Abstract
Primary human embryo lung fibroblasts and adult diploid fibroblasts infected by the human cytomegalovirus (HCMV) display β-galactosidase (β-Gal) activity at neutral pH (senescence-associated β-Gal [SA-β-Gal] activity) and overexpression of the plasminogen activator inhibitor type 1 (PAI-1) gene, two widely recognized markers of the process designated premature cell senescence. This activity is higher when cells are serum starved for 48 h before infection, a process that speeds and facilitates HCMV infection but that is insufficient by itself to induce senescence. Fibroblasts infected by HCMV do not incorporate bromodeoxyuridine, a prerequisite for the formal definition of senescence. At the molecular level, cells infected by HCMV, beside the accumulation of large amounts of the cell cycle regulators p53 and pRb, the latter in its hyperphosphorylated form, display a strong induction of the cyclin-dependent kinase inhibitor (cdki) p16INK4a, a direct effector of the senescence phenotype in fibroblasts, and a decrease of the cdki p21CIP1/WAF. Finally, a replicative senescence state in the early phases of infection significantly increased the number of cells permissive to virus infection and enhanced HCMV replication. HCMV infection assays carried out in the presence of phosphonoformic acid, which inhibits the virus DNA polymerase and the expression of downstream genes, indicated that immediate-early and/or early (α) genes are sufficient for the induction of SA-β-Gal activity. When baculovirus vectors expressing HCMV IE1-72 or IE2-86 proteins were inoculated into fibroblasts, the increase of p16INK4a (observed predominantly with IE2-86) was similar to that observed with the whole virus, as was the induction of SA-β-Gal activity, suggesting that the viral IE2 gene leads infected cells into senescence. Altogether our results demonstrate for the first time that HCMV, after arresting the cell cycle and inhibiting apoptosis, triggers the cellular senescence program, probably through the p16INK4a and p53 pathways.
Human cytomegalovirus (HCMV), a member of the beta-herpesvirus family, is widespread among humans, where it establishes a persistent infection that causes disease and mortality in immunocompromised individuals (44). HCMV remains the major viral cause of birth defects in humans, with an incidence of about 1% of live births. Among these congenital infections, the incidence of severe neurological complications is estimated to be at least 10%. Additionally, epidemiological data and pathological studies suggest a link between HCMV infection and atherosclerosis.
Following infection of permissive cells, i.e., human embryo fibroblasts (HELF), HCMV gene expression occurs in three temporal phases, designated immediate-early (IE), early, and late (41). The most abundant IE transcripts arise from the major IE region in the unique long region of the genome: differential-splicing events give rise to the 72-kDa IE protein (IE1) and the 86-kDa IE gene product (IE2), plus other isomers that regulate genes that are expressed during the S phase of the cell cycle. Studies from many laboratories have shown that these proteins act as transcriptional regulators of both virus and host cellular genes, such as DNA polymerase α, dihydrofolate reductase, thymidylate synthase, and ribonucleotide reductase (for a review see references 18 and 41).
Replication of HCMV, which encodes its own replication apparatus, appears to be favored in the absence of host cellular DNA synthesis (17). Fibroblasts experimentally infected in vitro by HCMV are halted in both the G1-S and G2-M compartments, depending on the cell cycle phase where infection occurs (5, 15, 28, 35). This cell cycle control is mediated by several viral proteins, such as IE2 and UL69, using mechanisms that have been determined (9, 24, 30, 43, 60). HCMV demonstrates a rather unique cell cycle arrest, preceded by a virally mediated mitogenic response of infected cells, with the transient induction of c-fos, c-jun, and c-myc (3). It induces the expression of proliferation markers such as proliferating cellular nuclear antigen (PCNA) and topoisomerase II (15, 26) and cellular enzymes involved in DNA synthesis (22, 31, 33, 38), but cellular activation does not seem to lead to cell division (1, 30). However, elevated levels of both cyclin E and B and their associated kinase activities are induced following infection (4, 40, 47), suggesting that HCMV has evolved mechanisms that support viral DNA synthesis but disallow cellular DNA replication by uncoupling cell cycle progression from the DNA synthesis process.
A number of reports have shown that HCMV inhibits apoptosis. Browne et al. (7), investigated the effect of HCMV infection on cellular mRNA accumulation by gene chip technology and found that it significantly regulated at least 25 mRNAs encoding proapoptotic and antiapoptotic proteins. Moreover, two immediate-early HCMV genes, namely, UL36 and UL37, encode two proteins designated vICA and vMIA, respectively, that protect infected cells by apoptosis induced by a variety of stimuli (19, 51).
Cell cycle progression is the result of the interaction between cyclins and their cyclin-dependent kinases (cdks) and several inhibitory proteins, the corresponding cyclin-dependent kinase inhibitors (cdki) (27, 49, 56). Cyclin D and the corresponding cdk4 and -6 activities govern the early progression through the G1 phase, ultimately leading to phosphorylation of the retinoblastoma (Rb) protein (pRb) and release of the Rb-mediated transcriptional block of the E2F promoter at the late G1 phase. Cyclin E can subsequently be expressed and, in association with cdk2, seems to be necessary for the G1-S transition. Cell cycle arrest is achieved by the negative regulation of cdks, exerted either by generally classified antiproliferative signals or by the specific activities of cdki. The Cip/Kip family of cdki, including p21CIP1/WAF (p21), p27, and p57 nonselectively targets several cyclin-cdk complexes, while the INK4 family, which includes p16INK4a (p16), p15, p18, and p14ARF, specifically inhibits cdk4 and -6 activity.
Complex organisms have evolved at least two cellular mechanisms to suppress the proliferation of cells at risk for oncogenic transformation: apoptosis or programmed cell death and cellular senescence or the senescence response (8, 11, 36, 37, 52). Senescent cells show an altered morphology—they are larger than normal cells and flattened—and display multiple molecular changes, namely, elevated levels of extracellular matrix proteins, such as fibronectin, and secreted or intracellular proteases or protease regulators, such as collagenase 1, stromelysin 1, and tissue plasminogen activator and its inhibitor (plasminogen activator inhibitor type 1 [PAI-1]), calpain (11). However, the most critical feature of senescence is the imposed cell cycle arrest. Recent studies have shown that senescent cells express a β-galactosidase (β-Gal) activity at pH 6, differing from normally expressed lysosomal β-Gal activity, which is measured at pH 4 (14). The neutral β-Gal activity has since been called senescence-associated β-Gal (SA-β-Gal) activity and is regarded as a useful marker of senescence (11). At the molecular level, accumulation of increased levels of cdki, such as p21 and p16, seems to be a general prerequisite, but differential roles in the induction of senescence and differentiation in human fibroblasts are attributed to cdki (54). Recently it was shown that the single overexpression of either p21, p16, or p57KIP2 can, in different cell lines and cellular systems, activate senescence programs (29, 57, 59). These results are substantiated by the findings that several kinds of tumor cells have genetically altered genes coding for cdks and by the fact that restoration of these pathways inhibits cell growth and induces tumor regression (59).
