Abstract
Hibiscus chlorotic ringspot virus (HCRSV) from naturally infected Hibiscus rosa-sinensis L. loses virulence in its experimental systemic host Hibiscus cannabinus L. (kenaf) after serial passages in a local lesion host Chenopodium quinoa. Here we report the genetic changes responsible for the loss of virulence at the molecular level. A remarkable covariation of eight site-specific amino acids was found in the HCRSV capsid protein (CP) after serial passages in C. quinoa: Val49→Ile, Ile95→Val, Lys270→Arg, Gly272→Asp, Tyr274→His, Ala311→Asp, Asp334→Ala, and Ala335→Thr. Covariation of at least three of the eight amino acids, Val49, Ile95, and Lys270, caused the virus to become avirulent in kenaf. Interestingly, the nature of the covariation was consistent and reproducible at each serial passage. These data indicate that the nonsynonymous substitutions of amino acids in the HCRSV CP after serial passages in C. quinoa are not likely to be random events but may be due to host-associated positive selection or accelerated genetic drift. The observed interdependence among the three amino acids leading to avirulence in kenaf may have implications for structural or functional relationships in this virus-host interaction.
Host-induced genome evolution allows for adaptation of plant and animal viruses to their hosts (20, 22, 23). Adaptive evolution has been found to be either random or selected, and it depends on the virus and its host species (1, 26). During the last few decades, viral genome evolution has been investigated at the molecular level in two different ways. (i) Viral populations are transferred in parallel to different host species or to the same host in different geographic areas, as evidenced in plant viruses such as tobacco mosaic virus (TMV) (14) and carnation mottle virus (2). (ii) Viral populations are transferred serially in a specific host or to different hosts (or cell lines) with serial passages, as shown in plant viruses TMV, cucumber mosaic virus (CMV), and cowpea chlorotic mottle virus (14, 24, 25) and animal viruses, including simian immunodeficiency virus (27) human immunodeficiency virus type 1 (30), and infectious bursal disease virus (29). Other than nucleotide sequencing, time-of-flight mass spectrometry has been used to determine amino acid variations of viral genomes (26). To date, the viral capsid protein (CP) is generally targeted for investigation of genome evolution.
Hibiscus chlorotic ringspot virus (HCRSV), a member of the genus Carmovirus in the family of Tombusviridae, possesses a positive single-stranded RNA genome that encodes seven open reading frames (ORF). A novel ORF p25 overlapping closer to the 5′ proximal part of the CP cistron has been found (12). In 1987, Hurtt reported that after HCRSV was isolated directly from hibiscus plants and subjected to five serial passages in Chenopodium quinoa, the amino acid composition of the progeny virus was altered and the virus became avirulent (13). In this study, we defined virulence as the ability of the virus to move systemically, to accumulate viral RNA to detectable levels, and to induce disease symptoms in the inoculated systemic host kenaf (Hibiscus cannabinus L.).
A rapid increase of virus-induced reduction of host fitness is the most general result of serial passage experiments (8). Such repeated bottleneck events are also expected to cause progressive changes of the original genome sequence (3, 6, 7). To further investigate the nature of the amino acid mutations in the CP and to determine the factors contributing to avirulence in kenaf, we isolated a wild-type HCRSV (wtHCRSV) from a naturally infected hibiscus plant (Hibiscus rosa-sinensis) for serial passage in C. quinoa and repeated the serial passage procedures as described previously (13).
Five individual local lesions were excised from C. quinoa initially inoculated with crude leaf extract from the wtHCRSV-infected hibiscus plant. Each local lesion was cut in half. Half of the local lesion was macerated in 0.1 M sodium phosphate buffer (pH 7.0) and used to inoculate a new kenaf plant and a new C. quinoa leaf for serial passage experiments. The other half of the local lesion was subjected to total RNA extraction, reverse transcription-PCR (RT-PCR), and DNA sequencing. RT-PCR was performed with primers flanking the whole CP region. The primers corresponded to viral sequence at nucleotides (nt) 2519 to 2536 and viral complementary sequence at nt 3794 to 3774, respectively. Primers 1 (nt 2519 to 2536), 2 (nt 3794 to 3774), 3 (nt 3007 to 3024), and 4 (nt 3138 to 3121) were used to sequence the CP gene (nt 2590 to 3627). Five individual samples, one from each of five different local lesions, were sequenced during each passage in C. quinoa. The CP sequences from wtHCRSV and progeny viruses obtained from serial passages were confirmed after the RT-PCR products were cloned into a biologically active cDNA clone of HCRSV p223. Each passage included five replicates, and a total of six serial passages were performed. All plants were maintained in a growth chamber under the conditions of 16 h of light and 8 h of dark at 25°C. The experiment was done twice.
