Abstract
Human immunodeficiency virus (HIV) entry is triggered by interactions between a pair of heptad repeats in the gp41 ectodomain, which convert a prehairpin gp41 trimer into a fusogenic three-hairpin bundle. Here we examined the disposition and antigenic nature of these structures during the HIV-mediated fusion of HeLa cells expressing either HIVHXB2 envelope (Env cells) or CXCR4 and CD4 (target cells). Cell-cell fusion, indicated by cytoplasmic dye transfer, was allowed to progress for various lengths of time and then arrested. Fusion intermediates were then examined for reactivity with various monoclonal antibodies (MAbs) against immunogenic cluster I and cluster II epitopes in the gp41 ectodomain. All of these MAbs produced similar staining patterns indicative of reactivity with prehairpin gp41 intermediates or related structures. MAb staining was seen on Env cells only upon exposure to soluble CD4, CD4-positive, coreceptor-negative cells, or stromal cell-derived factor-treated target cells. In the fusion system, the MAbs reacted with the interfaces of attached Env and target cells within 10 min of coculture. MAb reactivity colocalized with the formation of gp120-CD4-coreceptor tricomplexes after longer periods of coculture, although reactivity was absent on cells exhibiting cytoplasmic dye transfer. Notably, the MAbs were unable to inhibit fusion even when allowed to react with soluble-CD4-triggered or temperature-arrested antigens prior to initiation of the fusion process. In comparison, a broadly neutralizing antibody, 2F5, which recognizes gp41 antigens in the HIV envelope spike, was immunoreactive with free Env cells and Env-target cell clusters but not with fused cells. Notably, exposure of the 2F5 epitope required temperature-dependent elements of the HIV envelope structure, as MAb binding occurred only above 19°C. Overall, these results demonstrate that immunogenic epitopes, both neutralizing and nonneutralizing, are accessible on gp41 antigens prior to membrane fusion. The 2F5 epitope appears to depend on temperature-dependent elements on prefusion antigens, whereas cluster I and cluster II epitopes are displayed by transient gp41 structures. Such findings have important implications for HIV vaccine approaches based on gp41 intermediates.
Human immunodeficiency virus (HIV) entry occurs through a pH-independent mechanism involving the direct fusion of the virus and cell membranes. The viral envelope proteins that mediate this process include a soluble glycoprotein component, gp120, and a transmembrane subunit, gp41, which are assembled into trimeric spikes on the virion surface. In the currently accepted model of HIV infection, the entry process begins with the binding of gp120 to cell surface CD4. This interaction forms a gp120-CD4 complex that expresses a binding site for certain CC or CxC chemokine receptors on the gp120 component (71). The major chemokine receptor, or coreceptor, used by macrophage-tropic (or R5) HIV strains is CCR5 (1), while T-cell-tropic (or X4) viruses predominantly use CXCR4 (21). Contact between the coreceptor and the gp120-CD4 complex forms a tripartite intermediate that is thought to dislocate gp120 from gp41 (66). A series of conformational changes in gp41 occur in concert with these binding events, culminating in a structure that promotes membrane fusion and viral entry.
In order to complete the fusion process, gp41 transitions from a metastable conformation, maintained in the virion spike, into a highly stable “six-helix bundle” in which each monomer of the gp41 trimer takes on a “hairpin” conformation (6, 59, 66). Similar structures are formed by influenza virus hemagglutinin (HA) (4, 70), Moloney murine leukemia virus (19), human T-cell leukemia virus type 1 gp21 (40), Ebola virus GP2 (45, 65), simian parainfluenza virus 5 F (2), and respiratory syncytial virus (74). Formation of the gp41 hairpin structure is facilitated by interactions between two heptad repeat sequences in the gp41 ectodomain (HR1 and HR2) that form helical domains with hydrophobic faces (44). Crystallographic studies of gp41-derived peptides have revealed that the HR1 helices form a central trimeric “coiled coil,” which contains three highly conserved hydrophobic grooves into which the three HR2 helices are packed in an antiparallel orientation (6, 59, 66). In the complete gp41 molecule, a hydrophilic loop region containing two cysteines links HR1 and HR2, such that each monomer in the six-helix bundle folds into the hairpin conformation.
Recent evidence suggests that during viral replication the six-helix bundle is preceded by the formation of a trimeric “prehairpin” coiled-coil intermediate in which HR1 and HR2 are exposed (26, 27, 47, 49). Studies using gp41-derived peptides that bind HR1 or HR2 suggest that the prehairpin structure appears at some point after the gp120-CD4 complex forms and persists during a fusion lag phase that precedes lipid mixing (26, 27, 47, 49). CD4 alone is sufficient for inducing the putative prehairpin intermediate in some systems (26, 27, 47), although this may not be the case for all HIV isolates (26). Ultimately, coreceptor binding to the gp120-CD4 complex triggers a temperature-dependent conversion of the prehairpin structure to the six-helix bundle. Although some experiments suggest that this conversion occurs simultaneously with lipid mixing (47), more recent studies suggest that under certain conditions six-helix bundles may form and accumulate prior to membrane fusion (28).
Because of their unique structures, each gp41 intermediate is expected to possess a distinct antigenic character, possibly involving the selective exposure of sequences needed for viral fusion. Such sequences are predicted to be targets for HIV-neutralizing antibodies. However, it has been difficult to predict whether antibody responses raised against gp41 epitopes will be able to neutralize HIV infection. For example, the HR domains on the gp41 prehairpin are viewed as candidate targets for neutralizing antibody responses because they engage in interactions critical for viral fusion (26, 35, 37, 49, 67-69). Furthermore, these regions overlap immunogenic epitope clusters designated cluster I and cluster II (8, 30, 60). However, human monoclonal antibodies (MAbs) against these epitopes are either poorly neutralizing or noninhibitory (23, 41, 46, 55). Similarly, a conserved region in gp41 (3, 50, 53, 54) that is accessible in the envelope spike (58) has been viewed as a neutralizing antibody site because it contains a conserved epitope that is recognized by a potent, broadly neutralizing human MAb, 2F5 (10, 17, 62). However, antigens based on the putative 2F5 core epitope sequence elicit cognate antibodies that are not neutralizing (9, 14, 16, 18, 43, 72) and therefore may not precisely represent the MAb 2F5 binding site at the cell surface. In agreement, a recent study suggests that the 2F5 epitope is more complex than the known core epitope sequence (53).
In order to better define the antigenic nature of gp41 during HIV infection, we employed an assay system that was previously developed (22) to simultaneously detect cell-cell fusion and epitope exposure on intermediate HIV envelope structures. Here we present evidence that prehairpin intermediates with accessible, but nonneutralizing, cluster I and cluster II epitopes are formed rapidly upon HIV envelope-CD4 binding but disappear after membrane fusion has begun. In addition, we show that the broadly neutralizing 2F5 epitope is immunoreactive throughout the fusion process, although its immunoreactivity is distinctively temperature dependent.
MATERIALS AND METHODS
Cells.
The HeLa cell line provided by Richard Axel and the HeLa/CD4/MAGI and U373/CD4/MAGI cell lines provided by Michael Emerman were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). Both the HeLa/CD4/MAGI and U373/CD4/MAGI cells contain an HIV long-terminal-repeat (LTR)-driven β-galactosidase gene that is activated by HIV tat expression (38). The HeLa cells endogenously express CXCR4 (21). The cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, and the antibiotics G418 (0.1 mg/ml; Gibco BRL) and hygromycin B (0.05 mg/ml) (complete medium). The stable HeLa cell line (Env cells) producing replication-defective HXB2 virions was generated by cotransfection of HeLa cells with the genetic constructs HIV-gpt and HXB2-Env. The HIV-gpt construct contains the HXB2 proviral genome with the gp160-coding region removed and replaced with the bacterial gpt gene driven by the simian virus 40 promoter to allow selection by mycophenolic acid. These cells were maintained in DMEM containing 10% fetal bovine serum, 50 μg of mycophenolic acid/ml, and 50 μg of gentamicin/ml. To monitor consistent levels of HXB2 envelope expression, cells were labeled with the anti-gp120 MAb 2G12 and analyzed by flow cytometry.
Antibodies and reagents.
