at 4°C for 1 h with 30 μl of anti-BAF serum or preimmune rabbit serum in buffer C (20 mM HEPES-NaOH [pH 7.5], 5 mM MgCl2, 150 mM KCl, 10 mM DTT, 20 μg of aprotinin/ml, 6 mM EDTA, 400 μg of bovine serum albumin/ml) containing 0.05% Nonidet P-40 (NP-40). Then, 30 μl of protein A-G-agarose beads (Santa Cruz Biotechnology) was added to the mixture and incubation was continued at 4°C for 3 h. After a washing with the same buffer, viral DNA was recovered from the immune complex by proteinase K-sodium dodecyl sulfate treatment, phenol-chloroform extraction, and ethanol precipitation and detected by Southern blot analysis with a 32P-labeled probe for the MoMLV long terminal repeat (LTR) sequence (23). Figure 1 shows that initial PICs were recovered from the protein A-G-agarose after incubation with anti-BAF serum but that immunoprecipitation with preimmune serum yielded no recovery of PICs. Furthermore, the recovery of PICs with anti-BAF serum was significantly impaired by salt stripping (Fig. 1), indicating that BAF was dissociated from the salt-stripped PICs. Thus, we conclude that BAF is an authentic component of MoMLV PICs.
FIG. 1.
Immunoprecipitation analysis of MoMLV PICs with anti-BAF serum. Initial (lanes 1 and 3) and salt-stripped (lane 2) PICs were subjected to immunoprecipitation with anti-BAF rabbit serum (αBAF serum; lanes 1 and 2) or preimmune serum (lane 3) in the presence of 0.05% NP-40. Viral DNA was extracted from captured complexes and detected by Southern blotting with a 32P-labeled probe for the MoMLV LTR sequence. Lanes 4 and 5 show the input fraction of initial and salt-stripped PICs, respectively.
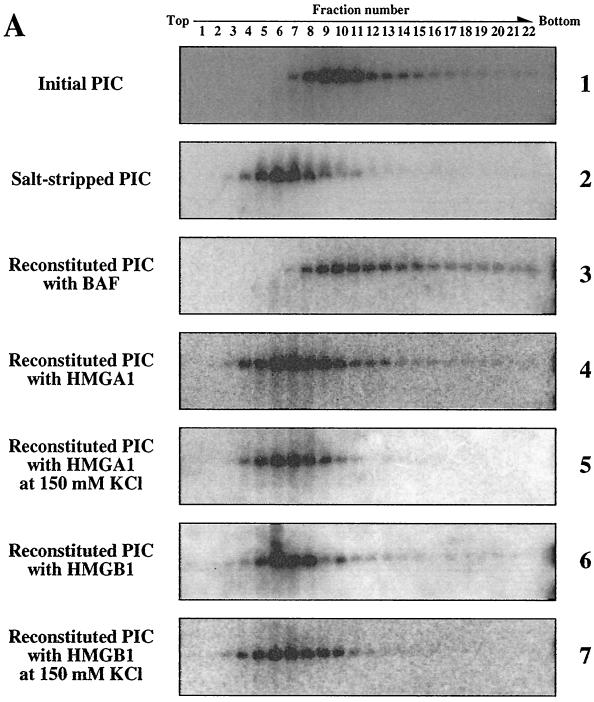
How does BAF block the autointegration of PIC? BAF binds dsDNA nonspecifically and bridges together dsDNA molecules (24, 31). This DNA-bridging property of BAF suggested that the association of BAF with viral DNA within the PICs might block autointegration by compacting the viral DNA, thereby making it inaccessible as a target (24). To test this hypothesis, we compared the sedimentation rates of initial, salt-stripped, and BAF-reconstituted PICs in sucrose gradients. To salt strip PICs, fraction I was incubated on ice with KCl to a final concentration of 750 mM for 1 h, and PICs were separated from free components with a Sephacryl S-1000 spin column equilibrated with buffer B containing 750 mM KCl, followed by a second spin dialysis with a column equilibrated with buffer B containing 150 mM KCl (24). Salt-stripped PICs were then incubated on ice for 1 h with human BAF protein (10 ng) (31) in a reaction mixture (100 μl) containing 20 mM HEPES-NaOH (pH 7.5), 5 mM MgCl2, 400 mM KCl, 6 mM EDTA, 1 mM EDTA, 0.04% bovine serum albumin, 1 mM DTT, 40% Nycodenz, and 10 mM (NH4)2SO4 (24). Integration activities of reconstituted PICs were evaluated by the previously described integration activity assay with φX174 replicative form I (RFI) DNA (Gibco BRL) as