Site-specific mutations of HCRSV appeared in C. quinoa plants after serial passages.
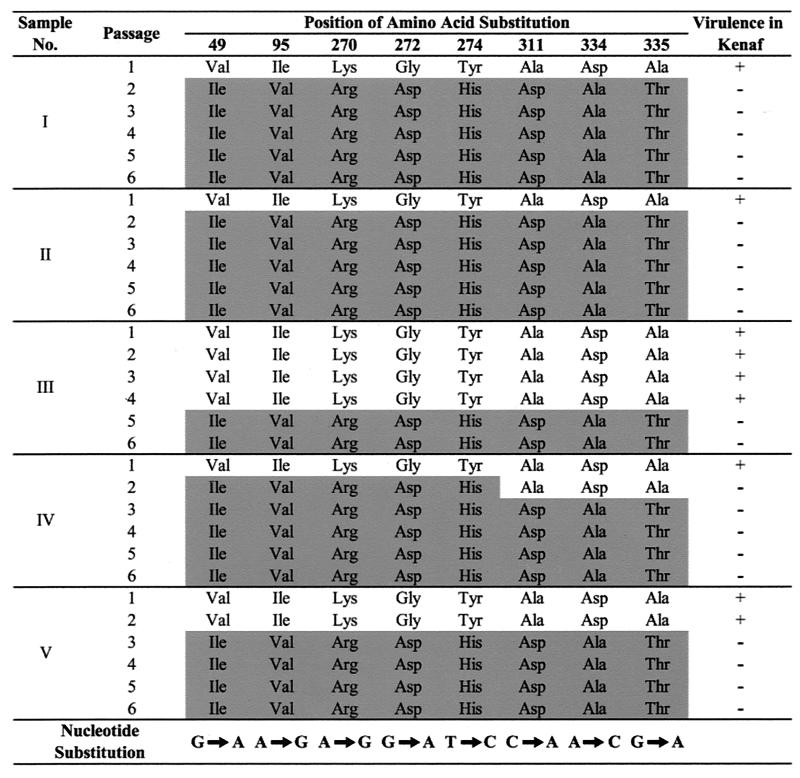
The wtHCRSV isolated from naturally infected H. rosa-sinensis induced chlorotic lesions on C. quinoa during all six serial transfers. The progeny viruses with more than four amino acid substitutions after serial passages in C. quinoa lost virulence in kenaf (Fig. 1, shaded substitutions). Northern blot analysis of those samples revealed no detectable level of genomic or subgenomic RNA accumulation in the inoculated and upper new leaves (data not shown). Analysis of the progeny viruses taken from each passage in C. quinoa showed the presence of specific mutations in the CP gene. Of the 18 nucleotide substitutions, eight resulted in nonsynonymous mutations. The nucleotide substitutions were predominantly from G-to-A and A-to-G. The bias of G-to-A transition has also been observed in CMV mutant populations (24). The covariation was found among eight site-specific amino acid positions at Val49→Ile, Ile95→Val, Lys270→Arg, Gly272→Asp, Tyr274→His, Ala311→Asp, Asp334→Ala, and Ala335→Thr, respectively (Fig. 1). The covariation occurred as early as passage 2 for some progeny isolates, and these mutations remained unchanged up to serial passage 6. The virus isolates that did not reach covariation in passage 2 were found to have a few identical mutations at the same amino acid residues at various passages, but subsequent passages brought them all to the same covariation of amino acids (Fig. 1). No other sequence variation was detected from 5 to 10 cloned RT-PCR fragments amplified from each passage after the observed equilibrium of covariation was attained. Of the eight amino acids, most were nonconserved changes and six were located between the residues 270 to 335, in the C terminus of the CP. The HCRSV CP has a total of 348 amino acids.
FIG. 1.
Representative samples of covariation of amino acids of wtHCRSV capsid protein with serial passages in C. quinoa. There were five replicates of each sample, and the entire experiment was repeated once. The amino acid substitutions are presented on a shaded background, and the nucleotide substitutions are shown at the bottom of the figure. Covariation of the eight amino acids was revealed at each passage until it reached equilibrium. Virulence (+) and avirulence (−) of the progeny viruses in kenaf plants are indicated. In a repeat experiment, four samples showed final covariation at passage 2 and one sample showed final covariation at passage 3. No other differences were observed.
