Abstract
Recently we showed that the membrane-proximal stem region of the vesicular stomatitis virus (VSV) G protein ectodomain (G stem [GS]), together with the transmembrane and cytoplasmic domains, was sufficient to mediate efficient VSV budding (C. S. Robison and M. A. Whitt, J. Virol. 74:2239-2246, 2000). Here, we show that GS can also potentiate the membrane fusion activity of heterologous viral fusion proteins when GS is coexpressed with those proteins. For some fusion proteins, there was as much as a 40-fold increase in syncytium formation when GS was coexpressed compared to that seen when the fusion protein was expressed alone. Fusion potentiation by GS was not protein specific, since it occurred with both pH-dependent as well as pH-independent fusion proteins. Using a recombinant vesicular stomatitis virus encoding GS that contained an N-terminal hemagglutinin (HA) tag (GSHA virus), we found that the GSHA virus bound to cells as well as the wild-type virus did at pH 7.0; however, the GSHA virus was noninfectious. Analysis of cells expressing GSHA in a three-color membrane fusion assay revealed that GSHA could induce lipid mixing but not cytoplasmic mixing, indicating that GS can induce hemifusion. Treatment of GSHA virus-bound cells with the membrane-destabilizing drug chlorpromazine rescued the hemifusion block and allowed entry and subsequent replication of GSHA virus, demonstrating that GS-mediated hemifusion was a functional intermediate in the membrane fusion pathway. Using a series of truncation mutants, we also determined that only 14 residues of GS, together with the VSV G transmembrane and cytoplasmic tail, were sufficient for fusion potentiation. To our knowledge, this is the first report which shows that a small domain of one viral glycoprotein can promote the fusion activity of other, unrelated viral glycoproteins.
Vesicular stomatitis virus (VSV) is an enveloped, nonsegmented, negative-strand RNA virus that is a prototypic member of the family Rhabdoviridae, genus Vesiculovirus. Due to its broad host range, simple genetic organization, and rapid growth in cell culture, VSV has been used widely to study various aspects of rhabdovirus entry, assembly, and release. The virus enters cells via receptor-mediated endocytosis. Approximately 1,200 VSV glycoprotein (G) molecules, which are organized as homotrimeric spikes that are anchored in the viral envelope, are responsible for virus attachment as well as for mediating fusion of the viral envelope with the endosomal membrane of the host cell following endocytosis. The low pH in the endosome causes a conformational change in G protein that drives the fusion of the viral envelope with the endosomal membrane (2, 9, 26). Fusion of the two membranes results in release of the viral nucleocapsid into the host cell cytoplasm, where viral replication occurs.
VSV G protein differs from influenza virus hemagglutinin (HA), the prototypic viral fusion protein, in that G protein does not require proteolytic processing to become fusion competent (17, 33). Also unlike influenza virus HA, the N terminus of G, apart from the signal sequence, is not particularly hydrophobic, and there is no obvious region in the amino acid sequence that can be defined as a “classical” fusion peptide (51). It was postulated that the VSV G fusion peptide is internal and that the region between amino acids 118 and 139 could be the putative fusion domain (33). Mutational analysis has provided evidence that the region between amino acids 118 and 136 corresponds to the G protein fusion peptide (14, 25, 53, 55).
Other regions of G protein have also been shown to be important for its fusion activity. Insertion of a 3-amino-acid linker in the membrane-proximal domain at positions 410 and 415 abolished membrane fusion activity, indicating that this region may be important for fusion (25). Substitution of amino acids in the region between 395 and 418 also affected the fusogenic activity of G protein (47). When mutations in the fusion peptide were combined with point mutations in the membrane-proximal region between amino acids 395 and 418, fusion activity was inhibited additively. However, one double mutation, G131A-G404A, was more fusogenic than the two individual mutations alone, suggesting that these regions may interact during fusion (46).
The membrane-proximal region of the VSV G protein is highly conserved among members of the genus Vesiculovirus (41). Structure predictions of the region between amino acids 385 and 444 of the VSV GIND (Indiana serotype) glycoprotein have shown that this region has a propensity to form α-helices, suggesting that this region may be capable of interacting directly with membranes (16). Recently, we showed that the membrane-proximal stem region of the VSV G protein ectodomain (G stem [GS]) mediates efficient VSV budding (41). In this report, we show that GS can potentiate the membrane fusion activities of several different heterologous viral glycoproteins. We postulate that the fusion potentiation activity results from the ability of GS to induce hemifusion.
MATERIALS AND METHODS
Plasmids and oligonucleotide-directed mutagenesis.
The construct pCAGGS-GSHA was obtained by subcloning an MluI-NheI fragment encoding GSHA from pVSV-ΔG-GSHA (41) into a modified form of the eukaryotic expression vector pCAGGS (32). The constructs pCAGGS-G124-E, -A133-K, -P127-D, -D137-L, and Qn-1 were obtained by subcloning MluI-NheI fragments from the appropriate GMMG minigenomes (13).
The constructs pCAGGS-CD4-G and -CD4-GS (CD4 ectodomain fused to GS at F421) were made by subcloning the KpnI-SphI restriction digest fragments encoding CD4-G and CD4-GS from VSV-ΔG-CD4-G/GS (41) into the pCAGGS vector previously digested with the same enzymes. To produce ΔG-F, the cDNA encoding the fusion protein (F) from the paramyxovirus simian virus 5 (SV5) (36) was subcloned into pVSV-ΔG-PL from pGEM-F (kindly provided by R. A. Lamb, Northwestern University) using the XhoI site, and clones were screened for proper orientation. The SV5 F cDNA was then moved from ΔG-F into pCAGGS by using the KpnI and NheI sites to produce pC-SV5-F.
Plasmids containing the measles virus F and H proteins (pCG-F and pCG-H5, respectively) and the corresponding antibodies were generously provided by Roberto Cattaneo, Mayo Clinic (5).
Epitope-tagged truncation mutants of GS.
Consecutive seven-amino-acid truncations of GS, beginning at amino acid Q427, that had an N-terminal Flag or HA epitope tag (Fig. 1) were generated as follows. The HA epitope tags were generated by PCR with a sense-strand oligonucleotide that overlapped the 3′ end of the M gene (MT-954) and an antisense oligonucleotide (5′AGGATGACCCGAGCCAGCGTAATCTGGTACATCATACGG-3′) that overlapped the HA epitope (in italics) in the template ΔG-GSHA (41) and contained the unique AvaI site (underlined) for cloning. The Flag epitope-tagged mutants were generated by PCR with the MT-954 sense primer and an antisense primer (5′GACCCGAGCCCTTATCGTCATCATCTTTGTAGTCGAACTTGCAATTCACCCCAATG-3′) that overlapped the signal sequence of G protein (indicated in bold type) and contained the unique AvaI site (underlined).
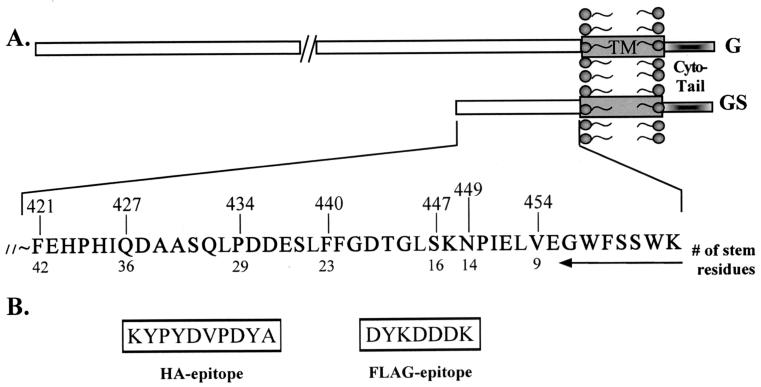
FIG. 1.
Schematic representation of GS truncations. (A) The amino acid sequence of the membrane-proximal ectodomain of G protein is shown. The numbers at the top of the sequence indicate the positions of the amino acids from the N terminus of G protein and correspond to the residues at which successive truncations of GS were made. The numbers below the sequence denote the number of amino acids from the transmembrane (TM) domain (box). (B) The amino acid sequences of the HA and Flag epitopes that were used to tag the various GS truncations are shown.
The PCR fragments were digested with the enzymes MluI and AvaI. The GS fragments corresponding to residues Q427, P434, F440, S447, and V454 were obtained by restriction digestion of the corresponding pBluescript CD4-GS constructs (41) with AvaI and SphI. The epitope tag and the fragments encoding the appropriate GS were cloned into the plasmid pVSV-ΔG-PL(+) (41), which had been previously digested with MluI and SphI in a three-way ligation.