Human MAbs 50-69, 98-6, 240-D, 246-D, and 126-6, derived from HIV-1-infected individuals in the United States, were provided by Susan Zolla-Pazner, New York University, New York, and through the AIDS Research and Reference Reagent Program. MAb 3D6 and MAb F240 were provided by L. Cavacini and Hermann Katinger through the AIDS Research and Reference Reagent Program. MAb 8F101 was provided by Ranajiit Pal, Advanced BioScience Laboratories Inc., Kensington, Md. MAb 2F5 was obtained from Hermann Katinger, IAM Pharmaceuticals, Inc., Vienna, Austria. Human MAb__17b was provided by James Robinson, Tulane University, New Orleans, La. The goat anti-mouse and goat anti-human secondary antibodies conjugated to Alexa 594 or Alexa 488, calcein-AM, CellTracker Green CMFDA, and CellTracker Blue CMF2HC were obtained from Molecular Probes, Eugene, Oreg. Bis-sulfosuccinimidylsuberate (BS3) was obtained from Pierce, Rockford, Ill. Soluble CD4 (sCD4) was obtained from Werner Meier at Biogen, Cambridge, Mass. The 17b Fab fragments were generated according to the manufacturer's protocols by using the ImmunoPure Fab Preparation kit from Pierce. 17b Fab fragments were directly conjugated to Alexa 594 by using an Alexa 594 protein labeling kit obtained from Molecular Probes. T20 was synthesized according to the published sequence (69).
Cell fusion assay.
Env cells (5 × 106 to 10 × 106) were labeled by suspension in prewarmed (37°C) serum-free DMEM, containing either 0.33 μM CellTracker Green CMFDA (absorption, 492 nm; emission, 516 nm) or 5 μM CellTracker Blue (absorption, 371 nm; emission, 464 nm), for 30 min at 37°C in a 5% CO2 incubator. Cells were then pelleted by centrifugation and resuspended in fresh prewarmed complete medium for 30 min at 37°C in a 5% CO2 incubator. Cells were washed three times with phosphate-buffered saline (PBS) and resuspended at a final concentration of 5 × 105 cells/ml in complete medium. In the standard fusion system, HeLa/CD4/MAGI cells (target cells) were seeded at maximal density on glass coverslips for 15 to 18 h. To perform experiments in the absence of a coreceptor, U373/CD4/MAGI cells, which do not express CXCR4, were used instead. Labeled Env cells (105 in 200 μl) were added to each coverslip and incubated for intervals of 0 to 120 min, as indicated below, at 37°C in a 5% CO2 incubator. Cell-cell fusion intermediates were arrested and fixed by adding BS3 to a final concentration of 1 mM. After 15 min at room temperature, the fixing process was stopped by addition of 20 mM Tris, pH 7.4, for 15 min at room temperature. Alternatively, the fusion intermediates were arrested by chilling cells to 4°C (24, 25, 31, 33, 47). The coverslips were rinsed three times in 4°C PBS and placed on ice. Nonspecific antibody binding was blocked by incubating the coverslips in 4% normal goat serum (NGS) for 30 min on ice. The coverslips were incubated with primary MAbs, at the indicated concentrations, in PBS containing 4% NGS for 1 h on ice. To investigate MAb 2F5 binding (15 μg/ml), primary antibody incubations were carried out at 37°C in the presence of 0.2% sodium azide, because such conditions produced optimal antibody binding. Double-staining experiments were carried out by adding combinations of murine and human MAbs to cells treated with the cytoplasmic dye CellTracker Blue. To fix both the primary antibody and cytoplasmic CellTracker dyes, coverslips were rinsed three times at 4°C with PBS, fixed in ice-cold 4% paraformaldehyde (PFA) for 1 min, and then permeabilized in chilled methanol for 10 min. After rehydration in PBS, the cells were incubated with goat anti-mouse and/or goat anti-human secondary antibodies coupled to Alexa 594 (absorption, 590 nm; emission, 617 nm) at 5 μg/ml for 30 min at room temperature. For double-labeled experiments, primary mouse MAbs were detected with Alexa 488-conjugated secondary antibodies (green), while primary human MAbs were detected with Alexa 594-conjugated secondary antibodies (red). Following three rinses in PBS, the cells were mounted in Vectashield (Vector Laboratories, Burlingame, Calif.) for microscopic analysis. Images were taken on a Zeiss LSM410 confocal microscope at a magnification of ×100. Nomarski images and images obtained with each fluorescent dye were acquired separately and later superimposed using Adobe Photoshop to provide a composite view of selected fields. Since membrane fusion in cell coculture systems is asynchronous (22, 28, 47, 64), images that appeared to represent the most advanced stage in the fusion process were selected for each time point.
Flow cytometric analyses.
Env cells (2 × 105) were washed twice with Flow Buffer (PBS, 5% fetal bovine serum, 0.2% sodium azide) and incubated in the presence or absence of 2 μg of sCD4/ml in DMEM for 60 min at 4°C. Following two washes with Flow Buffer, the cells were incubated with primary antibodies at the indicated concentrations for 60 min on ice. Cells were then washed twice with Flow Buffer and incubated with phycoerythrin (PE)-conjugated secondary antibodies (5 μg/ml) for 30 min at 4°C. Next, the cells were washed with Flow Buffer and fixed with 2% PFA. Appropriate isotype controls were used for each antibody to control for nonspecific binding.
Immunofluorescence analyses of sCD4-treated cells.
Env cells were allowed to adhere to 22-mm2 glass coverslips for 24 h at a density of no more than 7.5 × 105 cells per coverslip. sCD4 (2 μg/ml) in 200 μl of DMEM was added to the coated coverslips for 60 min at 4°C. sCD4-treated Env cells were rapidly cooled to 4°C without fixation and stained. The human anti-gp41 MAbs were added to the treated or untreated Env cells and incubated for 60 min on ice. Cells were rinsed three times in 4°C PBS and then incubated with goat anti-human secondary antibodies (5 μg/ml) coupled to Alexa 488 (Abs, 495 nm; EM, 519 nm) for 30 min on ice. Following three rinses in PBS, the coverslips were incubated with a human 17b Fab fragment (5 μg/ml) directly conjugated with Alexa 594 for 30 min on ice. Cells were then rinsed three times at 4°C with PBS, fixed in ice-cold 4% PFA for 1 min, and permeabilized in chilled methanol for 10 min. After rehydration in PBS, cells were mounted in Vectashield for microscopic analysis. Images were taken on a Zeiss LSM410 confocal microscope at a magnification of ×100.
Immunofluorescence analyses of cells in the absence of coreceptor binding.
CellTracker Green-labeled Env cells and target cells were treated with 5 μg of stromal cell-derived factor (SDF-1β)/ml for 1 h at 37°C and then cocultivated at 37°C for the indicated times as described above, but in the presence of 5 μg of chemokine/ml. Cell-cell fusion was arrested by fixation with BS3 or by rapid cooling to 4°C. Alternatively, CellTracker Green-labeled Env cells were cocultivated with U373 target cells that expressed only CD4. Following 120 min of coculture at 37°C, cell-cell fusion was arrested by fixation with BS3 or by rapid cooling to 4°C. For both assays, immunostaining was performed as described above by using MAbs at 1.5 μg/ml (MAb 50-69) and 1 μg/ml (MAb 98-6). Images were taken on a Zeiss LSM410 confocal microscope. Nomarski images and images for each fluorescent dye were acquired separately and were superimposed using Adobe Photoshop.
Temperature-dependent binding of MAb 2F5.
Env cells (2 × 105) were washed twice with DMEM and placed on ice in a final volume of 100 μl. The cells were then incubated with MAb 2F5 (15 μg/ml) for 30 min at various temperatures in a “Genius” unit (Techne Ltd.), which allows a temperature control precision of ±0.1°C. Following incubation with 2F5, the cells were washed twice with ice-cold Flow Buffer and fixed in 4% PFA for 10 min on ice. 2F5 binding was detected with an Alexa 594-conjugated secondary antibody (which appears red). Cells were subsequently mounted in Vectashield for microscopic analysis. Images were taken on a Zeiss LSM410 confocal microscope at a magnification of ×100.
Assays for inhibition of cell-cell fusion.