The aim of the work presented here was to examine the events that take place at cellular levels after inhibition of the cell cycle progression and apoptosis pathway. We show that primary HELF and human adult diploid fibroblasts (HDF) 48 h postinfection (p.i.) display intense SA-β-Gal activity and increased PAI-1 mRNA expression. These phenotypic changes, typical of senescent cells (8, 11, 14), are accompanied by changes of molecular markers such as increase of p16 and p53 protein together with cessation of bromodeoxyuridine (BrdU) uptake. Last, treatment of infected cells with phosphonoformic acid (PFA) and the use of baculoviruses carrying the HCMV IE1 or IE2 gene demonstrated that the IE2 protein is primarily responsible for premature senescence induction following HCMV infection.
MATERIALS AND METHODS
Cell culture.
Primary low-passage HELF and HDF, obtained from Istituto Zooprofilattico Sperimentale (Brescia, Italy) were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS) at 37°C in a 5% CO2 atmosphere and used at passages 6 to 12.
Virus stock preparation and virus infection.
The HCMV AD169 strain, purchased from the American Type Culture Collection (ATCC VR-538), and the Toledo strain, kindly provided by Ed Mocarski, Stanford University, were prepared in our laboratory by infecting semiconfluent layers of HELF at a multiplicity of infection (MOI) of about 0.001 PFU per cell. Virus stocks were collected when cell lysis was almost complete. Cells were scraped, briefly sonicated, filtered through 0.45-μm-pore-size Millipore filters, and stored at −80°C. Virus titers were measured as described elsewhere (32). Stock solutions containing approximately 1 × 106 to 5 × 106 PFU/ml were used in all experiments. Mock-infecting medium was prepared from uninfected cells according to the same procedure used for the preparation of HCMV.
For infection experiments, confluent cells were trypsinized and plated in DMEM-10% FCS at about 80% confluence 24 h before infection. Alternatively, to synchronize cells in G0 before infection, freshly plated cells were grown for 72 h in DMEM-0.2% FCS. In both cases, virus adsorption was performed for 2 h at 37°C (time zero) at MOIs varying from 1 to 5. Virus inocula were then discarded and replaced with fresh DMEM-10% FCS or DMEM-0.2% FCS. Mock-infected control cultures were exposed to an equal volume of mock-infecting medium. For virus inactivation, virus stock aliquots and mock-infecting fluid were irradiated with one pulse of 0.6 J/cm2 from a UV linker (PBI International), as previously described (33). In some experiments PFA was added after virus inoculation at a final concentration of 200 μg/ml. At various time points after infection (see figure legends) cells were washed with warm phosphate-buffered saline (PBS) and either scraped for total protein extraction or fixed for histochemical assays.
β-Gal assay.
Levels of SA-β-Gal were determined essentially as described by Dimri et al. (14). Cells were washed in PBS and fixed for 3 to 5 min in 0.25% glutaraldehyde. After extensive washing in PBS, cells were incubated overnight at 37°C in a solution containing 40 mM citrate-Na2HPO4 buffer (pH 6.0), 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 150 mM NaCl, 2 mM MgCl2, and 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml. Staining was evident in 2 to 4 h and was maximal in 12 to 16 h.
Northern blot analysis.
Total RNA (15 μg) from either mock- or HCMV-infected cells was extracted at 24 and 48 h p.i. with Eurozol (Euroclone), fractionated on a 1% agarose-formaldehyde gel, and then blotted onto a nitrocellulose membrane (Hybond C-Super; Amersham). The filters were baked for 2 h at 80°C and prehybridized for 4 h at 42°C in 50% formamide-750 mM NaCl-48.5 mM Na2HPO4-5 mM EDTA (pH 7.4)-2× Denhardt's solution-0.1% sodium dodecyl sulfate (SDS)-50 μg of denatured salmon sperm DNA per ml. Hybridization was carried out at 42°C overnight with a denatured 32P-labeled probe (106 cpm/ml) corresponding to PAI-1, kindly provided by D. J. Loskutoff (La Jolla, Calif.). The filters were then washed twice for 30 min at room temperature with 2× SCC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS and twice for 30 min at 42°C with 0.5× SSC-0.1% SDS and exposed to film with an intensifying screen at −80°C. Northern blots were also analyzed by PhosphorImager (Molecular Dynamics) to verify normalization and to determine fold differences in signal between the various samples.
BrdU labeling and immunofluorescence.
For BrdU double-labeling assays, either mock- or HCMV-infected cells were serum starved in DMEM-0.2% FCS for 48 h, synchronized into S phase in DMEM-10% FCS for 24 h as evaluated by fluorescence-activated cell sorter analysis (data not shown), and pulsed with 50 μM BrdU (Sigma) for 1 h. Following a washing with PBS, cells were fixed with 3.7% paraformaldeyde for 30 min at room temperature and extensively washed with PBS. Nucleic acids were denatured with 2 N HCl for 1 h and neutralized for 20 min in 0.1 M Na2B4O7, pH 8.5. Cells were then permeabilized with 0.2% Triton X-100 in PBS and incubated with a sheep anti-BrdU antibody (Ab; Sigma) together with Abs against HCMV antigens, such as anti-IEA (Argene Biosoft), and mouse monoclonal Ab anti-UL44 (Goodwin Institute, Plantation, Fla.) for 1 h at room temperature. Anti-IEA recognizes both IE HCMV antigens, IE1 and IE2. Secondary Abs were a donkey anti-sheep Ab labeled with fluorescein isothiocyanate and a goat anti-mouse Ab labeled with Texas Red, both at 1:100 (Molecular Probes). A final incubation of 3 min in 0.1 μg of DAPI (4′,6′-diamidino-2-phenylindole) in PBS was performed before mounting coverslips onto glass slides with PBS-glycerol (1:9). Immunofluorescence analysis was performed on an Olympus 1X70 laser confocal microscope, and images were captured with Fluoview, version 2.0.
Immunohistochemical analysis.
Cells were briefly washed with PBS, fixed at room temperature for 10 min in 90% acetone-1% H2O2, permeabilized with 0.1% Triton X-100 in PBS on ice for 5 min, and finally incubated in 50% methanol-0.6% H2O2 in PBS for 30 min in the dark. Cells were then incubated with anti-IEA, anti-pp65 (Argene Biosoft), and anti-UL44, all in 1% bovine serum albumin. The immunoperoxidase reaction was developed with a streptavidin-biotin universal detection kit (Immunotech) and visualized with diaminobenzidine (Fluka) in PBS containing 0.1% H2O2.
Western blot analysis.