The mutants N449, L453, and E452 were constructed by PCR-based mutagenesis with individual sense-strand oligonucleotides that had a common 5′ sequence (5′-ATGGCCTCGGGT) that contained an AvaI site (underlined) followed by the sequence coding for amino acid residue N449, L453, or E452, respectively, and an antisense oligonucleotide (5′-CCAAACATGAAGCTTCTGTTGTGCATGCTTTGAGTTAC-3′), which introduced an SphI site (underlined) within the 3′ untranslated region of the G in pVSVFL-2(+) (23). The amplicons were digested with AvaI and SphI and cloned into pVSV-ΔG-PL(+) together with the Flag or HA epitope-encoding fragments by three-way ligation. The sequences of the PCR-amplified fragments were confirmed by dideoxynucleotide sequencing. The constructs were then subcloned into pCAGGS-MCS as MluI-SphI fragments.
The construct gp160W3IGS was made by a PCR-based mutagenesis approach. This glycoprotein is composed of the gp120-gp41 ectodomain from human immunodeficiency virus type 1 (HIV-1) clone BH10 (39) through residue 671, which is connected by a linker encoding Ile-Ser-Gly to the C-terminal portion of VSV-GIND at residue 454. The gp160W3IGS cDNA was constructed by amplifying the sequence encoding the transmembrane-ectodomain junction of HIV-1 gp160 from plasmid pBH10 with a sense-strand oligonucleotide (5′-TGGATGGAGTGGGACAG-3′) (BH10-CR1+) and the noncoding primer (5′-GTTATACCCGAGATATTCCACAAACTTGCCCATTTATC-3′) (BH10-W3IAva), which introduced an AvaI site (underlined) and replaced the codon for tryptophan at position 672 of gp160 with an isoleucine codon (boldfaced) (44). The amplicon was digested with HindIII and AvaI and then ligated together with an EcoRI-HindIII fragment from wild-type BH10 into the EcoRI-AvaI-digested vector pGEM-CD4-V454 (41). The resulting construct was subcloned as an EcoRI-SphI fragment into pCAGGS-MCS, resulting in pC-gp160W3IGS (also called pC-W3IGS). This chimera expresses the HIV Env protein at high levels on the cell surface and retains the same chemokine receptor tropism as wild-type BH10 Env (unpublished data).
Transient transfections and syncytium assays.
Approximately 7 × 105 baby hamster kidney (BHK-21) cells were transfected with a DNA-liposome suspension containing 2 μg of the appropriate plasmid and 10 μl of Lipofectamine (Gibco-BRL) according to the manufacturer's instructions. Three hours posttransfection, the medium was replaced with Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), and the cells were incubated for 36 h at 37°C. Cells were then fixed with 3% paraformaldehyde and processed for indirect immunofluorescence microscopy.
Syncytium formation with HIV Env-expressing cells was done as follows. Approximately 7 × 105 BHK-21 cells were transfected with pCAGGS plasmids encoding GSHA, CD4, CD4-G, or CD4-GS alone or cotransfected with plasmids expressing CD4-G and GSHA. After 24 h, the cells were removed from the dish with trypsin-EDTA and then replated either alone or mixed with cells expressing gp160W3IGS. The cultures were incubated at 37°C for an additional 24 h, and then cells were fixed with 3% paraformaldehyde and probed with a monoclonal antibody (Sim.2) specific for the CD4 molecule or with a monoclonal antibody specific for the HA epitope.
Antibodies.
The following antibodies were used for staining the cells in the syncytium assays. The F1a monoclonal antibody, which is specific for the SV5 F protein, was a kind gift of R. A. Lamb, Northwestern University, and Rick Randall, St. Andrews University (38). The Sim.2 monoclonal antibody (28, 34), which binds to the CD4 ectodomain, and the goat anti-gp160 serum HT3 (27) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The other antibodies that were used included monoclonal antibody VIII, which is specific for the VSV GNJ (New Jersey serotype) glycoprotein (3), 12CA5 and HA.11 (Covance), which are specific for the HA epitope, and M2 (Sigma-Aldrich), which is specific for the Flag epitope.
Recovery of GS viruses from cDNA.
Recovery of the viruses was done as described previously (49) with a few modifications. Briefly, confluent monolayers of BHK-21 cells in 35-mm plates were infected with a recombinant vaccinia virus encoding the T7 RNA polymerase (vTF7-3) (15) at a multiplicity of infection of 5 for 1 h at 31°C. The cells were then transfected with a DNA-liposome suspension consisting of 5 μg of pVSV-ΔG-GSHA/Flag, 3 μg, 5 μg, 8 μg, and 1 μg of plasmids containing the N, P, G, and L genes from VSVIND, respectively, and 90 μl of TransfectACE (43, 52). After 3 h, the transfection mix was replaced with DMEM containing 10% FBS, and cells were incubated at 37°C. The supernatants were collected after 48 h and filtered through a 0.2-μm-pore-size filter (Millipore; Millex-GS) to remove vaccinia virus. The filtrates were then applied to BHK-21 cells which had been transfected with 2 μg of pCAGGS-GIND 24 h earlier. Recovery of the virus was assessed by examining the cells for cytopathic effects typical of a VSV infection after 24 to 36 h. The recovered viruses were plaque purified and then passaged on BHK-21 cells expressing G protein to make high-titer G-complemented virus stocks. Expression of the G-stem proteins in virus-infected cells was confirmed by immunofluorescence microscopy with antibodies specific for the HA and Flag epitopes.
Preparation of non-G-complemented GS viruses.
To produce GS virus stocks that had no full-length G protein in their envelopes (non-G-complemented viruses), BHK-21 cells were infected with the G-complemented stock viruses at a multiplicity of infection of 10. One hour postinfection, the inoculum was removed, and the cells were washed twice with serum-free DMEM and once with medium containing G-protein-specific monoclonal antibody I1 (24) to remove any residual G-complemented virus inoculum that might be present in the supernatants. At 16 h postinfection, the supernatants were collected, and the virus particles were pelleted through a 20% sucrose cushion by ultracentrifugation at 100,000 × g for 35 min. The viral pellet was resuspended in sterile Dulbecco's phosphate-buffered saline (PBS) and stored at −80°C until further use.
Virus binding assay.
Virus binding was done as described previously (12). Approximately 80,000 cpm of [35S]methionine-labeled wild-type VSV, non-G-complemented GSHA, or ΔG virus was added to 500 μl of binding medium (2 mM Na2HPO4, 2 mM NaH2PO4, 30 mM NaCl, 2 mM 2-[N-morpholino]ethanesulfonic acid [MES], 2 mM HEPES, 0.1% bovine serum albumin, and DMEM, titrated to pH 7.0 or pH 5.9 with HCl) and incubated at room temperature for 30 min. Virus suspensions were cooled on ice for 10 min and then added to confluent monolayers of BHK-21 cells that had been washed with ice-cold binding medium at the appropriate pH. Virus adsorption was done for 3 h on ice. The inoculum was removed, and the amount of unbound virus in the supernatants was determined by scintillation counting. Cells were washed three times with ice-cold binding medium, and the amount of virus eluted in the wash was determined as above. Cells were then solubilized with PBS containing 1% Triton X-100, and the amount of bound virus was determined.
Cell-cell fusion assays.
The cell-cell fusion assay was based on the redistribution of fluorescent probes between donor cells and target cells upon fusion. The fluorescent probes DiIC18(3)-DS (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine-5,5′-disulfonic acid), calcein-AM, and CMAC (7-amino-4-chloromethylcoumarin) were obtained from Molecular Probes (Eugene, Oreg.). The assay was done as described previously (30) with a few modifications.
(i) Preparation of donor cells.
BHK-21 cells (7 × 106) were infected with wild-type VSV, GSHA, or ΔG virus at a multiplicity of infection of 10. At 2 h postinfection, the cells were washed twice with serum-free DMEM and once with I1 monoclonal antibody to remove any unbound G-complemented virus. At 8 h postinfection, the cells were incubated with 10 μM calcein-AM in Dulbecco's PBS for 30 min at 37°C, washed twice with serum-free DMEM to remove excess dye, and then incubated in fresh DMEM with 5% FBS. After 2 h, the cells were labeled with DiI by incubating the cells in Dulbecco's PBS containing 5 μM DiI-DS for 20 min at 37°C, according to the manufacturer's instructions. Cells were washed three times with serum-free DMEM to remove the unincorporated label. Labeled cells were then removed from the plates with PBS containing 50 mM EDTA, washed two times with Dulbecco's PBS (changing tubes each time), and resuspended in 1 ml of serum-free DMEM.