Env cells (2 × 104) were added to wells of a 96-well microtiter plate (Falcon, Lincoln Park, N.J.) in 200 μl of complete medium and allowed to adhere for 15 to 18 h at 37°C. Target cells (2 × 104) were then added and cocultured with Env cells for 10 min at 37°C. The cells were then washed with PBS to remove unattached target cells, and anti-gp41 antibodies or T20 was added at the indicated concentrations at 23°C, which maintains a temperature-arrested state of fusogenicity (47). Control assays were carried out in the absence of a test inhibitor or with equivalent amounts of immunoglobulin G (IgG) isotype controls. After a 120-min incubation at the 23°C temperature-arrested state (47), the treated cells were returned to a fusogenic temperature (37°C) and cocultured 15 to 18 h. Syncytium formation was then quantified as a function of HIV Tat-mediated β-galactosidase production in the target cells. β-Galactosidase was quantified as a function of enzymatic activity by using a chemiluminescence reagent (Galactostar; Tropix, Bedford, Mass.) according to the manufacturer's protocol. The resulting chemiluminescence was quantified using a Victor2 fluorescence plate reader (EG&G Wallac, Gaithersburg, Md.). To estimate neutralization sensitivity after triggering with sCD4, Env cells (2 × 104) were added to wells of a 96-well microtiter plate (Falcon) and incubated for 15 to 18 h at 37°C in 200 μl of complete medium. Fifty microliters of a saturating concentration of sCD4 (2 μg/ml) in DMEM was added per well for 60 min at 4°C. The cells were then washed with ice-cold PBS to remove unbound sCD4, and anti-gp41 antibodies or T20 was added for 60 min at 37°C. Target cells (2 × 104) were then added and cocultured 15 to 18 h in the presence of anti-gp41 antibodies or T20. β-Galactosidase was quantified as a function of activity by using a chemiluminescence reagent (Galactostar; Tropix) according to the manufacturer's protocol. The resulting chemiluminescence was quantified using a Victor2 fluorescence plate reader (EG&G Wallac). Cells were also viewed microscopically after 120 min of incubation at 37°C.
RESULTS
In this study, we used a well-characterized in vitro coculture system to investigate the antigenic nature of gp41 during HIV fusion. As we reported earlier (22), the system is based on cocultures of two HeLa cell phenotypes, one expressing human CD4 (target cells) and the other expressing stable and consistent levels of the HIVHXB2 envelope on the cell surface (Env cells). Since the HXB2 envelope uses the CXCR4 coreceptor endogenously expressed on HeLa cells, the system provides all the surface components necessary for HIV-mediated fusion. Env cells are loaded with a fixable cytoplasmic dye, CellTracker Green, and cocultured with a monolayer of target cells. Ensuing cell-cell interactions are then allowed to progress for various lengths of time and arrested either by cooling the system to 4°C or by fixing cells with the homobifunctional cross-linker, BS3. Although HIV-mediated membrane fusion in cell coculture systems is asynchronous (22, 28, 47, 64), we were able to locate representative fields at each time point that marked the advancement of the fusion process in the cocultures. As we previously reported (22), cocultures arrested within 10 min at 37°C contain Env cells that are attached to target cells but without evidence of cytoplasmic dye transfer (Fig. 1). However, following 30 min in coculture, a portion of arrested cells (approximately 5%) begins to exhibit signs of membrane fusion, marked by the appearance of diffusely staining cell clusters experiencing cytoplasmic mixing (Fig. 1). The number of such fusion events accumulates over time such that by 120 min most fields contain large, diffusely staining syncytia with multiple areas of nuclear dye exclusion. Thus, a temporal change in the appearance of the cells, reflecting the progression of cell-cell attachment and fusion, is clearly visualized by this system.
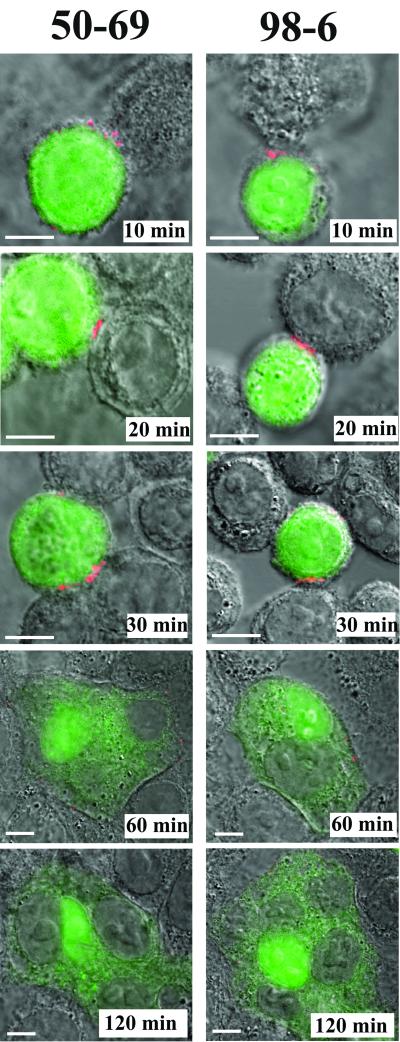
FIG. 1.
Anti-gp41 MAbs against cluster I and cluster II epitopes react with transient gp41 structures during cell-cell fusion. CellTracker Green-labeled Env cells were cocultivated with HeLa/CD4 target cells for the indicated times as described in Materials and Methods. Immunostaining was performed with BS3-fixed cells (shown) or with unfixed cells cooled to 4°C. Results are shown with the anti-cluster I MAb 50-69 (1.5 μg/ml) and the anti-cluster II MAb 98-6 (1 μg/ml), used at concentrations that produced the optimum signals. Other anti-cluster I MAbs, F240, 240-D, 3D6, and 246-D, produced identical results (data not shown), as did the anti-cluster II MAb 126-6 (data not shown). Cell nuclei appear as gray areas in stained syncytia. Fusion of Env and target cells produces the light green cytoplasmic staining surrounding the nuclei. All antibodies were tested in parallel along with human isotype control immunoglobulin, which produced no binding signal (data not shown). Images representative of at least three experiments are shown. Bars, 10 μm.
Exposure of cluster I and cluster II epitopes during cell-cell fusion.
A series of experiments was carried out using a panel of human MAbs directed against two immunogenic regions located within the gp41 ectodomain (29, 51, 52, 73). One region, identified as epitope cluster I, spans amino acid residues 598 to 604 and includes a nonhelical hydrophilic region that forms a disulfide loop in the six-helix bundle (73). The second region, designated epitope cluster II, includes residues 644 to 663 and comprises a portion of HR2 (32, 73). Anti-cluster I antibodies included MAbs 50-69, F240, 240-D, 3D6, and 246-D; the anti-cluster II MAbs were 98-6 and 126-6 (5, 20, 73). All anti-cluster I antibodies produced identical staining patterns in our fusion assay (data not shown) irrespective of whether they recognized linear versus conformational epitopes in protein or peptide-based assays (8, 30, 60, 73). Further, the binding patterns were identical to those observed with the two anti-cluster-II MAbs (data not shown).
Given these similarities, additional studies were carried out using only MAb 50-69 (anti-cluster I) and MAb 98-6 (anti-cluster II). Both MAbs were shown to recognize helix-stabilized structures in peptide-peptide or protein-peptide binding assays (8, 30, 60) and to exhibit enhanced exposure on HIV envelope-expressing cells treated with sCD4 (56). In particular, the 50-69 epitope is highly dependent on stabilization by HR2-derived peptides in peptide-peptide model systems (8, 30, 60). Notably, the two MAbs produced essentially identical staining patterns despite the distal locations of their cognate epitopes (Fig. 1). In each case, staining appeared in 10-min cocultures and was localized to Env cell interfaces in contact with target cells arrested by either cooling (data not shown) or cross-linking (Fig. 1). Approximately 6% of the attached Env cells demonstrated such staining. The MAbs exhibited the same binding patterns with a greater fraction of cells in cocultures arrested at 30 min (approximately 17% of the population), which is the time when membrane fusion events first appear (22). However, such staining was limited to attached cells that did not exhibit signs of cytoplasmic dye transfer. Notably, syncytia and actively fusing cells arrested in 60- or 120-min cocultures exhibited little or no staining with either MAb, although a few syncytia demonstrated a randomly localized, albeit weak, membrane staining pattern (Fig. 1). A minority of Env cells were attached to target cells without evidence of fusion at these later times and were reactive with the MAbs at contact interfaces. In control assays, the MAbs did not stain Env cells cocultured with CD4-negative HeLa cells at any time point (data not shown). Therefore, exposure of cluster I and cluster II epitopes required the expression of CD4 on the target cells.