Cells were washed with PBS, and cell pellets were resuspended in SDS lysis buffer (125 mM Tris-HCl [pH 6.8], 10 mM dithiothreitol, 3% SDS) containing 1 mM phenylmethylsulfonyl fluoride plus pepstatin (1 μg/ml) and aprotinin and leupeptin (both 4 μg/ml). Lysed cells were briefly sonicated, boiled, and cleared by centrifugation (15,000 × g). Protein concentrations were evaluated with protein determination kit Dc (Bio-Rad Laboratories). Samples (20 to 40 μg per lane) were separated in denaturing SDS-8.5 or 12.5% polyacrylamide gels. Proteins were then transferred to Immobilon-P membranes (Millipore) with a semidry apparatus (Schleicher & Schuell). Membranes were blocked in 5% nonfat dry milk in 10 mM Tris-Cl (pH 7.5)-100 mM NaCl-0.1% Tween 20. Western blot analysis was conducted by using Abs against pRb, p53, p21, and p16 (all from Santa Cruz Biotechnology), pRbSer780 (Cell Signaling Technology), actin (Chemicon International), and anti-HCMV IEA, which recognizes the IE proteins (IE-1 and IE-2) of HCMV. Appropriate secondary Abs conjugated with horseradish peroxidase were used (Sigma), and the chemiluminescence reaction was visualized by enhanced chemiluminescence (Pierce Supersignal), according to the manufacturer's instructions. Densitometry was performed by scanning the radiographs and then analyzing the bands with Quantity One software (Bio-Rad).
Baculovirus expression vectors.
To construct recombinant baculovirus expressing HCMV IE1 and IE2 in mammalian cells, cDNAs encoding IE1-72 and IE2-86 were excised from pCDNA3IE72 and pCDNA3IE86 with SmaI and XbaI and cloned into the BaculoGold (Pharmingen) VL1392 baculovirus replacement vector. Recombinant baculoviruses were generated as described by the manufacturer (Pharmingen) and tested for ability to express IE1 and IE2 proteins by indirect immunofluorescence and Western blot analysis using IE1- and IE2-specific Abs. Baculoviruses were titrated on SF9 insect cells, and equivalent numbers of viral particles were used to infect mammalian cells.
Routinely, mammalian cells were infected at approximately 2,000 PFU/cell for 3 h. Insect media containing the baculovirus were then washed off and replaced with DMEM-10% FCS, and cells were incubated for various times.
Flow cytometry.
To determine the cell cycle profiles of virus-infected cultures, cells were fixed with 70% ethanol and stained with propidium iodide, according to standard procedures. Cells were analyzed with a FACScan flow cytometer (Becton Dickinson) equipped with an argon laser tuned at 488 nm for fluorescence excitation. Propidium iodide fluorescence was measured with a band-pass filter at 585/44 nm. Approximately 5 × 105 cells were analyzed for each sample. Computer statistical analysis and graphic representation were performed.
RESULTS
HCMV induces senescence in primary human fibroblasts.
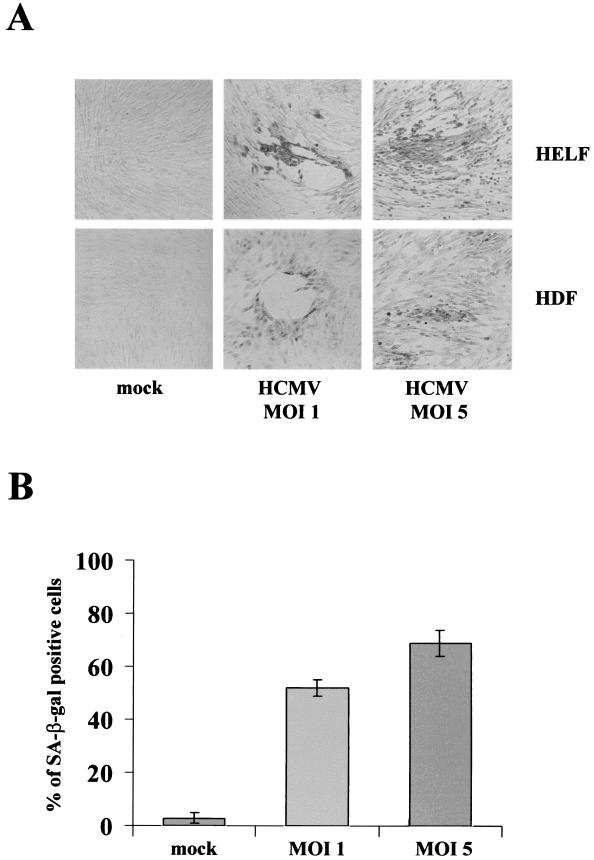
HCMV infection of permissive HELF has been reported to induce a cell cycle halt together with arrest of cell growth, enlarged cell volume, and decreased saturation density (17, 41, 44). These features are remarkably similar to those of cells that have surpassed their proliferative capacity and become senescent. To explore further the potential relationship between HCMV-induced cell cycle arrest and senescence, HELF were serum starved for 72 h, inoculated with HCMV strain AD169, and then analyzed for senescence-associated markers, namely, neutral β-Gal (SA-β-Gal at pH 6.0) and overexpression of the PAI-1 gene. SA-β-Gal activity was assessed at several times p.i. The percentage of SA-β-Gal-positive cells in HCMV-infected populations (MOI, 1) was initially low but increased at 48 h and reached 60% by 96 h, as shown in the representative pictures of Fig. 1A, top. At an MOI of 5, the highest intensity of SA-β-Gal staining was reached already at 72 h p.i. In contrast, less than 3% of the mock-infected cells were SA-β-Gal positive (Fig. 1A and B).
FIG. 1.
Senescent features of primary fibroblasts infected with HCMV. (A) Histochemical staining of primary fibroblasts infected with HCMV for SA-β-Gal activity. HELF and HDF were serum starved for 72 h, infected with HCMV, stained at 96 h, and photographed. All panels are shown at equal magnification. (B) Percentage of HELF positive for SA-β-Gal at 96 h p.i.. Cells (200) from each cell population were scored; the averages ± standard deviations of data from at least three separate experiments are shown. (C) Expression of PAI-1 in HELF infected with HCMV or mock infected at 24 h p.i.. The two transcripts of PAI-1 (3.2 and 2.2 kb in length) are indicated. (D) Confocal immunofluorescence assays demonstrating that HCMV-infected cells do not incorporate BrdU. After synchronization in the G0 phase by serum starvation, cells were infected with HCMV at an MOI of 1, stimulated with 10% FCS for 24 h, and then pulsed with BrdU for 1 h. Coverslips were processed for immunofluorescence as described in Materials and Methods. Double labeling consisted of a polyclonal anti-BrdU Ab coupled with monoclonal anti-HCMV Abs, either anti-IE antigens (a to c) or anti-UL44 (d to f). BrdU was decorated with a fluorescein isothiocyanate-conjugated secondary Ab (a and d; green), while a Texas-Red-conjugated secondary Ab was used to visualize anti-HCMV Abs (b and e; red). (c and f) Merged images obtained after confocal analysis. Magnification, ×20.
In addition, we analyzed the overexpression of PAI-1, whose upregulation during senescence has been reported (11), possibly due to the reorganization of the extracellular matrix proteins, and for this reason is often used as senescence marker. As shown in Fig. 1C, HCMV-infected HELF displayed a significant accumulation of PAI-1 mRNA at 24 h p.i., compared with mock-infected cells (2.8-fold for the large transcript and 2.5-fold for the small transcript). PAI-1 regulates cell-extracellular matrix interactions and is translated from two mRNAs, whose transcription is upregulated during senescence. These results, together with the strong induction of SA-β-Gal activity, indicate that the cell cycle arrest induced by HCMV infection may be identical to replicative senescence by every criterion tested.