the target DNA (24). For the sedimentation assay, PIC fractions were gel filtrated with a spin column equilibrated with buffer B containing 150 mM KCl to remove the Nycodenz from the solution, loaded on 15 to 30% sucrose gradients in buffer A containing 6 mM EDTA, and centrifuged in a Beckman TLS55 rotor at 30,000 rpm for 1 h at 4°C. The gradient was fractionated from the top into 22 fractions, and the viral DNA extracted from each fraction was detected by Southern blotting. Figure 2A shows that peak fractions of initial MoMLV PICs were numbers 8 to 12 (panel 1). In contrast to initial PICs, salt-stripped PICs, which only autointegrate (Fig. 2B), sedimented more slowly, with peak fraction numbers being 4 to 9 (Fig. 2A, panel 2). However, when the salt-stripped PICs were reconstituted with BAF, the intermolecular integration activities were restored (Fig. 2B) and the complexes sedimented like the initial complexes in sucrose gradients, with peaks in fractions 8 to 12 (Fig. 2A, panel 3). These results demonstrate that salt treatment alters the sedimentation of PICs in a manner consistent with the adoption by viral DNA of a more open structure and that reconstitution with BAF restores the native sedimentation behavior, together with an efficient intermolecular integration activity.
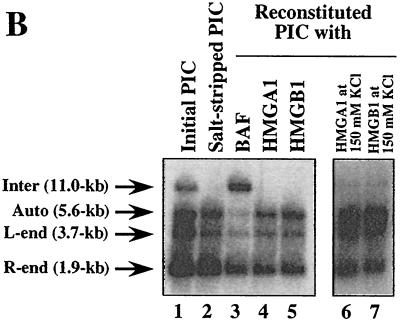
FIG. 2.
MoMLV PIC is compacted by BAF. (A) Sedimentation properties of initial (panel 1), salt-stripped (panel 2), BAF-reconstituted (panel 3), HMGA1-reconstituted (panels 4 and 5), and HMGB1-reconstituted (panels 6 and 7) PICs. The MoMLV PIC fraction was salt stripped with 750 mM KCl and then incubated with BAF, HMGA1, or HMGB1 protein in the presence of 400 mM KCl (panels 1, 2, 3, 4, and 6) or 150 mM KCl (panels 5 and 7). After gel filtration, each reconstituted PIC fraction was layered on a 15 to 30% sucrose gradient and centrifuged at 30,000 rpm for 1 h with a Beckman rotor. The gradient was fractionated from the top to bottom into 22 fractions, and viral DNA in each fraction was detected by Southern blotting. (B) Intermolecular integration activities of initial, salt-stripped, and reconstituted PICs. Salt-stripped PICs were incubated with BAF (lane 3), HMGA1 (lanes 4 and 6), or HMGB1 (lanes 5 and 7) in the presence of 400 mM KCl (lanes 1 to 5) or 150 mM KCl (lanes 6 and 7) and then added to the integration reaction mixture containing φX174 RFI DNA as the target DNA to check integration activities. After incubation, DNA products from the reaction were digested with BamHI and detected by Southern blotting with a 32P-labeled probe for the MoMLV LTR sequence. The 11.0-kb band results from intermolecular integration of the viral DNA into φX174 RFI DNA (Inter). The 5.6-kb band and the smear below it result from autointegration of the viral DNA (Auto). The 3.7- and 1.9-kb bands are the unreacted viral DNAs containing the 5′ LTR (L-end) and the 3′ LTR (R-end), respectively (23).