To better determine whether these staining patterns were induced by envelope-receptor interactions, the MAbs were tested with Env cells in the presence and absence of sCD4. Flow cytometric analyses revealed that MAb reactivity with untreated Env cells was very low to negligible (Fig. 2A). Similar results were obtained by indirect immunofluorescence microscopy (data not shown). In contrast, Env cells treated with sCD4 for 60 min at 4°C were strongly reactive with both cluster I and cluster II MAbs as determined by flow cytometry (Fig. 2A) and indirect immunofluorescence microscopy (Fig. 2B). The same staining profile was obtained when the cells were fixed with BS3 after sCD4 treatment (data not shown). Such results are in strong agreement with previous reports (56-58) indicating that cluster I and cluster II epitopes are hidden within constrained metastable envelope structures (66) and become exposed in response to sCD4 binding to the HIV envelope.
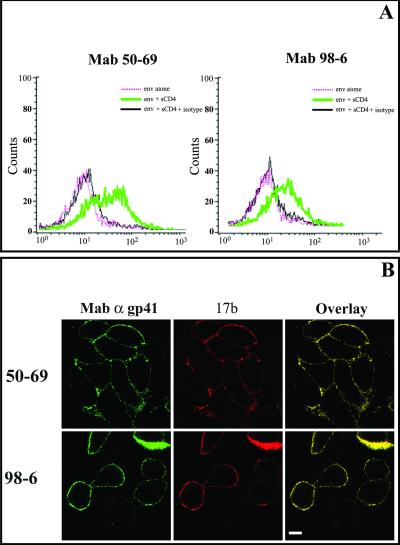
FIG. 2.
MAbs 50-69 and 98-6 bind to HIV envelope-expressing cells after treatment with sCD4. (A) Env cells were either left untreated or incubated with 2 μg of sCD4/ml in DMEM for 60 min at 4°C and were subsequently stained with MAb 50-69 (1.5 μg/ml) or 98-6 (1 μg/ml). Antibody staining was detected by PE-conjugated goat anti-human IgG. Human isotype control immunoglobulin was tested with treated cells (shown as an overlay on each histogram) for comparison. Control experiments repeated with untreated cells produced the same results as with treated cells. (B) Env cells were incubated with 2 μg of sCD4/ml in DMEM for 60 min at 4°C and subsequently double stained with the gp41 MAb 50-69 (1.5 μg/ml) or 98-6 (1 μg/ml) and an Alexa 594-labeled 17b Fab fragment (red). gp41 MAb staining was detected by an Alexa 488-conjugated goat anti-human secondary antibody (green). Superimposition of gp41 and 17b images produces a yellow stain indicating colocalized antibody binding. All antibodies were tested in parallel along with human isotype control immunoglobulin, which produced no detectable binding signal (data not shown). Images representative of at least three experiments are shown. Bar, 10 μm.
Given these results, we next attempted to characterize in greater detail the nature of the antigens recognized by MAbs 50-69 and 98-6. We first attempted to determine whether immunoreactivity early in the fusion process was due to CD4-induced conformational changes within intact envelope spikes or to gp120 shedding from gp41. sCD4-treated Env cells were double stained with either MAb 50-69 or MAb 98-6 combined with a Fab fragment of MAb 17b, which recognizes a CD4-induced epitope in the coreceptor binding domain of gp120 (Fig. 2B). Surface staining with the anti-gp41 MAbs and an Alexa 594-labeled 17b Fab was readily apparent and colocalized on CD4-treated Env cells (seen as yellow due to the overlap of green and red signals), indicating that gp120 was not noticeably dissociated from immunoreactive gp41 antigens as a result of envelope-CD4 interactions.
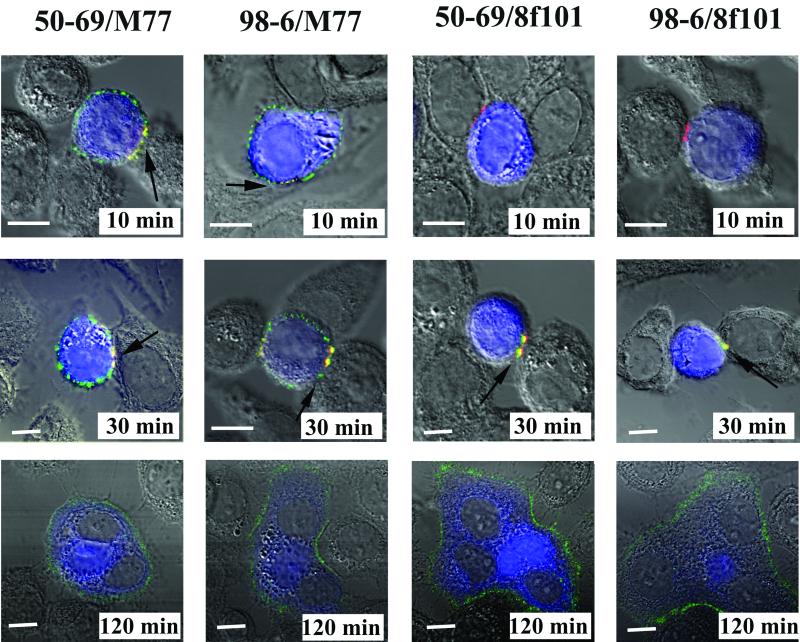
We next investigated the exposure of gp120 epitopes relative to the appearance of cluster I and cluster II epitopes during cell-cell fusion. MAb 17b was not suitable for these assays, because exposure of its cognate epitope is not observed during cell-cell fusion (22). Instead, two murine anti-gp120 MAbs were employed: M77, which binds to the HIVIIIB V3 loop (11, 13, 63), and MAb 8F101, which selectively stains gp120-CD4-coreceptor tricomplexes once they dissociate from gp41 (22). The murine anti-gp120 MAbs and human anti-gp41 MAbs were detected with secondary antibodies conjugated to Alexa 488 (green) and Alexa 594 (red), respectively. Accordingly, an alternative intracellular dye, CellTracker Blue, was used to visualize cytoplasmic mixing under these staining conditions. As shown in Fig. 3, both 50-69 and 98-6 staining colocalized with an M77 signal at the interfaces of attached Env and target cells in 10-min cocultures arrested by either BS3 fixation (Fig. 3) or rapid cooling to 4°C (data not shown). Unlike that of the anti-gp41 antibodies, MAb M77 staining was not concentrated at the cell contact interfaces but was instead evenly dispersed over the surfaces of target cells. Such results were in accordance with the analyses of sCD4-treated Env cells (Fig. 2). However, cells engaged in fusion at later times (i.e., 120 min) were stained only by M77 and not by the anti-gp41 MAbs (Fig. 3). In contrast, MAb 8F101 did not stain cells arrested earlier than 30 min in coculture, in agreement with previous results (22). However, 8F101 and gp41 MAb signals colocalized (seen as yellow due to the overlap of green and red signals) at fusing cell interfaces (Fig. 3) in 30-min cocultures. Fusing cells and syncytia arrested after more extended coculture times were stained only by 8F101 and not by the gp41 MAbs, except in rare instances where weak anti-gp41 reactivity was seen on a few syncytia.
FIG. 3.
gp41 structures that express cluster I and cluster II epitopes colocalize with gp120-CD4-coreceptor tricomplexes at contact interfaces during cell-cell fusion. CellTracker Blue-labeled Env cells were cocultivated with target cells for the indicated times as described in Materials and Methods. The cocultured cells were then fixed with BS3 and treated with the anti-cluster I MAb 50-69 (1. 5 μg/ml), the anti-cluster II MAb 98-6 (1 μg/ml), the anti-gp120 MAb M77 (5 μg/ml), and the anti-gp120-CD4-coreceptor tricomplex MAb 8F101 (5 μg/ml) as described in Materials and Methods. All MAbs were used at concentrations that produced optimum binding signals. Anti-gp41 MAb staining appears as a red signal; M77 or 8F101 binding appears as a green signal. Cell nuclei appear as gray areas in the stained syncytia. Fusion of Env and target cells produces the light blue cytoplasmic staining surrounding the nuclei. Arrows indicate the colocalization of MAbs at various time points. All antibodies were tested in parallel along with human isotype control immunoglobulin, which produced no binding signal (data not shown). Representative images are shown. Each experiment was repeated at least three times with the same results. Bars, 10 μm.