To support the notion that the mutations observed resulted from virus passages in C. quinoa, instead of a pool of quasispecies, wtHCRSV-infected crude sap was obtained from five different leaves of one hibiscus plant. Individual CP genes were amplified from separately prepared crude sap samples by RT-PCR and cloned into the biologically active clone of HCRSV p223. All five CP sequences determined were identical when aligned. In addition, in a separate experiment, 20 individual cDNA clones of the wtHCRSV CP gene were obtained from RT-PCR products from the same hibiscus plant. DNA sequencing results showed that all 20 individual CP genes were identical to the previously determined sequence of wtHCRSV. These data indicate that the nucleotide sequence of wtHCRSV CP from the viral population in the HCRSV-infected hibiscus plant is predominantly homogeneous. Serial passages of wtHCRSV in C. quinoa protoplasts were also attempted, but no mutation was found in the CP gene (data not shown). These data suggest that the initiation of wtHCRSV CP site-specific mutations and covariation appear to be associated with the whole plant host. This is consistent with a recent genetic diversity study on CMV that mutation patterns in whole plants were different from those in protoplasts after serial passages, as both cell-to-cell and systemic functions in whole plants are absent in protoplasts (25).
Although viruses exhibit rapid sequence divergence over periods of time and mutations occur in variable positions of viral genomes (28), genetic stability has been revealed in plant viruses over time, space, and host species separations (2, 14, 26). This agrees with the view that host-associated selection results in decreased diversity in a viral genome (10). It seems that the covariation leads to reduction in fitness (ability to replicate infectious progeny) (6) of the progeny HCRSV in kenaf. The genetic diversity for CMV, TMV, and cowpea chlorotic mottle virus remained relatively constant after 10 serial passages, and none of the mutations became fixed during the passage experiments (24). In another study, only one mutation was found in the TMV sequence after more than 10 serial passages in a local lesion host (14). The phenomenon that eight specific amino acid mutations occurred at the same residues of HCRSV CP within five passages in C. quinoa is indeed remarkable. Covariation has also been observed in the envelope protein of human immunodeficiency virus type 1 (15), and the relative roles of selection and genetic drift have been proposed in virus evolution (9). An unknown evolutionary mechanism may operate between the virus and its specific host to drive a selection force or to accelerate the process of natural genetic drift to arrive at the covariation equilibrium observed in this study.
Covariation of at least three amino acids in HCRSV CP leads to avirulence in kenaf.
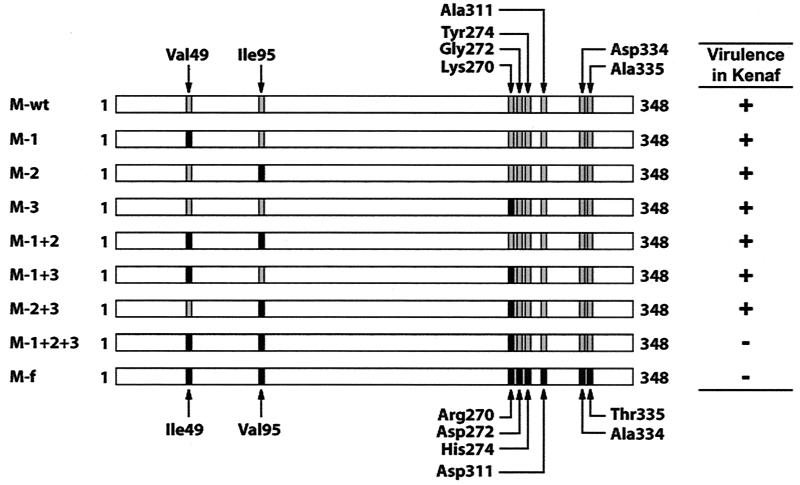
To investigate which of the eight amino acid substitutions are crucial in leading the progeny HCRSV to avirulence in kenaf, we analyzed each progeny virus from different serial passages. We observed that the progeny virus with the smallest number of amino acid substitutions that became avirulent in kenaf possessed five residue changes at Val49→Ile, Ile95→Val, Lys270→Arg, Gly272→Asp, and Tyr274→His (Fig. 1, sample IV, passage 2). Our initial approach was to construct and test mutant progeny viruses representing each of the five amino acid substitutions individually and add them to include all possible combinations. After testing the mutant virus generated with the first three amino acid changes of Val49→Ile, Ile95→Val, and Lys270→Arg, the resulting virus showed an avirulent phenotype in kenaf. We then narrowed down the choices and concentrated on investigating whether all three amino acids were required for avirulence. Viruses with various single and/or combined amino acid mutations were generated (Fig. 2). To generate the various mutant viruses, the CP gene of wtHCRSV was isolated from H. rosa-sinensis by RT-PCR. The PCR fragment was digested with PvuI and HpaI and cloned into the biologically active cDNA clone of HCRSV p223. This generated the M-wt clone in which the complete CP gene was derived from wtHCRSV. M-1+2+3 and M-f were made in the same way as the M-wt, except that the CPs were amplified from the progeny viruses containing the intermediate and final CP mutations after serial passage 6, respectively. Six constructs were generated by site-directed mutagenesis, namely, M-1, M-2, M-3, M-1+2, M-1+3, and M-2+3 (Fig. 2), to determine which amino acid or combination of amino acids contribute to avirulence of HCRSV in kenaf. All cDNA constructs were sequenced to verify the accuracy of each clone.