To rule out the possibility that virus adhesion and entry were themselves sufficient to induce a cultural shock, cells were infected with a UV-inactivated stock of AD169 and tested for SA-β-Gal activity. No SA-β-Gal activity was observed (data not shown), indicating that viral adsorption is not sufficient per se to trigger the senescence program.
Earlier studies showed that HELF can sustain in vitro more population doublings than HDF, revealing either an intrinsically different susceptibility to in vitro growth or a different replicative life span (20, 21). Their susceptibilities to senescence induction by HCMV could thus be different. To find out if this is the case, HDF were analyzed for SA-β-Gal induction after HCMV infection in the experimental conditions described above. Consistent with the results obtained with HELF, 3 to 4 days after virus infection strong SA-β-Gal staining was observed in the HDF-infected cells, as shown in the representative pictures of Fig. 1A, bottom. As expected, no SA-β-Gal activity was detected in mock-infected cells. Together, these results show that normal human fibroblasts after HCMV challenge undergo a senescence program irrespective of their replicative life span.
HCMV strains differ in terms of gene organization, replication, and ability to cause disease. To exclude the possibility that the observed SA-β-Gal induction was a strain-specific event, HELF infected with the HCMV Toledo strain were assayed as described above. The results were similar to those obtained with the AD169 strain. An intense SA-β-Gal activity reaction 3 days p.i. showed that it is not strain specific but a general property of HCMV (data not shown).
One of the defining characteristics of replicative senescence is a stable, essentially irreversible arrest of cell proliferation with a G1 DNA content (8, 37). To prove this, a double BrdU-labeling and immunofluorescence procedure using anti-BrdU and anti-HCMV antibodies was performed and the results were analyzed by using a confocal microscope. Cells synchronized in G0 upon serum starvation for 48 h were infected at an MOI of 1 with HCMV, incubated in DMEM-10% FCS for 24 h, and pulsed with BrdU for 1 h. The cells were then fixed and simultaneously stained with anti-HCMV Abs and a mouse monoclonal anti-BrdU Ab to detect nuclei that had incorporated BrdU during the labeling interval. As shown in Fig. 1D, cells expressing both IEA and UL44, which displayed a bright green fluorescence, were completely negative, whereas cells not expressing them showed the typical BrdU staining of S-phase cells. A few cells displayed an irregularly shaped BrdU labeling that colocalized with UL44 (data not shown). We believe that this kind of BrdU staining is indicative of viral DNA replication compartments rather than of cellular DNA, as already suggested by Ahn et al. (2).
The phase of the cell cycle at the beginning of the infection influences the induction of the senescence program.
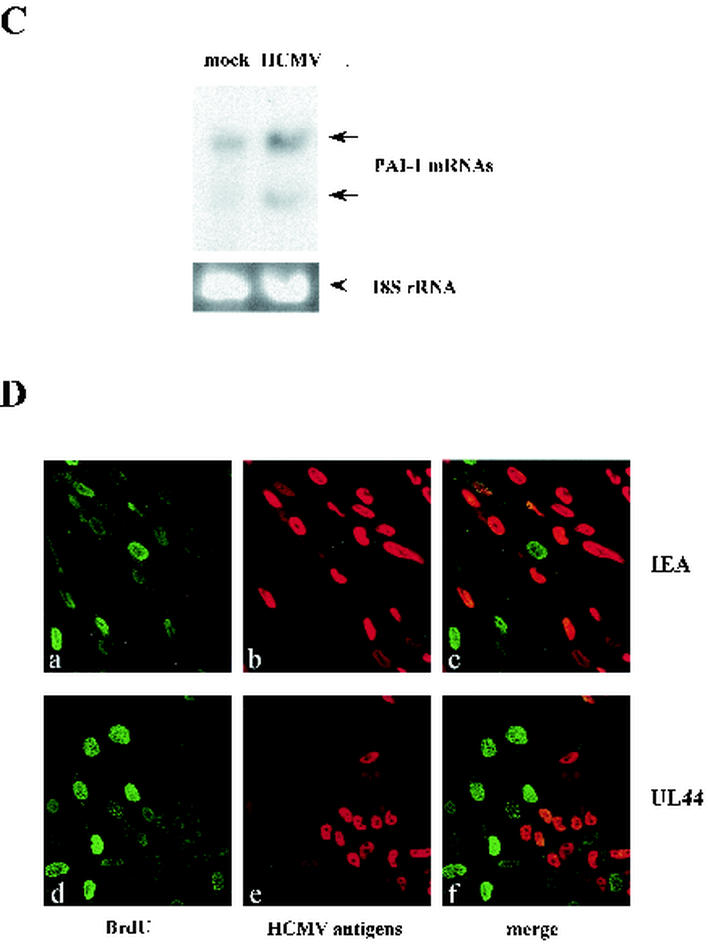
Cells infected in a quiescent state rapidly initiate viral protein synthesis and arrest before dividing. By contrast, when cells are infected near or during S phase, many cells are able to pass through S phase and undergo mitosis prior to cell cycle arrest (47). S-phase infection also produced a delay in the synthesis of IE and early proteins. Since a cardinal feature of senescence is the arrest of the cell cycle, we extended our observations by performing HCMV infections in different culture conditions: cells were (i) maintained and infected in DMEM-10% FCS, (ii) synchronized in G0 by serum starvation, infected with HCMV, and then stimulated with 10% FCS, or (iii) synchronized in G0 by serum starvation, infected with HCMV, and maintained in DMEM-0.2% FCS after infection. As expected an intense SA-β-Gal reaction was always detected, although with some variations, 3 to 4 days after infection (Fig. 2A). In cells synchronized before infection by serum starvation (columns 4 and 5), the reaction appeared earlier than in cells maintained in DMEM-10% FCS (column 3). This delay of SA-β-Gal induction in exponentially growing cells is consistent with the observation that HCMV infection during S phase significantly delays the onset of viral replication compared to viral infection of G1 cells (47).
FIG. 2.
Differential SA-β-Gal staining as a result of infection of cells at different phases of the cell cycle. (A) HELF maintained in different culture conditions were infected with HCMV (MOI, 1) and stained for SA-β-Gal activity at the indicated time points after infection. Column 1, mock-infected cells cultured in DMEM-10% FCS; column 2, mock-infected cells cultured in DMEM-0.2% FCS; column 3, HCMV-infected cells cultured in DMEM-10% FCS; column 4, cells synchronized in G0 by serum starvation, infected with HCMV, and then cultured in DMEM-10% FCS; column 5, cells maintained in DMEM-0.2% FCS after infection. For each type of infection, one representative flask of four was used for photography. Magnification, ×20. (B) Percentages of cells positive for SA-β-Gal at 96 h p.i.. Cells (200) from each cell population were scored; the averages ± standard deviations of data from at least three separate experiments are shown.
HCMV-infected HELF accumulate high levels of p16.