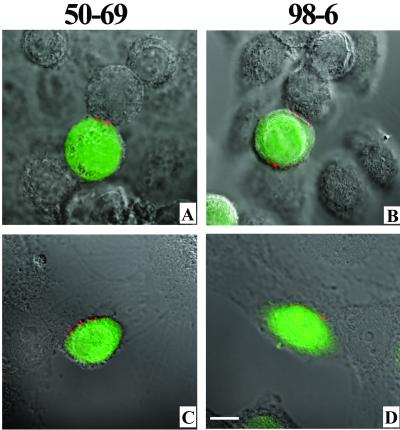
This staining pattern suggested that the antigens recognized by MAbs 50-69 and 98-6 were generated upon envelope-CD4 engagement. These results are consistent with some earlier findings (27, 47) but conflict with the findings of one study (36) in which conformational changes in the envelope protein were observed only if the appropriate coreceptor was expressed in conjunction with membrane-anchored CD4. To investigate this further, we repeated the fusion experiments, this time after treating the target cells with SDF-1 to prevent envelope interactions with CXCR4 (61). Analyses of the SDF-treated cells by immunofluorescence and flow cytometry confirmed that CXCR4 was down-regulated on the target cells, whereas CD4 expression levels remained unaltered (data not shown). As shown in Fig. 4A and B , SDF-1 clearly inhibited cell-cell fusion, as evidenced by the absence of apparent dye transfer or syncytium formation over a 120-min cocultivation period. However, anti-gp41 MAb binding was not influenced as a result of chemokine treatment. MAb binding was also observed at the interfaces of Env cells and CXCR4-negative U373 cells expressing only CD4 (Fig. 4C and D). Taken together, these results indicate that in the system used here, CD4 interactions at cell-cell interfaces are sufficient to generate the gp41 antigens recognized by anti-cluster I and anti-cluster II MAbs.
FIG. 4.
Exposure of cluster I and cluster II epitopes during cell-cell fusion is induced by gp120-CD4 complex formation. (A and B) CellTracker Green-labeled Env cells and target cells were pretreated with 5 μg of SDF-1/ml for 1 h at 37°C prior to cocultivation. The cells were then incubated for 120 min as described in Materials and Methods in the presence of 5 μg of SDF-1/ml. Interacting cells were stained with the indicated MAbs at 1.5 μg/ml (MAb 50-69) and 1 μg/ml (MAb 98-6). (C and D) CellTracker Green-labeled Env cells were cocultivated for 120 min with U373/CD4 target cells, which express CD4 in the absence of a coreceptor, as described in Materials and Methods. Interacting cells were stained with the indicated MAbs at 1.5 μg/ml (MAb 50-69) and 1 μg/ml (MAb 98-6). All antibodies were tested in parallel along with human isotype control immunoglobulin, which produced no binding signal (data not shown). Representative images are shown. Each experiment was repeated at least three times with the same results. Bar, 10 μm.
MAb binding to cluster I and cluster II epitopes does not inhibit cell-cell fusion.
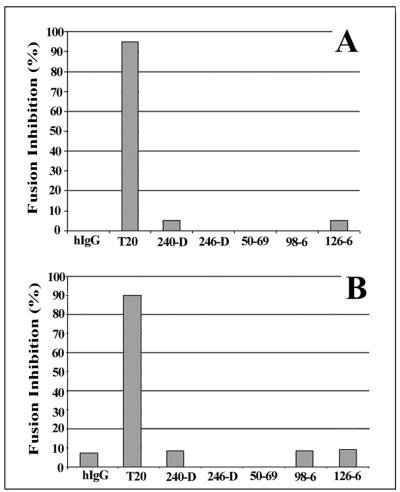
MAbs against cluster I and cluster II epitopes have been characterized as being either weakly neutralizing or noninhibitory in standard HIV infectivity assays (23, 41, 46, 55). However, it remained possible that the MAbs might be able to inhibit syncytium formation if allowed to react with the antigenic structures that appeared before membrane fusion began (Fig. 1 and 3). To explore this possibility in our system, the antigens were allowed to form on Env-target cell pairs for 10 min at 37°C (Fig. 1 and 3). The cocultures were then arrested prior to lipid mixing (47) by reducing the incubation temperature to 23°C in order to stop the fusion process. The arrested cultures were then reacted with the MAbs at 23°C. Reactivity with the temperature-arrested cells was confirmed by immunofluorescence analyses of the MAbs (data not shown). The temperature was then raised to 37°C so that fusion could proceed in the presence of the MAbs. As shown in Fig. 5A, under these conditions none of the MAbs inhibited cell-cell fusion, as measured by HIV Tat-driven β-galactosidase expression in the target cells. In contrast, T20 inhibited fusion as expected (26, 35, 37, 49, 67-69) when used at a molar concentration equivalent to the MAb concentrations. Such results demonstrated that prehairpin structures, which are the expected targets for T20 (7), were present on the arrested cells. Direct microscopic examination following 120 min of cocultivation confirmed the inability of the MAbs to suppress fusion. As in the untreated control assays, approximately 55% of the attached Env-target cell pairs fuse within 2 h (data not shown). In other experiments, the MAbs were added to cocultures that had been preincubated under temperature-arrested conditions for 120 min at 23°C. Previous studies using similar systems showed that such conditions permit the formation of prehairpin intermediates (47). However, after the temperature was raised to 37°C, no inhibition of fusion was observed, although T20 again potently blocked the formation of syncytia (data not shown). Thus, it appears that the binding of MAbs to their cognate antigens, either following 10 min of coculture or after prolonged culture at 23°C, does not adversely affect the fusogenic potential of the cells.
FIG. 5.
MAbs against cluster I and cluster II epitopes do not inhibit cell-cell fusion. (A) Env and target cells were cocultivated at 37°C for 10 min as described in Materials and Methods. Unbound target cells were removed by washing, and the remaining attached Env and target cells were temperature arrested at 23°C. The arrested cells were then incubated for 120 min at 23°C with MAb 240-D (55 nM), 246-D (79 nM), 50-69 (266 nM), 98-6 (66 nM), or 126-6 (84 nM). Normal human IgG (66 nM) and T20 (66 nM) were tested as controls. The treated cells were then returned to a fusogenic temperature (37°C) and cocultured for 15 to 18 h. (B) Env cells were incubated with 2 μg of sCD4/ml in DMEM for 60 min at 4°C. The treated Env cells were washed to remove unbound sCD4 and then incubated for 60 min at 37°C with the same MAbs and controls as for panel A. Target cells were then mixed with Env cells and cocultivated at 37°C. In both assay formats, cell-cell infection was determined after incubation for 18 h at 37°C by a quantitative β-galactosidase assay. Percent inhibition of fusion for each test condition was calculated relative to the level of fusion observed in control assays carried out in the absence of antibodies. Averages from triplicate assays are shown. The results shown are from a representative experiment repeated several times with the same results.
One possible explanation for these results was that the target antigens were shielded from antibody binding by the physical orientation of interacting Env-target cell membranes. To investigate this possibility, the assays were repeated, this time using Env cells that were treated with sCD4 for 60 min at 4°C to induce presentation of the immunoreactive antigen (Fig. 2). The triggered Env cells were then incubated with cluster I and cluster II MAbs for 1 h at 37°C and then cocultivated with target cells for 18 h at 37°C. As seen in Fig. 5B, the MAbs again failed to inhibit fusion, as determined by Tat-driven β-galactosidase expression in the target cells, whereas molar equivalent concentrations of T20 were strongly inhibitory. Direct microscopic observation of cell-cell fusion in 120-min cocultures confirmed that the MAbs had no discernible effect on syncytium formation. As in untreated control assays, approximately 40% of the attached target cells fused with the Env cells by 2 h in coculture (data not shown).
MAb 2F5 exhibits temperature-dependent binding to fusing cells.