(ii) Preparation of target cells.
Three hours prior to the assay, 4 × 106 BHK-21 cells in 6-cm dishes were labeled with the blue fluorescent dye CMAC by incubating them for 30 min at 37°C with Dulbecco's PBS containing 20 μM CMAC. Cells were washed two times with serum-free DMEM and then incubated with fresh DMEM-5% FBS at 37°C for 3 h.
The donor cells were overlaid on the CMAC-labeled target cells, and the culture was incubated at 4°C for 1 h to allow the donor cells to settle and make contact with the target cells. Unbound cells were then removed by washing twice with warmed serum-free DMEM, and the attached cells were incubated at 37°C for 15 min in 2 ml of serum-free DMEM. Fusion was triggered by incubating cells for 1 min at room temperature with fusion medium (10 mM Na2HPO4, 10 mM NaH2PO4, 150 mM NaCl, 10 mM MES, 10 mM HEPES) buffered to pH 7.0 or pH 5.9. The fusion medium was immediately replaced with serum-free DMEM, and fusion was allowed to proceed for 30 min at 37°C. The cells were then incubated on ice to arrest fusion. Phase-contrast and fluorescence micrographs were acquired with a 40× water immersion lens on a Zeiss Axiophot microscope equipped with a Zeiss Axiocam digital camera and accompanying AxioVision software. Images were adjusted for brightness and contrast with Adobe Photoshop 6.0 after conversion to .tiff files. Original .zvi images are available upon request.
CPZ rescue assay.
Stocks of VSV-green fluorescent protein (GFP) virus (19), non-G-complemented GSHA-GFP (41), and non-G-complemented ΔG-GFP virus (41) were made in BHK-21 cells. Virions were concentrated by ultracentrifugation, and an aliquot of the viral pellet was loaded on a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gel and stained with Coomassie blue (GelCode Blue; Pierce Chemical Company). Virus quantitation was based on the l-protein concentration determined with ImageQuant software (Molecular Dynamics).
The chlorpromazine (CPZ) rescue assay was done as described previously (54) with a few modifications. An equivalent amount of each virus, based on viral protein content, was added to 500 μl of binding buffer (pH 7.0), and the virus suspension was incubated at room temperature for 30 min. The virus suspension was cooled on ice for 10 min and then added to chilled BHK-21 cells on ice. Virus adsorption was done on ice for 3 h. The inoculum was removed, and cells were rinsed with warmed DMEM containing 10% FBS and incubated at 37°C for 10 min. Cells were then treated with DMEM-10% FBS containing CPZ (0.4 mM) for 1 min, washed twice, returned to drug-free medium immediately and incubated at 37°C for 2 h to allow the cells to recover from the effect of the drug. Cells were then overlaid with DMEM containing 5% FBS and 0.5% methylcellulose. Twelve hours later, the cells were examined with a fluorescence microscope (Zeiss Axiophot), and the number of GFP-positive cells was counted.
RESULTS
GS can potentiate the fusion activity of heterologous viral glycoproteins.
GSHA (G stem containing an N-terminal hemagglutinin epitope tag) is a truncation mutant of G protein from which amino acids 1 to 420 of the mature protein have been deleted. Figure 1A shows the amino acid sequence of the membrane-proximal ectodomain of GS. We have shown previously that GSHA is a trimer, that it is transported to the cell surface, and that it is important for efficient assembly and budding of VSV particles (41).
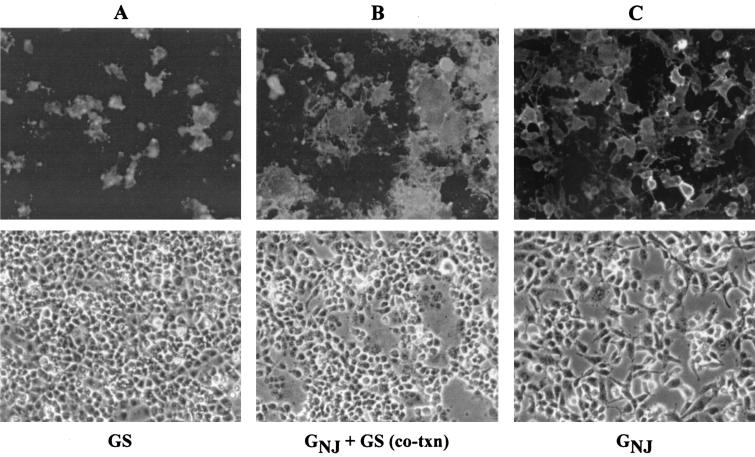
During our studies of GS-mediated virus assembly, we noticed that cells coexpressing GS and another membrane fusion protein produced extensive and large syncytia but that when GS was expressed alone, no syncytia were seen. This phenomenon is illustrated in Fig. 2. Figure 2A shows an example of cells expressing GSHA only and the absence of syncytia. For comparison, when we coexpressed GSHA with the VSV GNJ glycoprotein and maintained the culture at neutral pH, large, well-defined areas of cell-cell fusion could be seen (Fig. 2B). The same results were obtained when we coexpressed GSHA with the VSV GIND protein (data not shown). Normally, when cells expressing VSV G are maintained at neutral pH, very little to no syncytium formation occurs, as was seen when GNJ was expressed alone (Fig. 2C). These results suggested that GS could relieve the normal low-pH activation step required for VSV G-mediated membrane fusion and could functionally potentiate the fusion activity of G protein.
FIG. 2.
GS enhances membrane fusion induced by GNJ. BHK-21 cells were transfected with plasmids encoding GSHA (A), GSHA and GNJ (B), or GNJ alone (C). At 36 h posttransfection, cells were fixed with 3% paraformaldehyde and stained. The cells were not pretreated with fusion medium, and therefore, the pH was maintained at 7.2 to 7.4 throughout the incubation period. Cells expressing GNJ and GNJ plus GSHA were probed with monoclonal antibody VIII, which is specific for the GNJ glycoprotein. Cells expressing GSHA were stained with the 12CA5 monoclonal antibody, which is specific for the HA epitope. Representative epifluorescence and phase-contrast micrographs of the cells are shown.
Although it is well established that VSV G requires a low-pH trigger to obtain the fusion-competent conformation (2, 11, 35), we have found that when G protein is overexpressed at very high levels in BHK-21 cells, the G proteins of both the Indiana and New Jersey serotypes can induce some cell-cell fusion at neutral pH (42). The ability of VSV G to cause cell-cell fusion without prior exposure to low pH and the possible mechanism by which this occurs in the polarized endometrial cell line HEC-1A have been described previously (40). Therefore, it is more likely that the GS-mediated fusion potentiation that we observed after coexpression of GS with GNJ or GIND is due not to activation of G protein, per se, but to an enhancement of the low-level fusion activity normally present in cells expressing VSV G protein.
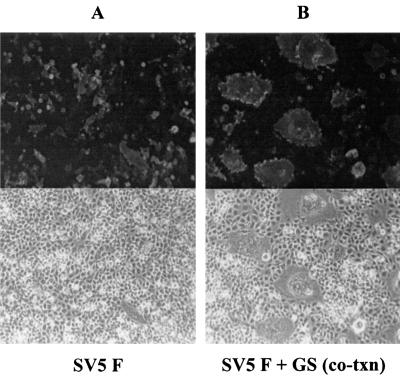
To determine if GS could potentiate the membrane fusion activity of other viral glycoproteins, we coexpressed GSHA with the SV5 F protein. SV5 F (strain W3A) protein can directly induce membrane fusion in the absence of its cognate HN protein (18). When SV5 F was expressed alone in cells, a few small syncytia containing four or five nuclei were observed (Fig. 3A). However, when GSHA was coexpressed with SV5 F, there was a dramatic increase in the number of syncytia formed and the size of the syncytia (Fig. 3B). By 36 h after transfection, giant polykaryons having 100 to 150 nuclei per syncytium were observed.
FIG. 3.