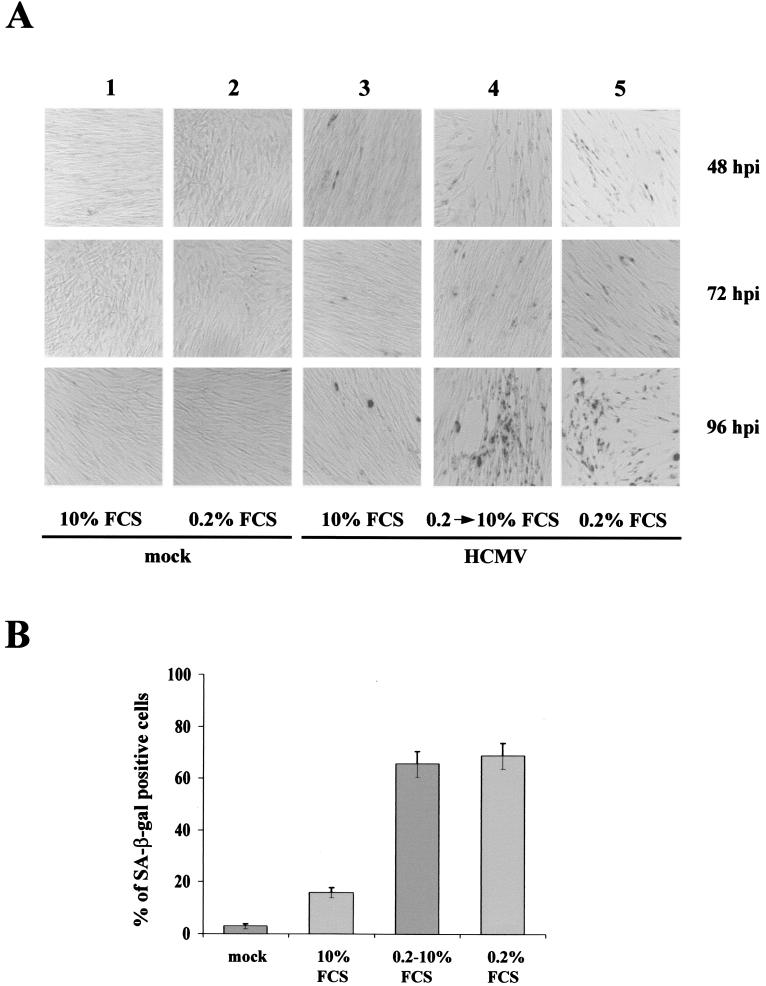
Cellular senescence is accompanied by a series of changes that, together, distinguish senescence from quiescence or differentiation (8, 11). These changes include altered expression of cell cycle proteins; upregulation of p53, p16, and p21; and the accumulation of SA-β-Gal. Earlier reports showed that by 24 h after HCMV infection the steady-state levels of p53 increase dramatically but its p21 and MDM2 targets are not activated (17, 42). This functional inactivation of p53 could be partly the result of its sequestration into viral replication centers in infected-cell nuclei. To define the molecular events underlying these effects of HCMV infection on proliferation, we assayed by Western blot analysis the steady-state levels of the tumor suppressor proteins p53 and pRb and the cdki p16 and p21 at different time points after infection or mock infection. In accord with previous reports, we also observed increases in p53 steady-state levels in HCMV-infected cells (42). Its levels were maximal at 48 h p.i. and then declined but were always higher in the infected cells (Fig. 3); highly phosphorylated pRb, accompanied by a notable increase in Rb steady-state levels, was induced at the same time point. Moreover, as reported by Chen et al. (12), the abundance of p21 declined over the next 48 h p.i. and remained low through 96 h p.i. compared to that in mock-infected cells. Interestingly, significant increases in the amount of p16 protein in HCMV-infected cells at 72 and 96 h p.i. (4.3- and 4.6-fold, respectively) were observed, while its levels in mock-infected cells remained relatively low. This protein may thus be critical for premature senescence induced by HCMV infection.
FIG. 3.
Effect of HCMV infection on cell cycle regulatory proteins. Immunoblots of cellular lysates corresponding to mock- and HCMV-infected HELF are shown. Cells were growth arrested by serum starvation and subsequently infected with HCMV (MOI, 3) or mock infected with conditioned medium containing 10% FCS as described in Materials and Methods. At different time points after infection, cell lysates were prepared, and equal amounts (30 μg) were separated by gel electrophoresis and transferred to Immobilon membranes. The proteins were then probed with anti-IE1/IE2, anti-pRb, anti-ppRb (phosphorylated form of pRb), anti-p53, anti-p16, and anti-p21 Abs. The membrane was then incubated with the appropriate secondary Ab conjugated with horseradish peroxidase and visualized with an ECL kit (Amersham). Actin immunodetection was performed as an internal control.
Infection of senescent cells enhances HCMV replication.
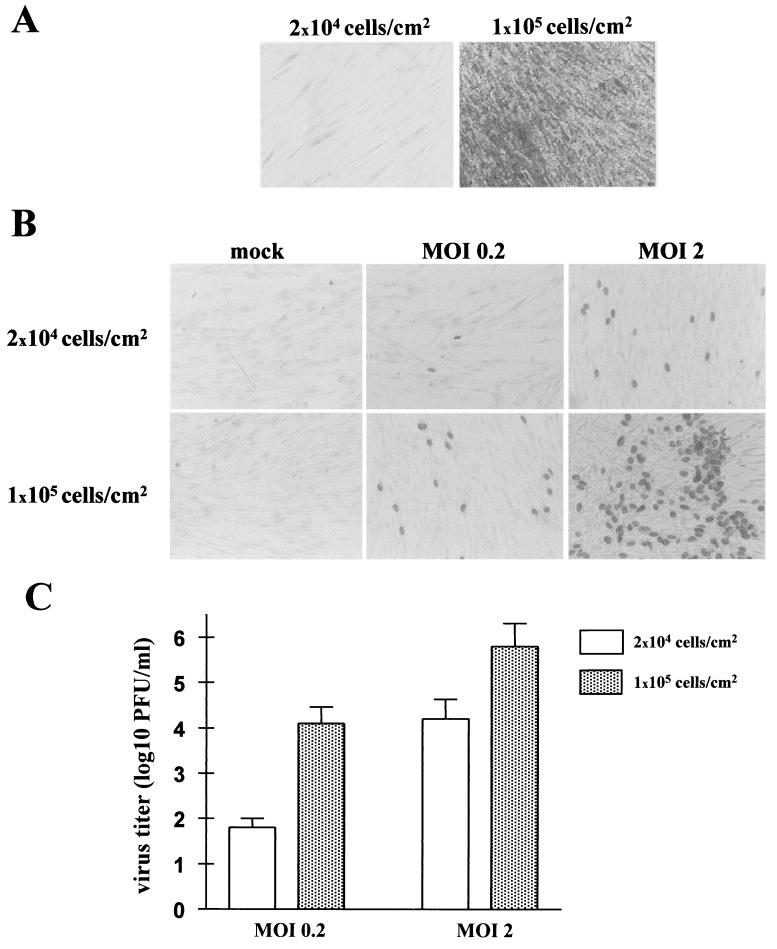
The extent to which HCMV infection is influenced by these changes was assessed by evaluating the ability of HCMV to grow in cells in which replicative senescence was artificially induced. HELF were seeded at low and high (2 × 104 and 1 × 105 cells/cm2) densities, serum starved for 72 h, and first evaluated for SA-β-Gal activity. As expected, the high-density and low-density cells were >90 and <3% positive, respectively (Fig. 4A), showing that the former undergo replicative senescence and can be used to illustrate the role of senescence in the early phases of HCMV infection. Thus, cells seeded at the two densities were serum starved for 72 h and infected at MOIs of 0.2 and 2, respectively. IEA staining of the low-density cells showed that the 1 or 2 positive cells per field at an MOI of 0.2 rose to 15 per field at an MOI of 2 (Fig. 4B). By contrast, high-density cells revealed 15 IEA-positive cells/field at an MOI of 0.2; this percentage strongly increased (≅50%) at an MOI of 2. Comparison of the virus yields demonstrated that at an MOI of 0.2 the titers of the senescent cells were 2.5 log units higher than those of the young cells (Fig. 4C). Interestingly, this difference decreased when cells were infected at an MOI of 2 (>1 log PFU), indicating that virus yield was not directly related to the smaller number of cells. It is nonetheless clear that replicative senescence at the start of infection benefits HCMV replication.