FIG. 2.
Schematic diagrams of HCRSV CP constructs cloned into p223. M-wt is the wild-type HCRSV CP sequence. M-f is the final serially passaged HCRSV CP sequence with eight amino acid substitutions. M-1+2+3 is an intermediate mutant with three amino acid substitutions. M-1, M-2, M-3, M-1+2, M-1+3, and M-2+3 are mutants generated with single or double amino acid changes by site-directed mutagenesis. Mutated residues (black boxes) and wild-type residues (shaded boxes) are shown. Virulence (+) or avirulence (−) in kenaf is shown to the right of the construct diagrams.
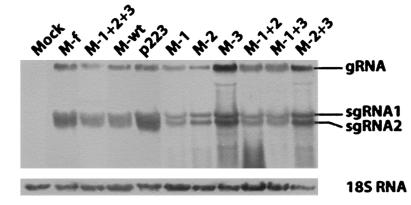
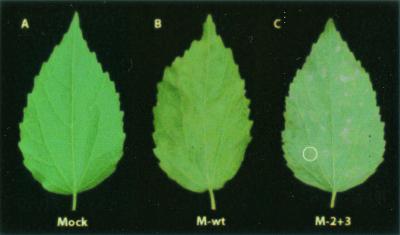
In vitro transcripts of the constructs were inoculated to kenaf protoplasts and whole plants. Analysis of the RNA profiles of HCRSV by Northern blot hybridization (17) showed that accumulation of genomic RNA and two subgenomic RNAs in kenaf protoplasts reached similar levels for all CP mutant viruses compared to that derived from the biologically active clone of HCRSV p223 at 24 h postinoculation (Fig. 3). This indicated that all mutants could replicate in kenaf protoplasts. When inoculated in whole kenaf plants, except for mutants M-2+3, M-1+2+3, and M-f, the virulence of mutant viruses was similar to that of the wtHCRSV. The number and size of local lesions at 7 days postinoculation and the severity of small systemic necrotic spots at 14 days postinoculation were similar. The CP mutant M-2+3 with two amino acid substitutions at Val95 and Arg270 caused symptoms later (20 days later) and yielded milder chlorotic lesions but eventually induced large systemic necrotic lesions on the leaves (Fig. 4C, circled lesion). This is different from the normal chlorotic and necrotic spots on kenaf inoculated with wtHCRSV (Fig. 4B). Since both mutant viruses M-1+2+3 with three mutations and M-f with eight mutations were avirulent in kenaf, covariation of at least three amino acids at Val49→Ile, Ile95→Val, and Lys270→Arg is sufficient to cause avirulence.
FIG. 3.
Northern blot analysis of the HCRSV CP mutants in kenaf protoplasts. Kenaf protoplasts were inoculated with in vitro transcripts of HCRSV CP mutant constructs. The biologically active cDNA clone of HCRSV p223 was used as a positive control, and mock inoculation with transcription buffer was used as a negative control. gRNA, genomic RNA; sgRNA, subgenomic RNA.
FIG. 4.