Some members of the HMG protein family have been implicated as being involved in retroviral DNA integration because they have been reported to increase the efficiency of in vitro integration reactions with salt-stripped HIV-1 or MoMLV PICs (12, 25) and to stimulate concerted integration reactions with purified HIV-1 integrase protein (19). We therefore subjected salt-stripped PICs to the same reconstitution procedure, replacing BAF with 1 μg of HMGA1 [formerly HMG I(Y)] (12); the concentration of HMGA1 was 100-fold greater than that of the BAF used in reconstitution reactions, since such concentrations of HMGA1 are reported to be necessary to stimulate intermolecular integration (12, 25). However, when reconstitution with HMGA1 was carried out according to the same protocol used for reconstitution with cell extract, we observed no significant stimulation of intermolecular integration (Fig. 2B) or recompaction of the DNA (Fig. 2A, panel 4) in salt-stripped PICs. We also tested whether HMGA1 could substitute for BAF when reconstitution reactions were carried out at lower salt concentrations (150 mM KCl) but again observed no recompaction of the viral DNA (Fig. 2A, panel 5). Similarly, 1 μg of HMGB1 (formerly HMG1) failed to recompact the viral DNA (Fig. 2A, panels 6 and 7) or stimulate intermolecular integration (Fig. 2B). Since reconstitution with cytoplasmic extract (23) or purified BAF by the same procedure efficiently restored intermolecular integration (Fig. 2B), we conclude that BAF in the cell extract is solely responsible for the intermolecular integration preference and that it modulates the compaction of MoMLV PICs. Our results concerning the relative efficiencies of BAF and HMGA1 at stimulating intermolecular integration of salt-stripped PICs parallel the findings of Chen and Engelman (7) with HIV-1 PICs; even a >2,000-fold excess of HMGA1 over BAF in otherwise identical reconstitution reaction mixtures resulted in only minimal stimulation of intermolecular integration.
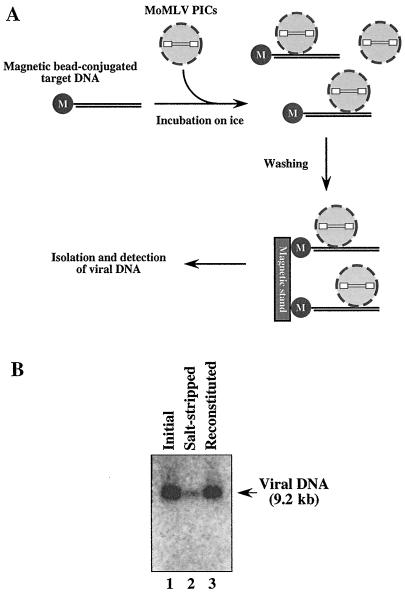
The DNA-bridging property of BAF raises the possibility that BAF may play a role in recruiting PICs to target DNA. We therefore tested this hypothesis by analyzing the activities of initial, salt-stripped, and BAF-reconstituted PICs in binding to target DNA. To prepare target DNA, a 5′-end-biotinylated 2-kb fragment (1 μg) of φX174 RFI DNA was amplified by PCR, conjugated with Streptavidin MagneSphere paramagnetic particles (Promega) in buffer C at room temperature for 30 min, and washed twice with the same buffer. To check the binding activities, initial, salt-stripped, and BAF-reconstituted PIC fractions were gel filtered with a spin column equilibrated with buffer B containing 150 mM KCl and incubated on ice for 1 h with the magnetic bead-conjugated target DNA in buffer C; the low temperature prevents the integration reaction from occurring during the binding assay (data not shown) (4). The PICs that associated with target DNA were then recovered by washing the beads three times with cold buffer C and deproteinized with proteinase K-sodium dodecyl sulfate. Viral DNA was extracted by phenol-chloroform and ethanol precipitation and detected by Southern blotting (Fig. 3A). Figure 3B shows that the binding activities of initial MoMLV PICs were significantly impaired by salt stripping. However, when the salt-stripped PICs were reconstituted with BAF, their target DNA binding activities were restored (Fig. 3B), consistent with our hypothesis that BAF promotes the association of PICs with target DNA. Although the architecture of chromatin, the target for viral DNA integration in infected cells, is different from that of naked DNA, our result suggests the possibility that BAF may play a role in anchoring PICs to DNA prior to the integration reaction. Thus, prevention of autointegration and promotion of intermolecular integration by BAF may involve one mechanism (DNA bridging) with two different outcomes: (i) intramolecular bridging by BAF may compact the viral DNA, making it less accessible as a target for integration, and (ii) anchoring of PICs to other DNA by BAF may promote intermolecular integration.
FIG. 3.