FIG. 4.
Infection of senescent cells enhances HCMV replication. (A) HELF seeded at both low (2 × 104 cells/cm2) and high (105 cells/cm2) densities were serum starved for 72 h and then stained for SA-β-Gal activity. (B) HELF were infected at an MOI of either 0.2 or 2 and, 24 h p.i., immunohistochemically stained for IE antigens with commercial anti-IEA Abs. Magnification, ×20. (C) HELF were infected at an MOI of either 0.2 or 2 and 8 days p.i. viral titers in the culture supernatants were evaluated and virus yields were plotted.
IE gene expression specifically induces a senescence phenotype in HELF cells.
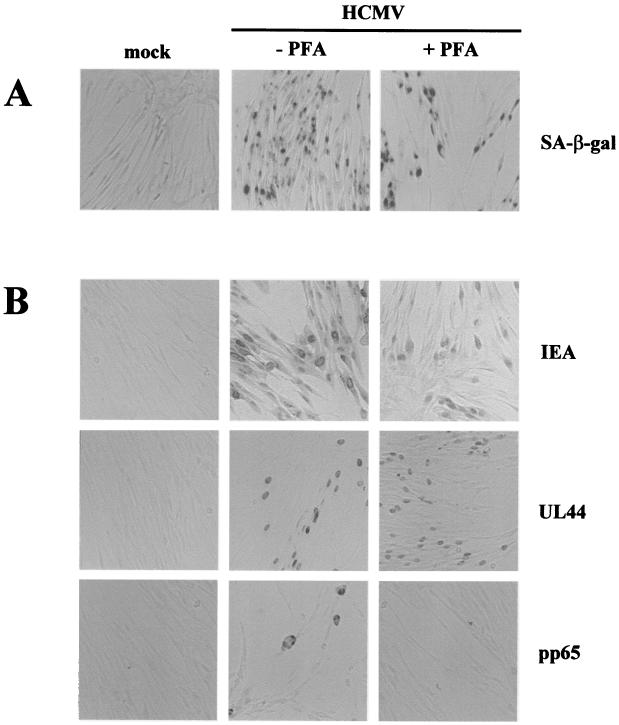
Kinetics experiments showed that senescence induction in infected cells may precede viral DNA replication. Moreover, the finding that UV-inactivated virus did not trigger the senescence program suggested that IE and/or early viral gene transcription may be responsible. SA-β-Gal induction in HCMV-infected cells treated with a viral DNA replication inhibitor was therefore assessed. Serum-starved HELF cells were infected with HCMV (MOI, 5) and maintained in the presence of the selective HCMV polymerase inhibitor PFA (58), which inhibits the expression of delayed early and late genes such as the pp65 gene. As expected, immunohistochemical staining for pp65 was negative in PFA-treated cells at 72 h p.i., whereas positive cells were observed in untreated cultures (Fig. 5B). By contrast, both IE1 and -2 and UL44 genes were expressed in infected cells at the same time point regardless of PFA treatment. Remarkably, SA-β-Gal staining was equally strong in both untreated and PFA-treated infected cells, indicating that the senescence program is evidently propelled by an early-phase mechanism (Fig. 5A).
FIG. 5.
IE and early (E) genes are per se sufficient to induce SA-β-Gal activity in infected cells. HELF were synchronized in G0 by serum starvation and treated with PFA immediately after HCMV infection (MOI, 5) or were not treated. (A) Histochemical staining for SA-β-Gal activity at 72 h p.i. (B) Immunohistochemical staining of IE (IE72 and IE86), E (UL44), and late (pp65) viral genes at 72 h p.i.. Magnification, ×20.
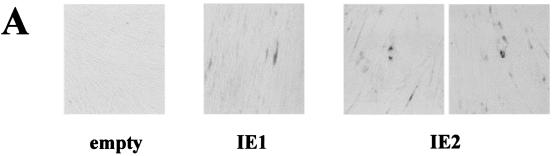
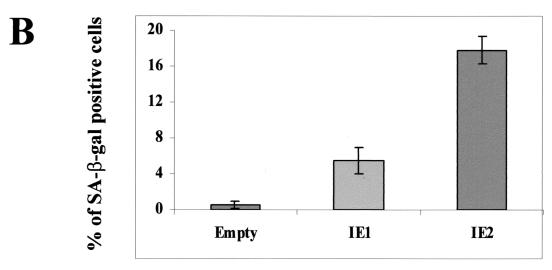
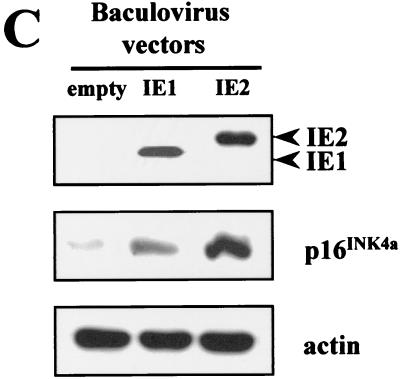
IE1-72 and IE2-86 proteins are widely thought to be the central regulators of the cascade of viral and cellular gene expression during lytic HCMV infection (41). The possibility that IE gene expression can mimic an HCMV-induced senescent state was therefore investigated. Since transfection of HELF cells is inefficient (less than 5%), we used recombinant baculovirus vectors that can transduce more than 45% of the HELF cells, as detected by immunohistochemical analysis with anti-IEA Abs (data not shown). Cells transduced with either bac-IE1 or bac-IE2 were stained for SA-β-Gal activity at different time points. The percentage of positive cells in bac-IE2-transduced cells was initially low (0.5%), started to increase at 48 h p.i. (6%), and peaked at 96 h p.i. (18%) (Fig. 6A). The induction pattern of SA-β-Gal activity in bac-IE1-transduced cells was similar to that in bac-IE2-transduced cells, though significantly lower (5 versus 18% at 96 h p.i.). In mock-transduced cells, positivity was always lower than 1% (Fig. 6A and B). Baculovirus-transduced cells expressed IE1 and IE2 at comparable levels at 48 h p.i., as measured by Western blotting analysis (Fig. 6C). Similar to the results obtained with HCMV, p16 expression in both cell populations was upregulated. Of note, IE2 induced a 4.9-fold-higher level of p16 expression than the empty vector, whereas IE1 induced a 2.3-fold-higher level. Thus, senescence induction through the upregulation of p16 expression seems to be at least in part mediated by HCMV IE2 protein expression.