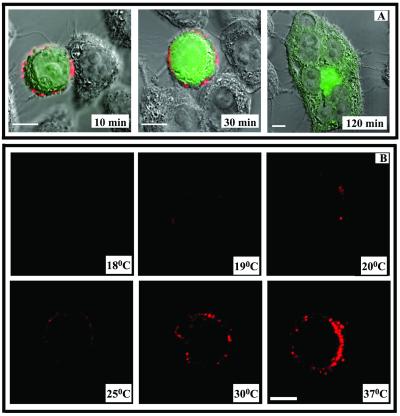
A second series of experiments was carried out to examine the binding of an anti-gp41 MAb (2F5) that is directed against an antigenic structure accessible within the HIV envelope spike (58, 75). MAb 2F5 was derived from an HIV-infected individual and is thought to react with a continuous epitope (amino acids 656 to 671) overlapping the C-terminal end of cluster II, proximal to the transmembrane domain of gp41 (50). As shown in Fig. 6A, MAb 2F5 reacted with attached Env cells in cocultures that were arrested by cross-linking. The MAb also reacted with free Env cells in the absence of target cells, in agreement with previous findings (34, 58, 75). In either case, the binding signal was distributed over the entire Env cell surface and was not noticeably altered at Env-target cell interfaces. However, the MAb did not react with fusing cells that exhibited cytoplasmic mixing (Fig. 6A, 120-min time point).
FIG. 6.
MAb 2F5 demonstrates temperature-dependent binding to Env cells. (A) CellTracker Green-labeled Env cells were cocultivated with target cells for the indicated times as described in Materials and Methods. Immunostaining was performed as described with MAb 2F5 (15 μg/ml) on BS3-fixed cells at 37°C. MAb 2F5 binding was tested in parallel along with a human isotype control immunoglobulin, which produced no binding signal (data not shown). Representative images from at least three separate experiments are shown. Bars, 10 μm. (B) Env cells were incubated with MAb 2F5 (15 μg/ml) for 30 min at various temperatures as described in Materials and Methods. Cells were then fixed, and MAb binding was detected with an Alexa 594-conjugated secondary antibody (red). MAb binding was tested in parallel along with a human isotype control immunoglobulin, which produced no binding signal (data not shown). Images representative of at least three experiments are shown. Bar, 10 μm.
Notably, a 2F5 signal was not detected on attached Env cells or syncytia under conditions where the cocultured cells were cooled to 4°C in order to arrest fusion (data not shown). One explanation for such findings was that MAb 2F5 binding to the Env cells was temperature dependent, in accordance with earlier work demonstrating poor MAb binding to virus-infected cells at 4°C (58). To further investigate this possibility, the MAb was incubated with unfixed Env cells at increasing temperatures ranging from 15 to 37°C. As shown in Fig. 6B, MAb 2F5 reactivity became evident at 19°C and increased steadily as the incubation temperature increased to 37°C. MAb reactivity was not detected at temperatures below 19°C. In contrast, a number of other anti-HIV envelope MAbs, including 2G12, M77, and IgG1B12, demonstrated strong reactivity with Env cells at all incubation temperatures ranging from 4 to 37°C (data not shown). Similarly, strong binding of MAb 98-6 to sCD4-treated Env cells was also observed at all of these temperatures.
DISCUSSION
HIV infection is inhibited in vitro and in vivo by peptides that bind to the prehairpin intermediates of gp41 (26, 35, 37, 49, 67-69). Therefore, it seems possible that antibody responses raised against gp41 intermediates could also neutralize HIV infection. However, a means to deliberately raise anti-gp41 neutralizing antibody responses has been elusive (12) and may not be realized without a more extensive understanding of the antigenic character of gp41 during cell-cell fusion. Accordingly, we used a panel of MAbs in combination with a previously characterized cell-cell fusion system (22) to analyze the antigenic changes in gp41 that occur during the process of syncytium formation.
Experiments with two MAbs, 50-69 and 98-6, demonstrated that cluster I and cluster II epitopes become exposed upon Env-target cell attachment, that such epitope exposure is confined to cell contact interfaces, and that the epitopes remain exposed until membrane fusion begins. We could eliminate the possibility that the localization of gp41 epitope exposure at cell contact sites was caused by envelope clustering because a gp120 epitope, M77, was not concentrated in the same manner (Fig. 3). If envelope clustering had occurred, MAb M77 would have exhibited its strongest reactivity at contact interfaces rather than showing the evenly dispersed binding that was observed (Fig. 3). Furthermore, another gp120-specific MAb, 2G12, exhibited the same staining pattern as M77 with no evidence of selective signal intensification at the Env-target cell interface (data not shown). We also could eliminate the possibility that the 50-69 and 98-6 epitopes were exposed by gp120 shedding from gp41 because the MAb M77 binding signal was detected at Env-target cell interfaces (Fig. 3). In agreement, a Fab fragment of MAb 17b, which recognizes a CD4-induced gp120 epitope, exhibited strong reactivity with sCD4-treated Env cells that colocalized with the anti-gp41 MAb signals (Fig. 2B). Taken together, the MAb binding data strongly suggested that cluster I and cluster II epitopes are exposed by a structural change in gp41 that is induced by target cell receptors.
The staining patterns of MAbs 50-69 and 98-6 also suggested that cluster I and cluster II epitopes are exposed on transitional structures that arise during the course of the fusion process. The prehairpin form of gp41 is one such structure that behaves in a manner consistent with the MAb binding data. Previous studies have shown that prehairpin structures with peptide-accessible HR domains appear in response to CD4-envelope interactions (26, 27, 36, 47) and persist on cell surfaces pending contact with coreceptors (27, 47, 49). In accordance, the antigens recognized by MAbs 50-69 and 98-6 were absent on free Env cells (Fig. 2) but became evident on Env cells exposed to CD4+ CXCR4− target cells (Fig. 4B), to SDF-treated target cells (Fig. 4A), or to sCD4 (Fig. 2B), in agreement with previous studies (56-58). Moreover, the antigen recognized by MAbs 50-69 and 98-6 appeared within 10 min of coculture (Fig. 1 and 3), prior to detection of the MAb 8F101 signal, which marks the formation of gp120-CD4-coreceptor tricomplexes (Fig. 3). Thus, the MAb binding data suggest a scenario in which cluster I and cluster II epitopes are exposed on the prehairpin intermediate of gp41. It should be noted that the appearance of prehairpin antigens in the absence of coreceptor interactions has not been universally observed in all fusion systems (26, 36). However, in the system used here, short-term Env-target cell cocultures (e.g., those arrested at 10 min) were also potently inhibited by T20 (Fig. 5). Such sensitivity indicated that prehairpin structures were present in the short-term cocultures prior to the formation of gp120-CD4-coreceptor tricomplexes.
This scenario is not necessarily at odds with the results of previous studies, which indicated that MAb 50-69 and 98-6 binding is dependent on the formation of coiled-coil structures. In peptide-based systems, MAb 50-69 bound only to N-terminal gp41 peptides that were associated with peptides derived from HR2 (30, 60). However, other experiments have suggested that the specific requirement for MAb binding to cluster I epitopes is a well-ordered (helical) and stabilized HR1 structure (8). In peptide-peptide systems such stability can be provided by HR1-HR2 interactions (8, 39, 60), although notably, 50-69 was able to react with a peptide known to form a helical structure (8, 42, 60) even in the absence of HR2-derived peptides (8). Therefore, it may be possible for MAb 50-69 to bind prehairpin antigens if the helical nature of HR1 is maintained during cell-cell fusion. Physical constraints such as envelope-receptor binding and/or cell-cell membrane bridging via gp41 could provide such stability. In the case of MAb 98-6, our results are in good agreement with those of peptide-based assays, which showed that binding to target antigens does not require the presence of HR1-derived peptides (30).