Effect of GS on SV5 F-mediated membrane fusion. BHK-21 cells were transfected with plasmids encoding SV5 F alone or cotransfected with plasmids encoding SV5 F and GSHA. Cells were fixed with 3% paraformaldehyde at 36 h after transfection and stained for surface expression of SV5 F with the monoclonal antibody F1a, which is specific for F protein. (A) Fusion activity of SV5 F alone. (B) Syncytium formation when SV5 F and GSHA were coexpressed.
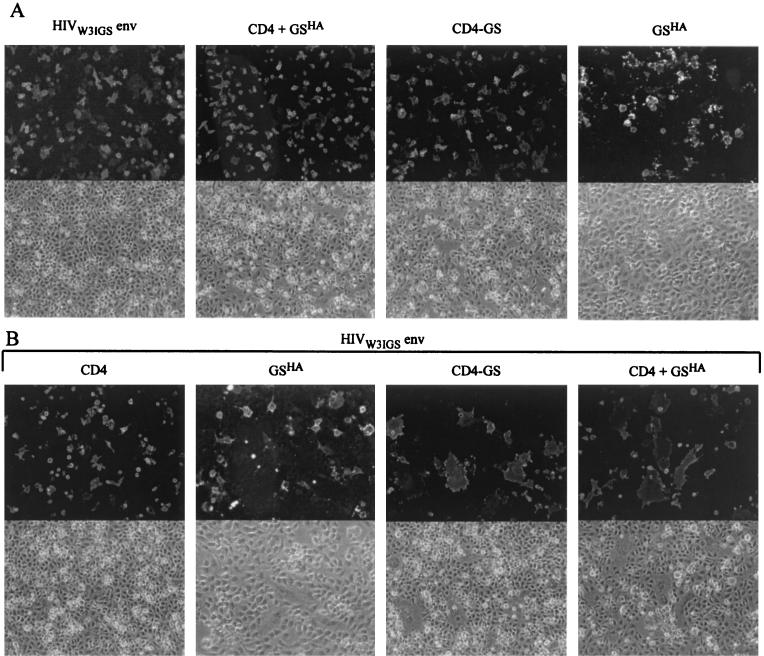
To determine the extent to which GS could enhance the membrane fusion activity of other viral glycoproteins, we next asked if GS had an effect on HIV-1 Env-mediated fusion. For these studies, we used a cell fusion assay in which nonhuman (BHK-21) cells expressing an X4-specific HIV envelope protein derived from the clone BH10 were mixed with cells expressing either human CD4 alone, CD4 and GS, or a CD4-GS chimera described previously (41). As expected, no syncytia were seen in the control cells expressing (i) HIV-1 Env alone, (ii) CD4 and GSHA together, (iii) a CD4-GS chimera alone, or (iv) GSHA alone (Fig. 4A). Likewise, when BHK-21 cells expressing HIV-1 Env were mixed with BHK-21 cells expressing CD4 (Fig. 4B, leftmost panel) no detectable cell-cell fusion occurred, which is consistent with previous studies demonstrating a requirement for chemokine coreceptor for HIV-1 Env-mediated membrane fusion (4, 10).
FIG. 4.
GS can function with HIV-1 Env to induce membrane fusion in a chemokine-independent manner. (A) BHK-21 cells were transfected with plasmids encoding either gp160W3IGS alone, CD4 and GSHA together, the chimeric protein CD4-GS alone, or GSHA alone. At 24 h posttransfection, the cells were removed from the plates, divided into two aliquots, and then replated. After an additional 24 h, the HIV-1 Env-expressing cells were stained with the gp160-specific goat serum HT3. Cells expressing CD4 or CD4-GS were stained with the CD4-specific monoclonal antibody Sim.2. Cells expressing GSHA were stained with an HA epitope-specific monoclonal antibody. (B) After 24 h of transfection, the cells expressing CD4 only, GSHA only, CD4 and GSHA together, or CD4-GS were mixed with cells expressing gp160W3IGS and replated. The mixed cultures were incubated for an additional 24 h and were then fixed, processed, and probed with the Sim.2 monoclonal antibody for CD4 expression or for GSHA with monoclonal antibody HA.11.
Similarly, when cells expressing HIV-1 Env were mixed with cells expressing GSHA only, no cell fusion occurred (Fig. 4B), demonstrating that GSHA is not sufficient to induce cell fusion on its own. However, when cells that were coexpressing CD4 and GSHA or cells expressing the CD4-GS chimera were mixed with cells expressing the HIV-1 envelope protein, a large number of syncytia were seen (Fig. 4B, two right-hand-most panels). These results confirmed that GS could function with a variety of different envelope proteins. More remarkable, though, was the observation that GS could apparently relieve the chemokine coreceptor requirement for CD4-dependent HIV-1 Env-mediated membrane fusion. As will be reported elsewhere, GS can function with a wide variety of different HIV-1 and HIV-2 envelope proteins, and therefore GS fusion potentiation is not restricted to enhancement of the gp160W3IGS envelope clone.
GS fusion enhancement requires an active fusion partner.
In contrast to the results described above, when GS was coexpressed with measles virus F protein, no discernible cell-cell fusion was observed. In contrast to the SV5 F protein, measles virus F requires coexpression with its cognate H protein for membrane fusion activity. However, some enhancement of syncytium formation was observed when measles virus F and H proteins were coexpressed with GS. A summary of the results obtained by coexpressing GSHA with various viral glycoproteins is shown in Table 1. Our results demonstrate that GS can potentiate the fusion activity of some but not all viral glycoproteins and that the fusion-potentiating activity can occur with glycoproteins that require a pH trigger as well as those that are pH independent. This suggests that GS acts at a step that is common to both fusion pathways.
TABLE 1.
Summary of fusion potentiation activity of GS upon coexpression with heterologous viral fusion proteinsa
| Glycoprotein | Fusion enhancement by GS |
|---|---|
| GIND | +/++ |
| GNJ | ++/+++ |
| SV5 F | ++++ |
| Moloney murine leukemia virus | + |
| HIVW3IGS | ++++ |
| Measles virus F | − |
| Measles virus F + H | +/− |
BHK-21 cells were transfected with plasmids encoding GSHA and the respective viral glycoprotein (1 μg each). After 24 h, the cells were removed from the dish, divided into three aliquots, and replated. After an additional 24 h, the cells were examined by phase-contrast microscopy to determine the number and size of the syncytia that were formed. Symbols: +, 2- to 10-fold increase in cell-cell fusion observed when GSHA was coexpressed compared to when the glycoprotein was expressed alone; ++, 10- to 20-fold increase; +++, 20- to 30-fold increase; ++++, 30- to 40-fold increase; −, no fusion potentiation was observed. CV-1 cells were used for the measles virus F and H fusion assays because the measles virus receptor is not present on BHK-21 cells.
We next asked if GS could relieve the fusion defects of some G protein mutants that have been described previously. The mutants that were used for these assays were A133-K (14) and QN-1 (53), which have no detectable fusion activity, and D137-L, G124-E, and P127-D (13, 14), which have a reduced pH threshold for fusion and can fuse as well as wild-type G protein at a pH below 5.7. All the mutants have wild-type G protein cell-binding activity. When we coexpressed these constructs with GS in BHK-21 cells, there was no enhancement of membrane fusion, indicating that GS did not rescue the fusion-defective phenotypes of these mutants when the cells were maintained at neutral pH or at pH 5.9 (data not shown). These results, together with the results from the measles virus F experiment (Table 1), indicate that fusion potentiation by GS requires a fully functional viral glycoprotein.
GS can bind membranes.
To understand the basis for the membrane fusion-enhancing activity of GS, we performed a series of experiments designed to examine various steps in the fusion process. The first step of glycoprotein-mediated membrane fusion requires binding of the glycoprotein to the target membrane. To determine whether GS has cell-binding activity, we incubated radiolabeled GSHA virus with BHK-21 cells in ice-cold medium buffered to pH 7.0 or pH 5.9 for 3 h. Wild-type VSV served as the positive control, while “bald” viruses lacking G protein (ΔG virus) served as the negative control for binding. Virus binding was done on ice to prevent endocytosis of the virus as well as to inhibit the fusion of the viral envelope with the host cell membranes following exposure to acid pH.