FIG. 6.
Senescent features of the IE gene-transduced HELF. (A) SA-β-Gal activity in HELF transduced with empty vector and with IE1- or IE2-expressing baculoviruses at 96 h p.i.. Photographs are at the same magnification. (B) Percentages of cells positive for SA-β-Gal at 96 h p.i. Cells (200) from each cell population were scored; the averages and standard deviations of data from at least three separate experiments are shown. (C) Immunoblots of cellular lysates corresponding to cells transduced with empty vector or with IE1- or IE2-expressing baculoviruses at 48 h p.i.
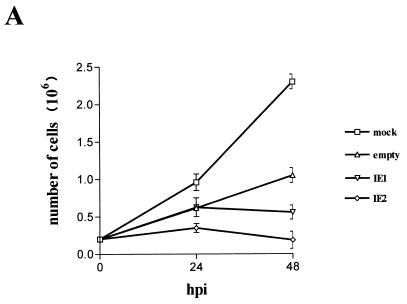
Since induction of cell senescence is strictly dependent on the arrest of cell growth and the cell cycle (8, 11, 37), a parallel investigation of the influence of bac-IE1 and bac-IE2 on both cell functions was also made. HELF were growth arrested by incubation in medium containing 0.5% FCS for approximately 72 h. They were then infected with bac-IE1, bac-IE2, or empty baculovirus with conditioned medium containing 10% FCS, and samples were collected at different times to determine the cell number and DNA content. By 48 h p.i., the number of cells in the empty-baculovirus-infected cultures had increased fourfold and approximately two rounds of cell division had been completed. The absence of a significant increase in the number of bac-IE2-transduced cells indicated that overexpression of the IE2 protein arrests cell growth (Fig. 7A). IE1 overexpression also affected cell proliferation, although to a lesser extent: by 48 h p.i. the cell population had completed only approximately one round of cell division.
FIG. 7.
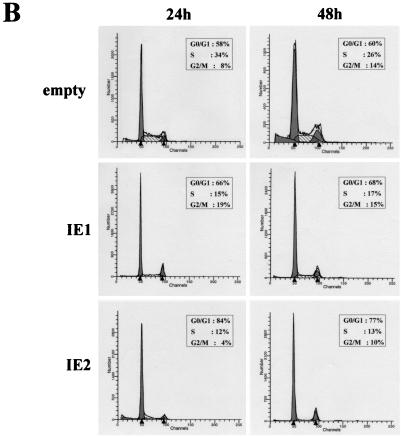
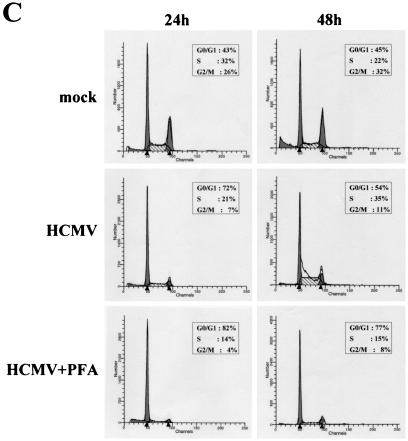
IE2 inhibits quiescent cells from entering S phase after serum restimulation. (A) Growth curve of HELF infected with IE1- or IE2-expressing baculoviruses. Cells were serum starved for 72 h, infected with IE1- or IE2-expressing baculoviruses or with empty baculovirus, and then stimulated with 10% FCS. Every 24 h p.i. cells were harvested and counted with a hemacytometer. The calculated densities versus the time points of harvest are shown. (B) Cell cycle distribution of IE1- or IE2-expressing baculoviruses at 24 and 48 h p.i. as measured by DNA content flow cytometry. Cells were serum starved for 72 h, infected with IE1- or IE2-expressing baculoviruses or with empty baculovirus, and then stimulated with 10% FCS. (C) Cell cycle distribution of HELF at 24 and 48 h after HCMV infection as measured by DNA content flow cytometry. Cells were serum starved for 72 h, infected with HCMV (MOI, 3) in the absence or presence of PFA at 200 μg/ml or mock infected, and then stimulated with 10% FCS.
To determine whether normal cell cycle progression is inhibited by IE1 and/or IE2 proteins, DNA content profiles were recorded from empty-bac-, bac-IE1-, and bac-IE2-infected cells after serum stimulation. The normal cell cycle distribution pattern of empty-bac-infected HELF is depicted in Fig. 7B. IE2 expression induced a dramatic change in this pattern. At 24 h p.i. the G1 population increased from 58% of cells infected with empty baculovirus to 84%, whereas S and G2/M populations were reduced from 34 to 12% and from 8 to 4%, respectively. No significant difference in this pattern of distribution was observed at 48 h p.i.. The cell cycle distribution upon transduction of the IE1 protein was similar, although the percentage of cells arrested at the G1/S phase was lower than that obtained with the IE2 protein. The cell cycle profile is therefore consistent with the impairment of cell growth observed in HELF transduced with bac-IE1 or bac-IE2. As expected, at 24 after serum stimulation and infection, HCMV-infected cells remained in the G0/G1 phase of the cell cycle, whereas mock-infected cells progressed to S and G2/M phases (Fig. 7C). At 48 h p.i. the DNA content in virus-infected cells started to increase, with a pattern similar to that for mock-infected cells. However, this increase at 48 h p.i. was completely blocked by PFA treatment, consistent with the interpretation that the DNA content at later times p.i. was of viral origin (Fig. 7C).
These results indicate that both HCMV IE proteins, especially IE2, can partially mimic the induction of the premature-senescence phenotype observed with HCMV, possibly through activation of the specific cdki p16 INK4a protein.
DISCUSSION
In the past few years it has become apparent that HCMV markedly dysregulates host cell functions (41, 44). As outlined in the introduction, HCMV arrests cells in G1 but at the same time induces several S-phase-promoting activities required for DNA synthesis (3, 5, 15, 28, 35). To create its replication factory, the virus must indeed activate the appropriate biosynthetic pathways for DNA synthesis (17, 18, 22, 31, 33, 41). Determining how the virus stimulates G0-arrested cells to enter an aborted cell cycle is important for understanding the biology of the virus-cell interaction.
Here we present data identifying HCMV as an inducer of a state that shares several features with that of classic premature senescence. This observation fits in well with HCMV physiology, since during viral infection the progression of human fibroblasts is blocked at multiple points, predominantly in G1 (5, 15, 28, 30, 35). HELF expressing viral IE genes, but not the bystander negative ones, become enlarged, acquire a flattened and irregular shape typical of senescent cells, and exhibit an increase of specific markers, such as SA-β-Gal activity and PAI-1 mRNA expression. Moreover, the onset of senescence in infected HELF is accompanied by a cellular DNA synthesis halt, as shown by lack of BrdU uptake.