However, we cannot rule out the possibility that the 50-69 and 98-6 binding patterns reflect the exposure of cluster I and cluster II epitopes on other types of gp41 intermediates. A recent report indicated that six-helix bundles are formed before membrane mixing (28) under temperature-arrested conditions at 31.5°C. Such conditions rendered fusing cells less sensitive to inhibition by T20 but more sensitive to neutralization by rabbit antibodies against six-helix bundles (28). Based on these data, it might be expected that six-helix bundles were available for MAb 50-69 and 98-6 interactions in our system prior to cytoplasmic dye transfer. Conversely, six-helix bundles may not be antigenic outside of temperature-arrested conditions, since anti-bundle antibodies were not inhibitory at 37°C (28). A different study (47) indicated that six-helix-bundle formation occurs simultaneously with, and not before, membrane mixing. In this case, six-helix bundles could have been present only on the Env cells that exhibited cytoplasmic dye transfer and/or MAb 8F101 staining, which signaled the detachment of gp120-CD4-coreceptor tricomplexes from gp41. Since such events were only observed following 30 min in coculture (Fig. 3), the MAb binding we observed at earlier times could not have involved six-helix bundles. However, these structures were almost certainly present on some attached cells in the 30-min cocultures and thus potentially available to interact with the anti-gp41 MAbs at later times. Thus, the colocalized 50-69, 98-6, and 8F101 (Fig. 3) signals seen in 30-min cocultures might have arisen from six-helix bundles located in the vicinity of tricomplexes at fusing cell interfaces. An alternative, albeit speculative, possibility is that the cluster I and cluster II epitopes were exposed on novel intermediate envelope structures that have not yet been characterized. For example, the envelope trimer might assume different conformations according to how many of its three CD4 binding sites have been occupied. In the simplest scenario, one CD4 binding event would be sufficient to trigger a prehairpin structure, whereas three such events would be needed to complete the formation of a six-helix bundle. An envelope trimer occupied by only two CD4 receptors might represent an intermediate structure formed midway through the process. The evaluation of this possibility awaits the development of MAbs that distinguish envelope trimers according to the number of bound CD4 molecules.
Irrespective of how the cluster I and cluster II epitopes appeared, MAbs 50-69 and 98-6 did not recognize syncytia exhibiting extensive cytoplasmic mixing (Fig. 1 and 3). It is unlikely that this was caused by the diffusion of the HIV envelope over a larger surface area, since the signals produced by MAbs M77, 8F101 (Fig. 3), and 2G12 (data not shown) were clearly visible on the majority of syncytia formed in extended cocultures. A similar preservation of MAb A32 staining on syncytia was seen in our previous studies (22). Therefore, the HIV envelope persists on syncytium membranes at a density sufficient to allow detection by anti-envelope MAbs. It is more probable that the 50-69 and 98-6 signals were lost as a result of six-helix bundle core complexes binding directly to syncytium membranes, as was demonstrated in a previous study (39). Such perturbation of the six-helix bundle could significantly occlude and/or disrupt cluster I and II epitopes, which are determined by ordered HR1 and HR2 domains. Thus, the exposure of cluster I and cluster II epitopes might have been eliminated by the conversion of prehairpin antigens to six-helix bundles, which were then bound up by the membranes of syncytia.
It is noteworthy that MAbs against cluster I and cluster II epitopes fail to neutralize HIV infection (23, 41, 46, 55), even though they bind sequences in or near HR1 and HR2. It was previously speculated that the anti-cluster I and anti-cluster II MAbs are incapable of preventing fusion because their cognate epitopes are presented mainly on “dead spikes” of six-helix bundles (15) that appear only after membrane fusion has begun. This scenario now seems unlikely given our results showing that MAb binding clearly precedes, rather than follows, fusion (Fig. 1). Accordingly, we attempted to promote neutralization by mixing the MAbs with cells that were temperature arrested as soon as the cognate antigen was exposed. For example, 10-min cocultures were temperature arrested and, at the same low temperature, treated with the MAbs (Fig. 5A). Under these conditions, prehairpin structures form but do not fold into six-helix bundles (47). Thus, T20 strongly inhibited fusion when the system was returned to the fusogenic temperature of 37°C (Fig. 5A). In contrast, the MAbs failed to suppress fusion (Fig. 5A), even though they reacted strongly with the arrested cells (data not shown). The MAbs also failed to block the fusion of Env cells that were triggered by sCD4, which were not subject to any spatial constraints on antibody binding that might be introduced by cell-cell contact (Fig. 5B). Taken together, these results could indicate that the structures recognized by the MAbs are mainly “off-pathway” and do not fold into active fusogenic structures. Such antigens, if present in sufficient numbers, might even act as decoys to lure the MAbs away from fusion-competent gp41 structures. However, this model is difficult to reconcile with the disappearance of MAb reactivity upon the initiation of fusion. Alternatively, it is possible that our cell-cell fusion system is highly efficient and therefore inherently resistant to interference by certain anti-gp41 antibodies. Such resistance may apply only to antibodies against cluster I or cluster II epitopes, since experiments in which we preincubated Env cells with 1.6 μM MAb 2F5 resulted in 80% inhibition of fusion (data not shown). More sensitive and/or less robust fusion systems and conditions, such as those recently described (28), may be needed to reveal the neutralizing activities of antibodies directed against prehairpin intermediates or six-helix bundles.
As expected, MAb 2F5 (10, 17, 30, 62) behaved differently from MAbs 50-69 and 98-6 in that it reacted with antigens presented by Env cells before (Fig. 6B) and after (Fig. 6A) attachment to target cells. However, like the other anti-gp41 MAbs, 2F5 failed to bind syncytia (Fig. 6A), in agreement with previous evidence that the 2F5 epitope is occluded once HR1 and HR2 interact (30) to form six-helix bundles. It is possible that the 2F5 binding site is also lost when six-helix bundles associate with fusing cell membranes. In any case, the pattern of MAb 2F5 immunoreactivity was consistent with the concept that MAb 2F5 neutralizes HIV by blocking the formation of hairpin structures (30).
The exposure of the 2F5 epitope on Env cell antigens was remarkably temperature dependent and imperceptible below 19°C (Fig. 6B). Such results agree with previous analyses of MAb 2F5 binding to virus-infected cells, which showed antigen binding at 37°C but not at 4°C (59). Since increasing the incubation temperature from 19 to 37°C did not diminish the immunoreactivity of anti-gp120 MAb 2G12, M77, or IgG1B12 with the free Env cells (data not shown), it is unlikely that the 2F5 epitope was exposed by temperature-induced shedding of gp120. It seems more likely that a certain amount of flexibility or relaxation in the HIV envelope spike occurs at 19°C and facilitates 2F5 epitope exposure. This concept is consistent with the results of previous studies, which showed that sCD4-induced shedding of gp120 from RF virions increases markedly at temperatures above 20°C (48). Alternatively, the 2F5 epitope may possess key temperature-dependent conformational elements that have not yet been characterized. Although the 2F5-binding site was originally mapped to a linear epitope (ELDKWA) in the gp41 ectodomain (50), more-recent evidence also suggests that it may be considerably larger and more complex than expected (53).
Overall, these experiments have two important implications for HIV vaccine development. First, the data suggest that it may be difficult to design effective subunit vaccines based on prehairpin gp41 antigens and/or epitopes overlapping their HR domains. Although prehairpins, or related intermediates, appear to be antigenic at Env-target cell interfaces (Fig. 1 and 3), the more immunogenic (71) cluster I and cluster II epitopes (the ones most likely to elicit antibodies in response to immunization) are not targets for neutralizing antibodies. Second, the data suggest that the antigenic nature of the 2F5 epitope region may be more complex than that of a simple linear epitope. It is already apparent that the 2F5 core epitope does not easily generate neutralizing humoral responses (9, 14, 16, 18, 43, 72), even though it is recognized by a broadly cross-reactive neutralizing MAb. The temperature-dependent nature of MAb 2F5 could indicate that the immunogencity of the 2F5 region also requires labile structural elements in addition to the core epitope sequence. Some of these elements may reside within a larger putative 2F5 epitope that was recently identified (53). Therefore, it may be necessary to develop immunogens that incorporate additional structural elements of the HIV envelope along with the ELDKWA sequence in order to successfully generate broadly neutralizing antibody responses against gp41.
Acknowledgments
We thank Lai-Xi Wang for the synthesis of T20. We also thank Robert Blumenthal and W. Jonathan Lederer for helpful discussions.