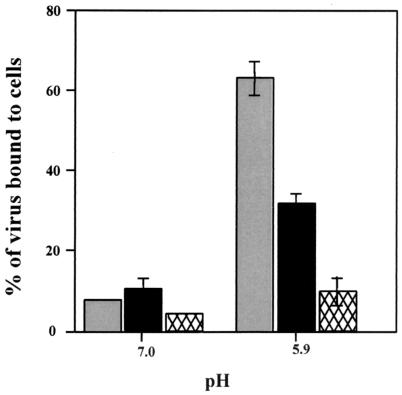
As shown in Fig. 5, GSHA virus bound to cells in a manner similar to that of wild-type VSV at pH 7.0. However, at acid pH, the binding of wild-type VSV increased by five- to sixfold, while the binding of GSHA increased by only two- to threefold. Similar results were obtained with a GS virus that lacked the HA epitope (data not shown), showing that the HA epitope tag did not contribute significantly to the binding properties of GS. The small amount of binding of ΔG virus to the cells at both the pH values examined was considered nonspecific background binding. These observations show that GS can mediate binding of the virus to target membranes. This binding could potentially bring two membranes in close proximity to initiate the fusion reaction.
FIG. 5.
Binding of wild-type VSV and GSHA virus to cells. Radiolabeled virions (∼80,000 cpm) were resuspended in binding medium adjusted to pH 7.0 or 5.9. The virus suspensions were added to cells and allowed to bind for 3 h on ice. The inoculum was removed (unbound fraction), and the cells were washed three times with ice-cold binding medium at the same pH to remove unbound virus. Cells were then solubilized with PBS containing 1% Triton X-100, and the lysates were collected (bound fraction). The amount of radioactivity in the bound and unbound fractions was determined by scintillation counting. The binding activity of the viruses is shown as a percentage of the total amount of input virus. Each bar represents an average of three separate experiments done in triplicate. Gray bars, wild-type VSV; solid bars, GSHA virus; hatched bars, ΔG virus.
GS induces hemifusion.
We next examined whether GS could mediate the next step in the fusion pathway, namely, the mixing of the outer leaflets of the two membranes. For this analysis, we used a three-color fusion assay similar to that described previously for other viral fusion proteins (30). In this assay we used BHK-21 cells infected with wild-type virus, GSHA virus, or ΔG virus as the donor cells. By 6 h postinfection, the virus-infected cells expressed high levels of either G protein or GSHA on the plasma membrane (data not shown). The virus-infected donor cells were then labeled with a red lipidic dye (DiI-DS) and a green cytoplasmic dye (calcein-AM). The target cells were labeled with a blue cytoplasmic dye (CMAC). The donor cells were then overlaid onto the target cells and incubated on ice for 1 h to allow cell attachment. After warming the culture to 37°C for 15 min, fusion was triggered by incubating the cells in fusion medium buffered to pH 5.9 or 7.0 for 1 min.
This assay is based on the redistribution of the dyes between the donor cell and target cell during the fusion process. When membrane fusion occurs, the donor cells redistribute the lipidic dye DiI first and then, upon opening of the fusion pores, calcein-AM is transferred to the cytoplasm of the target cell. The dye CMAC forms a glutathione adduct with a molecular mass of ∼600 Da within the cytoplasm of the target cells that cannot pass through the small fusion pores and thus helps in distinguishing the target cells from donor cells (31). If there is redistribution of DiI only to the target cells, this indicates that outer leaflet mixing has occurred but that the fusion reaction did not go to completion. Mixing of the outer leaflet only is known as the hemifusion stage. If both DiI and calcein-AM are transferred to the target cells, then complete fusion has occurred.
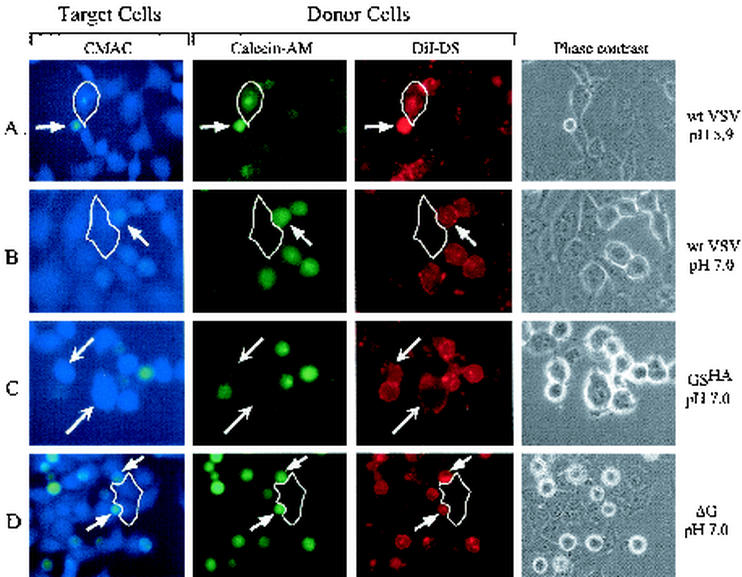
As shown in Fig. 6A, when cells infected with wild-type VSV were mixed with the target cells (blue) and exposed to pH 5.9, cell fusion occurred, as indicated by cells that were triple stained with DiI, calcein-AM, and CMAC (arrows). For comparison, when cells were maintained at a pH of 7.0 (Fig. 6B), there was no transfer of either DiI or calcein-AM to the target cells. These results agree with those obtained in previous studies with an octadecylrhodamine (R18) dequenching assay, which showed that VSV G protein requires a low-pH trigger to exchange the lipidic dye and initiate fusion (2, 37). In contrast to the pH-dependent fusion activity of G protein, cells expressing GSHA transferred DiI at pH 7.0 (Fig. 6C); however, no transfer of calcein-AM occurred even after exposure to pH 5.9 (data not shown). These data indicate that GSHA can induce hemifusion but not pore formation. The lipid-mixing and content-mixing events observed with wild-type VSV-infected cells or the hemifusion induced by GSHA were not seen in ΔG virus-infected cells at either pH 7.0 (Fig. 6D) or pH 5.9 (data not shown).
FIG. 6.
Three-color fusion assay. BHK-21 cells were infected with wild-type (wt) VSV (panels A and B), GSHA virus (panel C), or ΔG virus (panel D) at a multiplicity of infection of 10. After 8 h of infection, the cells were labeled with DiI (red) and calcein-AM (green) and then removed from the plates. The cells were then overlaid on BHK-21 cells that had been labeled with CMAC (blue). After the virus-infected donor cells had attached to the plate or settled onto the target cells, the medium was replaced with fusion medium buffered to pH 5.9 or 7.0. After 1 min, the fusion buffer was removed, and the cells were incubated for 20 min in growth medium at 37oC and then placed on ice to prevent any further lipid or content mixing. Phase-contrast and fluorescence images were obtained with filters for 4′,6′-diamidino-2-phenylindole (DAPI) (blue), rhodamine (red), and fluorescein isothiocyanate (FITC) (green). The DAPI filter set did not eliminate bleedthrough of the FITC fluorescence, and therefore the FITC-labeled cells appear greenish-blue in the micrographs. The target cells labeled with CMAC appear as larger, flat blue cells, while the donor cells appear as small, round cells and are labeled both red and green. Relevant target cells are highlighted by a thin white outline in panels A, B, and D. (A) Fusion mediated by wild-type G protein at pH 5.9. The phase-contrast image shows one small donor cell adjacent to two target cells. Transfer of both DiI and calcein-AM from the donor cell to the target cell (blue) can be seen (arrows). (B) A second plate identical to the one shown in panel A but maintained at pH 7.0 shows a lack of transfer of DiI or calcein-AM to the outlined target cell, consistent with the requirement for low pH in initiating VSV G-mediated membrane fusion. (C) Fusion mediated by GSHA at pH 7.0. Several donor cells in contact with target cells are shown. Only the transfer of DiI from the donor to the target cells was seen (arrows). There was no transfer of calcein-AM, showing that complete fusion had not occurred. (D) Multiple donor cells infected with ΔG-VSV which are in contact with an outlined blue target cell are shown, but no transfer of either DiI or calcein-AM occurred.
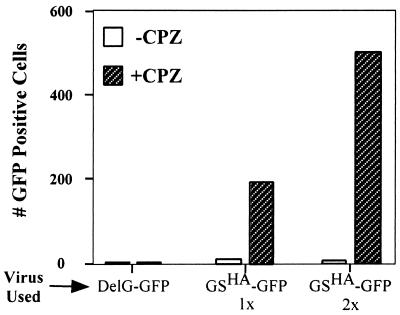
Rescue of GSHA virus infection by CPZ.