Enhancement by BAF of the association of MoMLV PICs with target DNA. (A) Schematic depiction of the PIC binding assay. The MoMLV PIC fraction was mixed with target DNA (2 kb) which had been conjugated with paramagnetic beads, and the binding reaction was carried out on ice to avoid the integration of PIC DNA. After the particles were washed with cold buffer, viral DNA was isolated from recovered PICs and detected by Southern blotting. (B) Activities of initial (lane 1), salt-stripped (lane 2), and BAF-reconstituted (lane 3) PICS in binding to target DNA.
Recent studies demonstrate that the structures and compositions of virion-derived nucleoprotein complexes change dynamically at early phases of retroviral infection (14, 15). During this stage, host factors such as BAF are expected to be recruited and utilized to form the appropriate nucleoprotein complex to establish the infection. At what step in the viral replication cycle is BAF incorporated into the PIC? Previous data show that BAF is not present in virions (24). Furthermore, BAF does not associate with single-stranded DNA or with RNA in vitro (24, 31). Therefore, it is likely that BAF is recruited into viral nucleoprotein complexes in the cytoplasm as the viral dsDNA is synthesized. Our immunoprecipitation data demonstrate the presence of BAF in cytoplasmic extract, and indirect immunofluorescence studies with antibodies specific to BAF show it to be localized predominantly in the cytoplasm during the G1 stage of the cell cycle, although a certain amount is also localized in the nucleoplasm and the nuclear membrane (T. Haraguchi, personal communication). Based on our data that a low concentration of BAF is able to reconstitute salt-stripped MoMLV PICs, the concentration of BAF in the cytoplasm may be sufficient to form integration-competent PICs in the infected cell. However, because BAF is essential for cell division (31) and its cellular distribution is regulated during the cell cycle, it is possible that the expression level and/or localization of BAF may play a regulatory role in the establishment of retroviral infection at a postentry step. Further work will be required to investigate the effects on viral infectivity of the dynamics of BAF in the host cell.
Acknowledgments
We thank Katherine Wilson for antibody specific to BAF, Raymond Reeves for the human HMGA1 expression plasmid, and Martin Gellert for human HMGB1 protein. We also thank Tokuko Haraguchi for communicating results prior to publication, Eric Greene and Kiyoshi Mizuuchi for their comments on the manuscript, and Jian-Yong Wang, Min Li, Tomokazu Yoshinaga, and Ying Huang for helpful suggestions.
This work was supported in part by the NIH Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Asante-Appiah, E., and A. M. Skalka. 1999. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 52:351-369. [DOI] [PubMed] [Google Scholar]
- 2.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 3.Brown, P. O. 1997. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 4.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. [DOI] [PubMed] [Google Scholar]
- 5.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1989. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA 86:2525-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman, F. D., and R. Craigie. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. USA 88:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craigie, R. 2001. HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 276:23213-23216. [DOI] [PubMed] [Google Scholar]
- 9.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 11.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211-1221. [DOI] [PubMed] [Google Scholar]
- 12.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 13.Farnet, C. M., and W. A. Haseltine. 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA 87:4164-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara, T., and K. Mizuuchi. 1988. Retroviral DNA integration: structure of an integration intermediate. Cell 54:497-504. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa, K. 1999. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112:2485-2492. [DOI] [PubMed] [Google Scholar]
- 18.Goff, S. P. 1992. Genetics of retroviral integration. Annu. Rev. Genet. 26:527-544. [DOI] [PubMed] [Google Scholar]
- 19.Hindmarsh, P., and J. Leis. 1999. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 63:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 21.Katz, R. A., G. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 22.Katzman, M., R. A. Katz, A. M. Skalka, and J. Leis. 1989. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 63:5319-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, M. S., and R. Craigie. 1994. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc. Natl. Acad. Sci. USA 91:9823-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, L., C. M. Farnet, W. F. Anderson, and F. D. Bushman. 1998. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J. Virol. 72:2125-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman, P. A., and J. A. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 87:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shumaker, D. K., K. K. Lee, Y. C. Tanhehco, R. Craigie, and K. L. Wilson. 2001. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 20:1754-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umland, T. C., S. Q. Wei, R. Craigie, and D. R. Davies. 2000. Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry 39:9130-9138. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, K. L. 2000. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 10:125-129. [DOI] [PubMed] [Google Scholar]
- 30.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng, R., R. Ghirlando, M. S. Lee, K. Mizuuchi, M. Krause, and R. Craigie. 2000. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. USA 97:8997-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]