Kenaf leaves showing different symptoms induced by different CP mutants of HCRSV. (A) No symptoms (mock inoculated). (B) Chlorotic spots induced by M-wt. (C) Necrotic lesions (circled) induced by HCRSV CP mutant M-2+3.
Since the ORF p25 overlaps with the coding region of the CP gene, we investigated the potential implications of mutations in the CP that might alter the amino acids of ORF p25. As most of the amino acid substitutions occurred at the C terminus of the CP, only two amino acids of concern fell into the shared nucleotide region of ORF p25. The first mutation of CP at Val49 did not change the amino acid in ORF p25. The other mutation of CP at Val95 changed the amino acid of ORF p25 from Ile91 to Met. The other six amino acid mutations of CP are located beyond the coding region of p25. The fact that mutant viruses M-2 and M-1+2 both contained Val95 and were as infectious and as virulent as M-wt rules out the possible contribution of ORF p25 to avirulence in kenaf.
In the genus Carmovirus, turnip crinkle virus (TCV) CP has been shown to modulate symptoms due to amino acid substitutions in the capsid surface (11, 16, 18). Also, direct interaction between TCV CP and an Arabidopsis protein designated TIP (TCV-interacting protein) has been demonstrated recently (21). Capsid proteins are also involved in virus-host interactions for potato virus X, TMV, alfalfa mosaic virus, and other morphologically diverse plant viruses (4, 5, 19). In HCRSV, the CP mutant virus M-2+3 induced larger necrotic lesions on systemically infected kenaf leaves (Fig. 4C). Since all progeny viruses could replicate to a level comparable to that of M-wt in kenaf protoplasts (Fig. 3) and only mutant virus M-2+3 induced large necrotic lesions on whole kenaf plants, Ile95→Val and Lys270→Arg might be part of a component that acts as an elicitor which can trigger the hypersensitive response pathway in kenaf and lead to severe necrosis. On the other hand, when all three amino acid substitutions are present, they render this virus avirulent in kenaf. This may have implications for structural or functional relationships in this virus-host interaction. It is reasonable to suggest that all eight amino acid residues may be crucial in the virus interaction with an unknown kenaf host factor(s). The dynamics of virus-host interactions in the HCRSV system is interesting and remains to be explored.
Acknowledgments
This research was supported in part by The National University of Singapore through research grant R-154-000-111-112.
We thank Peter Palukaitis from the Scottish Crop Research Institute for critical reading of this manuscript.
REFERENCES
- 1.Aldaoud, R., W. O. Dawson, and G. E. Jones. 1989. Rapid, random evolution of the genetic structure of replicating tobacco mosaic virus populations. Intervirology 30:227-233. [DOI] [PubMed] [Google Scholar]
- 2.Canizares, M. C., J. F. Marcos, and V. Pallas. 2001. Molecular variability of twenty-one geographically distinct isolates of Carnation mottle virus (CarMV) and phylogenetic relationships within the Tombusviridae family. Arch. Virol. 146:2039-2051. [DOI] [PubMed] [Google Scholar]
- 3.Clarke, D. K., E. A. Duarte, A. Moya, S. F. Elena, E. Domingo, and J. Holland. 1993. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. J. Virol. 67:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz, S. S., and D. C. Baulcombe. 1993. Molecular analysis of potato virus X isolates in relation to the potato hypersensitivity gene Nx. Mol. Plant-Microbe Interact. 6:707-714. [DOI] [PubMed] [Google Scholar]
- 5.Dawson, W. O. 1992. Tobamovirus-plant interactions. Virology 186:359-367. [DOI] [PubMed] [Google Scholar]
- 6.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 7.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 8.Ebert, D. 1998. Experimental evolution of parasites. Science 282:1432-1435. [DOI] [PubMed] [Google Scholar]
- 9.Frost, S. D. W., M. Nijhuis, R. Schuurman, C. A. B. Boucher, and A. J. L. Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Arenal, F., A. Fraile, and J. M. Malpica. 2001. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39:157-186. [DOI] [PubMed] [Google Scholar]
- 11.Heaton, L. A., and M. M. Laakso. 1995. Several symptom-modulating mutations in the coat protein of turnip crinkle carmovirus result in particles with aberrant conformational properties. J. Gen. Virol. 76:225-230. [DOI] [PubMed] [Google Scholar]