To determine if GS induced lipid mixing, as measured by DiI transfer, was a biologically relevant lipid-destabilization event, we examined whether the membrane curvature agent CPZ could rescue GSHA virus infection. CPZ is a lipid that can destabilize hemifusion diaphragms and drive the formation of a fusion pore (6). CPZ has been used previously to rescue hemifusion intermediates of influenza virus hemagglutinin (29) and Moloney murine leukemia virus glycoprotein (54). For this assay, we bound GSHA-GFP virus to cells at 4°C and then shifted the cells to 37°C to allow the formation of GS-dependent hemifusion diaphragms. The cells were then treated with 0.4 mM CPZ for 1 min and incubated overnight in drug-free medium. The number of GFP-positive cells in the presence or absence of CPZ was then quantified. GFP expression can occur only if the virus successfully entered the cells and started to replicate.
As shown in Fig. 7, when cells bound with GSHA virus were treated with CPZ (hatched boxes), there was a 200- to 500-fold increase in the number of GFP-positive cells compared to cells that were not treated with CPZ. Also, we found that increasing the amount of GSHA virus used for the assay resulted in a proportional increase in the number of GFP-positive cells. In contrast, CPZ treatment of cells bound with ΔG-GFP virus resulted in no GFP-positive cells. As expected, wild-type VSV-GFP virus was able to infect and spread throughout the culture in the presence or absence of CPZ (data not shown). These data indicate that GS can induce lipid mixing, which results in the formation of a biologically functional hemifusion diaphragm. Therefore, GS-mediated fusion potentiation likely results from the ability of GS to induce the formation of a hemifusion diaphragm, thereby reducing the energy barrier for membrane fusion.
FIG. 7.
Chlorpromazine rescues GSHA virus infectivity. Equivalent amounts of GSHA-GFP virus, ΔG (DelG)-GFP virus, and VSV-GFP virus were resuspended in binding medium buffered to pH 7.0 and incubated at room temperature for 30 min. Following the incubation, the virus suspension was cooled on ice for 10 min and then added to BHK-21 cells on ice. Following 3 h of binding on ice, the inoculum was removed, and the cells were washed with fresh medium to remove the unbound virus. Cells were then treated with medium containing 0.4 mM CPZ for 1 min. The drug-containing medium was removed immediately, and the cells were incubated in drug-free medium overnight. Twelve hours later, the number of cells expressing GFP was counted. The open boxes show the number of GFP-positive cells in the absence of CPZ, while the hatched boxes indicate the number of GFP-positive cells after CPZ treatment. VSV-GFP infected more than 90% of the cells in the presence or absence of CPZ, and the GFP cell count is not shown.
Deletion analysis of the GS fusion potentiation domain.
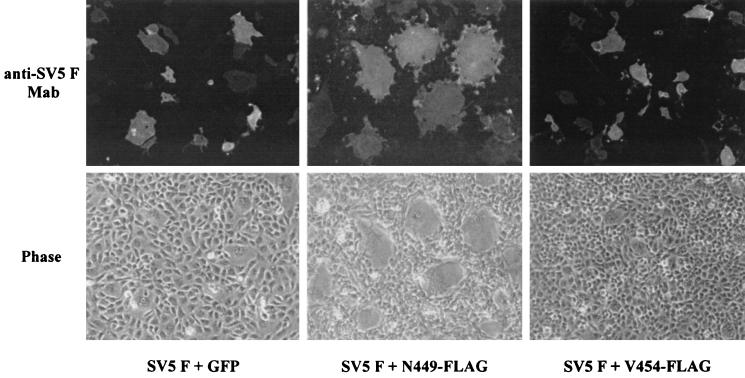
To determine how much of the ectodomain of GS is required for fusion potentiation, we made a series of deletion mutants, starting from residue F421 to V454 in GS (Fig. 1A). Each of the constructs was N-terminally tagged with two different epitopes (Flag or HA) to monitor surface expression. All the constructs that were examined were expressed on the cell surface, and there was no difference in the expression levels of any of the proteins regardless of the epitope tag used (data not shown). We found that GS having 14 amino acids (N449) was able to potentiate the fusion activity of SV5 F, while a construct having 9 amino acids (V454) resulted in no fusion potentiation (Fig. 8). These results show that as few as 14 amino acids of GS are sufficient for the fusion potentiation function of GS.
FIG. 8.
Fusion potentiation by GS deletion mutants. BHK-21 cells were cotransfected with plasmids encoding SV5 F and either GFP or the GS truncation mutants that were tagged with the Flag epitope. At 36 h posttransfection, the cells were fixed and probed with the F1a monoclonal antibody to detect SV5 F protein. Rhodamine-labeled goat anti-mouse immunoglobulin antibody was used to visualize the cells. Phase-contrast and epifluorescence images were obtained with a 10× water immersion lens with a Zeiss Axiophot microscope. At least 10 images were obtained for each sample. Representative photomicrographs of cells expressing GS with either 14 amino acids (N449) or 9 amino acids (V454) are shown.
DISCUSSION
Emerging models of viral glycoprotein-mediated membrane fusion propose that the membrane-proximal domains are important for the fusion activity of these proteins (48). The concept that the membrane-proximal regions of viral glycoproteins participate in the fusion process is supported by the following observations. Sequence comparisons of the membrane-proximal regions of retroviruses, filoviruses, orthomyxoviruses, rhabdoviruses, alphaviruses, and flaviviruses have revealed that this region contains an unusually high concentration of aromatic amino acids (48). Mutations in the membrane-proximal regions of HIV gp41, in particular the conserved tryptophan residues, reduced the amount of glycoprotein incorporated into the viral envelope and also inhibited the membrane fusion activity of the mutant proteins (44).
Mutations between amino acids 474 and 477 in the membrane-proximal region of the human parainfluenza virus type 2 F protein also severely affected the fusion activity of F protein (50). Insertions in the membrane-proximal region of the SV5 F protein have also been shown to abolish the fusion activity of the glycoprotein (56). Data from these studies show that mutations in the membrane-proximal regions affected the fusion activity without having an effect upon the oligomerization or transport of the glycoproteins. It is believed that the membrane-proximal regions may act as flexible tethers which help fusion proteins attain a fusion-competent conformation. These regions may also contribute to proper positioning of the protein molecules between the opposing lipid bilayers (50).
In this report, we provide evidence that the membrane-proximal stem region of the VSV G ectodomain can interact with target membranes and cause mixing of the outer leaflets of two apposing bilayers. We have also shown that the GS can act in trans and promote the fusion activity of unrelated glycoproteins. To our knowledge, this is the first report which shows that a small domain of one viral glycoprotein can promote the fusion activity of other, unrelated viral glycoproteins. GS fusion potentiation occurred only in the presence of fusion-competent glycoproteins and was not observed with fusion-defective mutants. This indicates that the receptor-binding activity of the fusion protein is not sufficient for GS-mediated membrane fusion enhancement; rather, GS functions like a catalyst, perhaps by reducing the activation energy required to initiate and drive membrane fusion in the presence of a fully active fusion protein.
Previously, it had been shown by others that a G protein peptide fragment extending from amino acids 392 to 490, which included the transmembrane domain, could be labeled by a hydrophobic photolabeling reagent (8). Whether the transmembrane domain alone was involved or whether membrane-proximal amino acid residues were also labeled was not determined. Based on our results showing that GS can induce hemifusion, it seems likely that at least some of the membrane-proximal residues can insert into the outer leaflet of a closely apposed lipid bilayer.
The ability of GS to cause lipid mixing and the formation of a functional hemifusion intermediate can best be explained by the proposed quantitative model for membrane fusion described by Kuzmin and colleagues (22). This model is based on the fact that fusion pores cannot form spontaneously between apposing membranes under physiological conditions. The model proposes that fusion proteins provide the energy to bend the two apposing membranes towards each other in the form of a “nipple.” Transient displacement of the lipid polar head groups at the tip of the nipple results in hydrophobic patches, which lead to the merger of the two outer leaflets of the apposing membranes. The merger of the two leaflets produces a hemifusion intermediate in which there is outer leaflet mixing but not mixing of the inner leaflet. If protein-assisted membrane fusion proceeds through a common series of steps as outlined above, then GS must have an inherent ability to cause membrane bending or some equivalent type of lipid perturbation which results in outer leaflet mixing.