This work was supported in part by grants NHLBI R01 03-5-20064, R21 03-5-21326, and PO1 03-5-21332 (project 2) to A.L.D.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 4.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 5.Cavacini, L. A., C. L. Emes, A. V. Wisnewski, J. Power, G. Lewis, D. Montefiori, and M. R. Posner. 1998. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res. Hum. Retrovir. 14:1271-1280. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 7.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. H., M. L. Greenberg, D. P. Bolognesi, and T. J. Matthews. 2000. Monoclonal antibodies that bind to the core of fusion-active glycoprotein 41. AIDS Res. Hum. Retrovir. 16:2037-2041. [DOI] [PubMed] [Google Scholar]
- 9.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 10.Conley, A. J., J. A. Kessler II, L. J. Boots, J. S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denisova, G. F., M. Zerwanitzer, D. A. Denisov, E. Spectorman, I. Mondor, Q. Sattentau, and J. M. Gershoni. 2000. Expansion of epitope cross-reactivity by anti-idiotype modulation of the primary humoral response. Mol. Immunol. 37:53-58. [DOI] [PubMed] [Google Scholar]
- 12.de Rosny, E., R. Vassell, P. T. Wingfield, C. T. Wild, and C. D. Weiss. 2001. Peptides corresponding to the heptad repeat motifs in the transmembrane protein (gp41) of human immunodeficiency virus type 1 elicit antibodies to receptor-activated conformations of the envelope glycoprotein. J. Virol 75:8859-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.di Marzo Veronese, F., R. Rahman, R. Pal, C. Boyer, J. Romano, V. S. Kalyanaraman, B. C. Nair, R. C. Gallo, and M. G. Sarngadharan. 1992. Delineation of immunoreactive, conserved regions in the external glycoprotein of the human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 8:1125-1132. [DOI] [PubMed] [Google Scholar]
- 14.Ding, J., Y. Lu, and Y. Chen. 2000. Candidate multi-epitope vaccines in aluminium adjuvant induce high levels of antibodies with predefined multi-epitope specificity against HIV-1. FEMS Immunol. Med. Microbiol. 29:123-127. [DOI] [PubMed] [Google Scholar]
- 15.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, X. N., Y. Xiao, and Y. H. Chen. 2001. ELNKWA-epitope specific antibodies induced by epitope-vaccine recognize ELDKWA- and other two neutralizing-resistant mutated epitopes on HIV-1 gp41. Immunol. Lett. 75:149-152. [DOI] [PubMed] [Google Scholar]
- 17.D'Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. AIDS Clinical Trials Group Antibody Selection Working Group. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 18.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 19.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 20.Felgenhauer, M., J. Kohl, and F. Ruker. 1990. Nucleotide sequences of the cDNAs encoding the V-regions of H- and L-chains of a human monoclonal antibody specific to HIV-1-gp41. Nucleic Acids Res. 18:4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, F., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 22.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forthal, D. N., G. Landucci, M. K. Gorny, S. Zolla-Pazner, and W. E. Robinson, Jr. 1995. Functional activities of 20 human immunodeficiency virus type 1 (HIV-1)-specific human monoclonal antibodies. AIDS Res. Hum. Retrovir. 11:1095-1099. [DOI] [PubMed] [Google Scholar]
- 24.Frey, S., M. Marsh, S. Gunther, A. Pelchen-Matthews, P. Stephens, S. Ortlepp, and T. Stegmann. 1995. Temperature dependence of cell-cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type 1. J. Virol. 69:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu, Y. K., T. K. Hart, Z. L. Jonak, and P. J. Bugelski. 1993. Physicochemical dissociation of CD4-mediated syncytium formation and shedding of human immunodeficiency virus type 1 gp120. J. Virol. 67:3818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion active conformation of gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 27.Gallo, S. A., A. Puri, and R. Blumenthal. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40:12231-12236. [DOI] [PubMed] [Google Scholar]
- 28.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 76:6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorny, M. K., V. Gianakakos, S. Sharpe, and S. Zolla-Pazner. 1989. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 74:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart, T. K., A. Truneh, and P. J. Bugelski. 1996. Characterization of CD4-gp120 activation intermediates during human immunodeficiency virus type 1 syncytium formation. AIDS Res. Hum. Retrovir. 12:1305-1313. [DOI] [PubMed] [Google Scholar]
- 32.Horal, P., B. Svennerholm, S. Jeansson, L. Rymo, W. W. Hall, and A. Vahlne. 1991. Continuous epitopes of the human immunodeficiency virus type 1 (HIV-1) transmembrane glycoprotein and reactivity of human sera to synthetic peptides representing various HIV-1 isolates. J. Virol. 65:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jernigan, K. M., R. Blumenthal, and A. Puri. 2000. Varying effects of temperature, Ca2+ and cytochalasin on fusion activity mediated by human immunodeficiency virus type 1 and type 2 glycoproteins. FEBS Lett. 474:246-251. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 36.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404-409. [DOI] [PubMed] [Google Scholar]
- 37.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 38.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kliger, Y., S. G. Peisajovich, R. Blumenthal, and Y. Shai. 2000. Membrane-induced conformational change during the activation of HIV-1 gp41. J. Mol. Biol. 301:905-914. [DOI] [PubMed] [Google Scholar]
- 40.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laal, S., S. Burda, M. K. Gorny, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1994. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J. Virol. 68:4001-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawless, M. K., S. Barney, K. I. Guthrie, T. B. Bucy, S. R. Petteway, Jr., and G. Merutka. 1996. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry 35:13697-13708. [DOI] [PubMed] [Google Scholar]
- 43.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 44.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 45.Malashkevich, V. N., B. J. Schneider, M. L. McNally, M. A. Milhollen, J. X. Pang, and P. S. Kim. 1999. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl. Acad. Sci. USA 96:2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDougal, J. S., M. S. Kennedy, S. L. Orloff, J. K. Nicholson, and T. J. Spira. 1996. Mechanisms of human immunodeficiency virus type 1 (HIV-1) neutralization: irreversible inactivation of infectivity by anti-HIV-1 antibody. J. Virol. 70:5236-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore, J. P., and P. J. Klasse. 1992. Thermodynamic and kinetic analysis of sCD4 binding to HIV-1 virions and of gp120 dissociation. AIDS Res. Hum. Retrovir. 8:443-450. [DOI] [PubMed] [Google Scholar]
- 49.Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nyambi, P. N., M. K. Gorny, L. Bastiani, G. van der Groen, C. Williams, and S. Zolla-Pazner. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nyambi, P. N., H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, A. Nadas, and S. Zolla-Pazner. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 74:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schulke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 55.Robinson, W. E., Jr., M. K. Gorny, J. Y. Xu, W. M. Mitchell, and S. Zolla-Pazner. 1991. Two immunodominant domains of gp41 bind antibodies which enhance human immunodeficiency virus type 1 infection in vitro. J. Virol. 65:4169-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sattentau, Q. J., and J. P. Moore. 1993. The role of CD4 in HIV binding and entry. Philos. Trans. R. Soc. Lond. B 342:59-66. [DOI] [PubMed] [Google Scholar]
- 58.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 59.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi, Y., S. Zolla-Pazner, Y. Xu, X. Zhang, S. Takeda, and T. Hattori. 2000. Human monoclonal antibody 98-6 reacts with the fusogenic form of gp41. Virology 273:333-340. [DOI] [PubMed] [Google Scholar]
- 61.Tarasova, N. I., R. H. Stauber, and C. J. Michejda. 1998. Spontaneous and ligand-induced trafficking of CXC-chemokine receptor 4. J. Biol. Chem. 273:15883-15886. [DOI] [PubMed] [Google Scholar]
- 62.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watkins, B. A., S. Buge, K. Aldrich, A. E. Davis, J. Robinson, M. S. Reitz, Jr., and M. Robert-Guroff. 1996. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J. Virol. 70:8431-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss, C. D., S. W. Barnett, N. Cacalano, N. Killeen, D. R. Littman, and J. M. White. 1996. Studies of HIV-1 envelope glycoprotein-mediated fusion using a simple fluorescence assay. AIDS 10:241-246. [DOI] [PubMed] [Google Scholar]
- 65.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 66.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 67.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retrovir. 9:1051-1053. [DOI] [PubMed] [Google Scholar]
- 68.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 71.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 72.Xiao, Y., Y. Zhao, Y. Lu, and Y. H. Chen. 2000. Epitope-vaccine induces high levels of ELDKWA-epitope-specific neutralizing antibody. Immunol. Investig. 29:41-50. [DOI] [PubMed] [Google Scholar]
- 73.Xu, J. Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65:4832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]