- 12.Huang, M., D. C.-Y. Koh, L.-J. Weng, M.-L. Chang, Y.-K. Yap, L. Zhang, and S.-M. Wong. 2000. Complete nucleotide sequence and genomic organization of hibiscus chlorotic ringspot virus, a new member of the genus Carmovirus: evidence for the presence and expression of two novel open reading frames. J. Virol. 74:3149-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurtt, S. S. 1987. Detection and comparison of electrophorotypes of hibiscus chlorotic ringspot virus. Phytopathology 77:845-850. [Google Scholar]
- 14.Kearney, C. M., M. J. Thomson, and K. E. Roland. 1999. Genome evolution of tobacco mosaic virus populations during long-term passaging in a diverse range of hosts. Arch. Virol. 144:1513-1526. [DOI] [PubMed] [Google Scholar]
- 15.Korber, B. T. M., R. M. Farber, D. H. Wolpert, and A. S. Lapedes. 1993. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc. Natl. Acad. Sci. USA 90:7176-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laakso, M. M., and L. A. Heaton. 1993. Asp → Asn substitutions in the putative calcium-binding site of the turnip crinkle virus coat protein affect virus movement in plants. Virology 197:774-777. [DOI] [PubMed] [Google Scholar]
- 17.Liang, X. Z., S. W. Ding, and S. M. Wong. 2002. Development of a kenaf (Hibiscus cannabinus L.) protoplast system for replication study of hibiscus chlorotic ringspot virus. Plant Cell Rep. 20:982-986. [Google Scholar]
- 18.Lin, B., and L. A. Heaton. 1999. Mutational analyses of the putative calcium binding site and hinge of the turnip crinkle virus coat protein. Virology 259:34-42. [DOI] [PubMed] [Google Scholar]
- 19.Neeleman, L., A. C. van der Kuyl, and J. F. Bol. 1991. Role of alfalfa mosaic virus coat protein gene in symptom formation. Virology 181:687-693. [DOI] [PubMed] [Google Scholar]
- 20.Ni, Y. H., M. H. Chang, P. J. Chen, H. H. Lin, and H. Y. Hsu. 1997. Evolution of hepatitis C virus quasispecies in mothers and infants infected through mother-to-infant transmission. J. Hepatol. 26:967-974. [DOI] [PubMed] [Google Scholar]
- 21.Ren, T., F. Qu, and T. J. Morris. 2000. HRT gene function requires interaction between a NAC protein and viral protein to confer resistance to turnip crinkle virus. Plant Cell 12:1917-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rima, B. K., J. A. Earle, K. Baczko, V. ter Meulen, U. G. Liebert, C. Carstens, J. Carabana, M. Caballero, M. L. Celma, and R. Fernandez-Munoz. 1997. Sequence divergence of measles virus haemagglutinin during natural evolution and adaptation to cell culture. J. Gen. Virol. 78:97-106. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Cerezo, E., A. Moya, and F. Garcia-Arenal. 1989. Variability and evolution of the plant RNA virus pepper mild mottle virus. J. Virol. 63:2198-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider, W. L., and M. J. Roossinck. 2000. Evolutionarily related Sindbis-like plant viruses maintain different levels of population diversity in a common host. J. Virol. 74:3130-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider, W. L., and M. J. Roossinck. 2001. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J. Virol. 75:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.She, Y. M., S. Haber, D. L. Seifers, A. Loboda, I. Chernushevich, H. Perreault, W. Ens, and K. G. Standing. 2001. Determination of the complete amino acid sequence for the coat protein of brome mosaic virus by time-of-flight mass spectrometry. J. Biol. Chem. 276:20039-20047. [DOI] [PubMed] [Google Scholar]
- 27.Spencer Valli, P. J., and J. Goudsmit. 1998. Structured-tree topology and adaptive evolution of the simian immunodeficiency virus SIVsm envelope during serial passage in rhesus macaques according to likelihood mapping and quartet puzzling. J. Virol. 72:3673-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss, J. H., and E. G. Strauss. 2001. Virus evolution: how does an enveloped virus make a regular structure? Cell 105:5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi, T., M. Ogawa, Y. Inoshima, M. Miyoshi, H. Fukushi, and K. Hirai. 1996. Identification of sequence changes responsible for the attenuation of highly virulent infectious bursal disease virus. Virology 223:219-223. [DOI] [PubMed] [Google Scholar]
- 30.Yuste, E., C. Lopez-Galindez, and E. Domingo. 2000. Unusual distribution of mutations associated with serial bottleneck passages of human immunodeficiency virus type 1. J. Virol. 74:9546-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]