Previously we reported that GS resulted in efficient, high-level budding of recombinant VSVs encoding either GS alone or chimeric envelope proteins fused to GS (41). We proposed that GS effected efficient virus budding by causing membrane curvature, which facilitated bud site formation. This idea was supported by the observation that virus budding can be inhibited by incubation of infected cells with some membrane curvature agents (20). Although no direct measure of membrane bending by GS has been performed, based on our results of GS-assisted virus budding and GS-mediated membrane fusion potentiation, it seems reasonable that GS has an inherent ability to modify membrane topology. In the context of a budding virus, GS may cause the formation of nipple intermediates which may provide the membrane curvature needed to initiate virus budding. In the context of two closely apposed membranes, the same GS-mediated nipple formation would result in establishment of a “stalk” where the cis monolayers merge. Subsequent lateral diffusion of GS molecules in the plane of the membrane would result in expansion of the stalk to a hemifusion diaphragm.
One of the unexpected outcomes of our studies was the observation that expression of GS with CD4 relieved the requirement for chemokine coreceptors in HIV envelope-mediated membrane fusion. Models of HIV Env fusion propose that upon binding of gp120 to CD4 on the target cell, a conformational change is induced that allows the V1/V2 loop to interact with the appropriate chemokine coreceptor (either CCR5 or CXCR4) (1). A series of subsequent conformational changes in gp41 take place that result in the exposure of the fusion peptide, followed by the formation of the six-helix bundle and membrane fusion. Recent evidence showed that the gp120 can undergo extensive conformational changes after binding to CD4 alone and that, under some conditions, the coreceptor may not be needed for these structural changes (reviewed in reference 7). It was also suggested that coreceptor association might possibly accelerate the formation of the six-helix bundle, leading to efficient membrane fusion (7).
Our results show that when we coexpressed CD4 and GS together in BHK-21 cells and overlaid the cells expressing both molecules onto HIV-1 Env-expressing cells, extensive syncytium formation occurred. Furthermore, we have found that GS can function with R5, X4, and several other Env proteins, suggesting that the fusion potentiation reported here is not HIV Env protein specific (42). Since BHK-21 cells are not known to express any endogenous chemokine coreceptors that can be used by the HIV-1 Env protein, our results indicate that the GS-mediated fusion potentiation is chemokine coreceptor independent. One explanation for this is that GS, by its ability to induce membrane curvature and hemifusion diaphragm formation, may provide an environment that allows the Env glycoprotein to undergo the necessary post-CD4 binding conformational changes needed to induce membrane fusion.
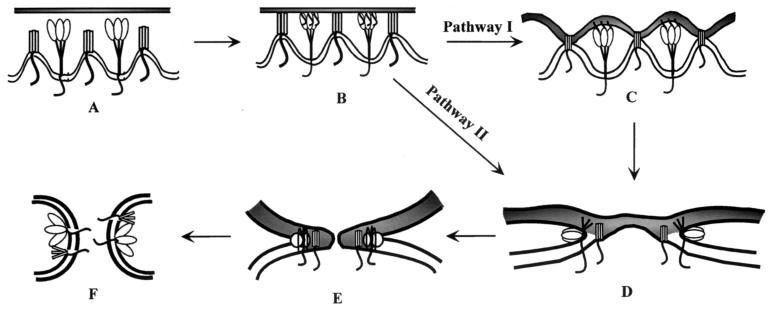
Although it is possible that GS may interact with different fusion proteins by independent mechanisms, we favor the hypothesis that GS brings about fusion potentiation through a common step in the fusion pathway. Based on our observations, we propose that GS trimers cause bending of membranes and result in the formation of nipples (Fig. 9A). Although we have depicted one GS trimer to be causing this deformation, multiple GS trimers are likely required for membrane bending. During fusion, when the two opposing membranes are brought close together, areas of contact are established between the two membranes, mediated by the interaction of the fusion protein with its receptor as well as by the binding of GS to the target membrane (Fig. 9B). These interactions could result in establishing multiple sites of contact and possibly tighter binding. In some cases, this binding may be sufficient to trigger conformational changes in the fusion protein and result in release of the fusion peptide (Fig. 9C, pathway I). Additional membrane destabilization by GS would lead to the formation of a hemifusion diaphragm (Fig. 9D).
FIG. 9.
Model for fusion potentiation of heterologous viral glycoproteins by GS. The membrane at the top of each step represents the target membrane, while the membrane at the bottom, which contains the fusion protein (depicted as a trimeric molecule with globular heads) and GS (depicted as trimeric rectangles), represents the donor membrane. Panels A to E illustrate the pathway towards complete fusion between the two membranes mediated by the viral fusion protein and GS. (A) Two individual membranes before the initiation of fusion. The microdomains where GS is located are shown as having increased membrane curvature. (B) GS alone or GS and the fusion protein bind to the target membrane, which results in close membrane apposition and the initiation of conformational changes in the fusion protein. In pathway I, the fusion protein undergoes additional conformational changes that result in exposure of the fusion peptide, which immediately inserts into the target membrane, as shown in panel C. (D) The glycoprotein undergoes further conformational changes that result in formation of the “hairpin” loop intermediate, which is the driving force for membrane merger. Destabilization of the target membrane by the interaction of GS as well as changes induced by the fusion protein cause the formation of a hemifusion diaphragm. In pathway II, the binding events are sufficient to cause GS-mediated hemifusion diaphragm formation (D), which then triggers activation of the fusion protein, release of the fusion peptide, and formation of a hemifusion diaphragm. The unequal forces exerted on the membranes by the lateral movement of protein molecules result in the formation of a fusion pore (E), which then enlarges and completes the fusion reaction (F).
Alternatively, we know that GS-mediated membrane destabilization can cause outer leaflet mixing, which would result in the establishment of a more hydrophobic environment that could drive conformational changes in the fusion protein. Subsequent exposure of the fusion peptide would drive additional conformational changes leading to membrane fusion (pathway II). In either case, lipid mixing mediated by the fusion protein and by GS would provide multiple contact sites between the two membranes where free transfer of lipid could occur. These sites would significantly reduce the free energy required for expansion of the stalks into a hemifusion diaphragm and result in an apparent enhancement of membrane fusion activity. Our data indicate that GS alone cannot mediate the formation of the fusion pore and that, therefore, the next stage, which involves opening of the fusion pores and mixing of cytoplasmic contents (Fig. 9F), must be mediated by a functional membrane fusion protein.
In conclusion, we have shown that the membrane-proximal stem region of the VSV G protein can potentiate the membrane fusion activity of heterologous glycoproteins. We suggest that membrane fusion potentiation is mediated by the ability of GS to induce lipid mixing, which probably enables the fusion proteins to overcome the energy barrier required to trigger the conformational changes needed to initiate and drive membrane fusion. In the case of some fusion proteins, such as measles virus F, which requires interaction with the measles virus HN protein to achieve a fully membrane fusion-active state, GS-induced lipid mixing is not sufficient to drive the necessary conformational changes needed for fusion activation.
We have also shown that at least 14 amino acids of the stem are required for this fusion potentiation function. Sequence analysis shows that this region is highly conserved among the glycoproteins of the vesiculoviruses (41). Additional studies to determine which of the conserved amino acids are important for the fusion potentiation function are in progress. It will also be interesting to determine if mutations in this region have an effect on membrane fusion activity in the context of the full-length G protein.
An exciting extension of these studies is the development of novel vectors that utilize the membrane fusion-potentiating activity of GS. For example, it has been shown that a VSV expressing CD4 and CXCR4 could specifically target and kill HIV-1-infected cells (45). Our observations that GS, in the absence of chemokine coreceptors, can potentiate the membrane fusion activity of HIV Env proteins suggest the possibility of developing an HIV-specific targeting vector that would be effective against HIV isolates independent of the coreceptor tropism and that could have potential application as an HIV therapeutic agent.
Acknowledgments
We thank Bob Lamb for the SV5 F cDNA, Rick Randall and Bob Lamb for the anti-SV5 F monoclonal antibody, and Roberto Cattaneo for the measles virus F and H cDNAs and antibodies. We also acknowledge the NIH AIDS Research and Reference Reagent Program as the source for antibody HT3, recognizing the HIV envelope protein, and the Sim.2 monoclonal antibody, specific for CD4. We greatly appreciate the encouragement and advice of Himangi Jayakar and the technical assistance of Carolyn Matthews.
This work was supported by NIH grant GM-53726 to M.A.W.
REFERENCES
- 1.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal, R., A. Bali-Puri, A. Walter, D. Covell, and O. Eidelman. 1987. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J. Biol. Chem. 262:13614-13619. [PubMed] [Google Scholar]
- 3.Bricker, B. J., R. M. Snyder, J. W. Fox, W. A. Volk, and R. R. Wagner. 1987. Monoclonal antibodies to the glycoprotein of vesicular stomatitis virus (New Jersey serotype): a method for preliminary mapping of epitopes. Virology 161:533-540. [DOI] [PubMed] [Google Scholar]
- 4.Broder, C. C., D. S. Dimitrov, R. Blumenthal, and E. A. Berger. 1993. The block to HIV-1 envelope glycoprotein-mediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s). Virology 193:483-491. [DOI] [PubMed] [Google Scholar]
- 5.Catomen, T., H. Y. Hussein, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernomordik, L., A. Chanturiya, J. Green, and J. Zimmerberg. 1995. The hemifusion intermediate and its conversion to complete fusion: regulation by membrane composition. Biophys. J. 69:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durrer, P., Y. Gaudin, R. W. H. Ruigrok, R. Graf, and J. Brunner. 1995. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J. Biol. Chem. 270:17575-17581. [DOI] [PubMed] [Google Scholar]
- 9.Fan, D. P., and B. M. Sefton. 1978. The entry into host cells of Sindbis virus, vesicular stomatitis virus, and Sendai virus. Cell 15:985-992. [DOI] [PubMed] [Google Scholar]
- 10.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 11.Florkiewicz, R. Z., and J. K. Rose. 1984. A cell line expressing vesicular stomatitis virus glycoprotein fuses at low pH. Science 225:721-723. [DOI] [PubMed] [Google Scholar]
- 12.Fredericksen, B. L., and M. A. Whitt. 1998. Attenuation of recombinant vesicular stomatitis viruses encoding mutant glycoproteins demonstrate a critical role for maintaining a high pH threshold for membrane fusion in viral fitness. Virology 240:349-358. [DOI] [PubMed] [Google Scholar]
- 13.Fredericksen, B. L., and M. A. Whitt. 1996. Mutations at two conserved acidic amino acids in the glycoprotein of vesicular stomatitis virus affect pH-dependent conformational changes and reduce the pH threshold for membrane fusion. Virology 217:49-57. [DOI] [PubMed] [Google Scholar]
- 14.Fredericksen, B. L., and M. A. Whitt. 1995. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J. Virol. 69:1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst, T. R., P. L. Earl, and B. Moss. 1987. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudin, Y., H. Raux, A. Flamand, and R. W. H. Ruigrok. 1996. Identification of amino acids controlling the low-pH-induced conformational change of rabies virus glycoprotein. J. Virol. 70:7371-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helenius, A. 1993. Influenza virus fusion: from models towards a mechanism, p. 89-111. In J. Bentz (ed.), Viral fusion mechanisms, vol. 1. CRC Press, Ann Arbor, Mich.
- 18.Horvath, C. M., R. G. Paterson, M. A. Shaughnessy, R. Wood, and R. A. Lamb. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 66:4564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeetendra, E., and M. A. Whitt. 2001. Characterization of the minimal budding domain in the vesicular stomatitis virus (VSV) glycoprotein, p. 96, W19-2. In Proceedings of the 20th Annual Meeting of the American Society for Virology, University of Wisconsin-Madison, Madison.
- 21.Kreis, T. E., and H. F. Lodish. 1986. Oligomerization is essential for transport of vesicular stomatitis virus glycoprotein to the cell surface. Cell 46:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzmin, P. I., J. Zimmerberg, Y. A. Chizmadzhev, and F. S. Cohen. 2001. A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci. USA 98:7235-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawson, N., E. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157-167. [PubMed] [Google Scholar]
- 25.Li, Y., C. Drone, E. Sat, and H. P. Ghosh. 1993. Mutational analysis of the vesicular stomatitis virus glycoprotein G for membrane fusion domains. J. Virol. 67:4070-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsh, M., and A. Helenius. 1989. Virus entry into animal cells. Adv. Virus Res. 36:107-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushita, S., M. Robert-Guroff, J. Rusche, A. Koito, T. Hattori, H. Hoshino, K. Javaherian, K. Takatsuki, and S. Putney. 1988. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J. Virol. 62:2107-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCallus, D. E., K. E. Ugen, A. I. Sato, W. V. Williams, and D. B. Weiner. 1992. Construction of a recombinant bacterial human CD4 expression system producing a bioactive CD4 molecule. Viral Immunol. 5:163-172. [DOI] [PubMed] [Google Scholar]
- 29.Melikyan, G. B., S. A. Brener, D. C. Ok, and F. S. Cohen. 1997. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza virus hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muñoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 33.Ohnishi, S.-I. 1988. Fusion of viral envelopes with cellular membranes. Curr. Top. Membr. Transp. 32:257-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oravecz, T., and M. A. Norcross. 1993. Costimulatory properties of the human CD4 molecule: enhancement of CD3-induced T cell activation by human immunodeficiency virus type 1 through viral envelope glycoprotein gp120. AIDS Res. Hum. Retrovir. 9:945-955. [DOI] [PubMed] [Google Scholar]
- 35.Paternostre, M. T., R. J. Lowy, and R. Blumenthal. 1989. pH-dependent fusion of reconstituted vesicular stomatitis virus envelopes with Vero cells. Measurement by dequenching of fluorescence. FEBS Lett. 243:251-258. [DOI] [PubMed] [Google Scholar]
- 36.Paterson, R. G., S. W. Hiebert, and R. A. Lamb. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc. Natl. Acad. Sci. USA 82:7520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puri, A., M. Krumbiegel, D. Dimitrov, and R. Blumenthal. 1993. A new approach to measure fusion activity of cloned viral envelope proteins: fluorescence dequenching of octadecylrhodamine-labeled plasma membrane vesicles fusing with cells expressing vesicular stomatitis virus glycoprotein. Virology 195:855-858. [DOI] [PubMed] [Google Scholar]
- 38.Randall, R. E., D. F. Young, K. K. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 39.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 40.Roberts, P. C., T. Kipperman, and R. W. Compans. 1999. Vesicular stomatitis virus G protein acquires pH-independent fusion activity during transport in a polarized endometrial cell line. J. Virol. 73:10447-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robison, C. S., IV. 2001. Ph.D. thesis. University of Tennessee Health Science Center, Memphis.
- 43.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 44.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnell, M. J., J. E. Johnson, L. Buonocore, and J. K. Rose. 1997. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell 90:849-857. [DOI] [PubMed] [Google Scholar]
- 46.Shokralla, S., R. Chernish, and H. P. Ghosh. 1999. Effects of double-site mutations of vesicular stomatitis virus glycoprotein G on membrane fusion activity. Virology 256:119-129. [DOI] [PubMed] [Google Scholar]
- 47.Shokralla, S., Y. He, E. Wanas, and H. P. Ghosh. 1998. Mutations in a carboxy-terminal region of vesicular stomatitis virus glycoprotein G that affect membrane fusion activity. Virology 242:39-50. [DOI] [PubMed] [Google Scholar]
- 48.Suárez, T., W. R. Gallaher, A. Agirre, F. M. Goñi, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong, S., F. Yi, A. Martin, Q. Yao, M. Li, and R. W. Compans. 2001. Three membrane-proximal amino acids in the human parainfluenza virus type 2 (HPIV 2) F protein are critical for fusogenic activity. Virology 280:52-61. [DOI] [PubMed] [Google Scholar]
- 51.White, J. M. 1990. Viral and cellular membrane fusion proteins. Annu. Rev. Physiol. 52:675-697. [DOI] [PubMed] [Google Scholar]
- 52.Whitt, M. A., L. Buonocore, J. K. Rose, V. Ciccarone, A. Chytil, and G. Gebeyehu. 1991. TransfectACE reagent: transient transfection frequencies greater than 90%. Focus 13:8-12. [Google Scholar]
- 53.Whitt, M. A., P. Zagouras, B. Crise, and J. K. Rose. 1990. A fusion-defective mutant of the vesicular stomatitis virus glycoprotein. J. Virol. 64:4907-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zavorotinskaya, T. 1999. Ph.D. thesis. University of Tennessee Health Science Center, Memphis.
- 55.Zhang, L., and H. P. Ghosh. 1994. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J. Virol. 68:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, J., R. E. Dutch, and R. A. Lamb. 1997. Proper spacing between heptad repeat B and the transmembrane domain boundary of the paramyxovirus SV5 F protein is critical for biological activity. Virology 239:327-339. [DOI] [PubMed] [Google Scholar]