The view that induction of the senescence program is of physiological significance during HCMV infection is supported by the following observations. (i) This program requires viral gene expression, since UV-inactivated virus did not induce senescence. (ii) Consistent with previous findings showing that infection of S-phase cells delays the expression of viral IE genes, we observed a delay of senescence onset when proliferating instead of quiescent cells were infected. (iii) HCMV gene products expressed during the initial 24 h of infection appear to mediate activation of the senescence program. (iv) Inhibition of delayed early and late gene expression by the selective HCMV DNA polymerase inhibitor, PFA, did not impair senescence induction. (v) Transduction of the HCMV IE2 is sufficient to stimulate HELF to undergo senescence. (vi) Premature senescence of HCMV-infected cells, following G1/S block, is an active process, since it requires fine tuning of cellular proteins, such as p16 and p53. (vii) Finally, a replicative senescence state in the early phases of infection significantly increased the number of cells permissive to virus infection and enhanced HCMV replication.
The term premature or replicative senescence describes the finite replicative capacity of somatic cells in culture, which eventually results in complete cessation of cellular division (8, 9, 25, 37). While cell cycle arrest marks the onset of senescence, it is not a necessary consequence thereof: additional signals activate and enforce the senescence program. Cellular senescence is indeed accompanied by a series of changes that combine to distinguish it from quiescence or differentiation (8, 9, 25, 37). These changes include a number of phenotypic traits: cells become enlarged, acquire a flattened and irregular shape, and exhibit increased acidic SA-β-Gal activity and PAI-1 mRNA expression. Senescence is also accompanied by a number of characteristic changes in protein expression. Among these, the upregulation of pRb in its hypophosphorylated form, p53, p16, and p21 appears to be functionally significant for the establishment and maintenance of the senescence state (13, 39, 48).
This report presents the first evidence of the induction of the p16 protein in HCMV-infected cells. Although we and others (17, 53) have detected increased steady-state levels of p53 protein after HCMV infection, this induction did not interfere with HCMV stimulation of Rb phosphorylation. Phosphorylation causes pRb to loose its growth-inhibitory properties and converts it into a state in which it is largely and perhaps totally inert. Moreover, HCMV can arrest cells in G1 (5, 15, 28, 35) but, at the same time, induces S-phase-promoting activities, such as increased cyclin E-associated kinase activity (4, 40, 47) and activation of PCNA (14), as well as other proliferation-coupled factors required for DNA synthesis (22, 31, 33, 38). Normally, these findings characterize a cell cycle state which lies beyond the restriction (R) point (45). The R point is the most important stage of growth control in mammalian cells and is typically activated during the G1 arrest imposed on cells by overexpression of cdki, such as p16 and p21. These findings are consistent with the view that HCMV-infected cells, although showing R point restriction characteristics, do not start cellular DNA replication. The virus may thus dissociate cell cycle progression from DNA synthesis and trigger the senescence program.
The present results do not clarify how p16 and p53 activate the senescence program upon HCMV infection. Lin et al. (34) have demonstrated that Ras is initially mitogenic but eventually induces premature senescence by involving p53 and p16 tumor suppressors. Thus, p53 and p16 act cooperatively to promote Ras-induced arrest, and disruption of either protein in rodent cells prevents arrest and is sufficient for transformation. In human cells, p53 and p16 also appear to cooperate to promote arrest, though inactivation of both pathways appears necessary to escape senescence (48). Consistent with these observations, U3T3MG glioblastoma cells, which are not subject to the replicative constraints of primary mammalian cells owing to mutations in the p16 and p53 pathways, do not undergo premature senescence upon HCMV infection (data not shown) and, although permissive to virus replication, display much lower virus yields (41).
The results obtained with the baculovirus vectors encoding either IE1 or IE2 proteins show that one of the genes responsible for senescence induction might be the IE2 gene. Its overexpression significantly increases staining for SA-β-Gal, causes the accumulation of HELF with a DNA content of 2 N, typical of cells arrested in G1 or G1/S phase, and arrests cell growth, together with strong induction of p16. Of note, the percentage of SA-β-Gal-positive cells upon bac-IE2 infection was always lower than that observed with HCMV-infected cells (18 versus 60%). In addition to IE2, other viral factors might contribute to the regulation of senescence induction in infected cells. During a 48-h time course after HCMV infection of HDF, several classes of genes, including interferon response genes and cell cycle regulators, were found to be upregulated or downregulated by threefold or greater at at least two consecutive time points (50, 61). Moreover, Tahara et al. (55) have shown increases in expression levels of interferon-inducible genes in senescent HDF. Finally, we have demonstrated that the interferon-inducible 204 gene is transcriptionally activated by mouse cytomegalovirus (MCMV) and is required for its replication (46). The 204 protein contains two pRb binding motifs and functions as a growth suppressor in sensitive cells by delaying G0/G1 progression into S phase. Inactivation of p204 by dominant-negative mutants deregulates the cell cycle and renders cells intrinsically resistant to MCMV replication. Altogether these observations suggest that in HCMV-infected cells progression of the cell cycle and consequently of the senescence process is the result of a complex interaction between viral and cellular proteins.
The IE86 protein is a strong, somewhat promiscuous transcriptional activator that interacts with a variety of basal (e.g., TATA binding protein, TFIID, and TAF 130) as well as promoter-specific transcription factors (e.g., SP1, CREB, and EGR1) (41, 44). Furthermore, IE2 can also bind both p53 (53) and pRb (23), potent regulators of cell cycle progression, and abolishes their ability to modulate transcription of artificial reporter genes. These findings, together with the observation that IE2 activates the cyclin E promoter (4), are consistent with the view that IE2 may be part of an HCMV activity that guides infected cells past the restriction point but at the same time prevents cells from entering S phase.
A variety of physiological stimuli can provoke a cell to enter senescence. Following extensive passage in culture (39) and exposure to oxidative damage (10) or activation of oncogenes (6, 48), primary cultures of mammalian cells enter into irreversible growth arrest and display the hallmarks of senescence. A few herpesviruses, such as herpes simplex virus type 1, HCMV, and Epstein-Barr virus elicit a cell cycle block and thereby actively prevent progression into S phase and cell growth (16). A number of reports have shown that apoptotic pathways are suppressed following CMV infection (7, 19, 51), leaving open the question about the fate of HCMV-infected cells. Our demonstration that upon infection HELF that halted at the G1/S or early S phase undergo senescence provides the first illustration of the events that take place after inhibition of the cell cycle.
Acknowledgments
We thank E. Mocarski and D. J. Loskutoff for reagents.
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro, Istituto Superiore di Sanità (PF AIDS), and MURST-CNR (Biotechnology Program L95/95 to S.L. and Program 40% to M.G.).
REFERENCES
- 1.AbuBakar, S., W. W. Au, M. S. Legator, and T. Albrecht. 1988. Induction of chromosomal aberration and mitotic arrest by cytomegalovirus in human cells. Environ. Mol. Mutagen. 12:409-420. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boldogh, I., S. AbuBakar, and T. Albrecht. 1990. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science 247:561-564. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., T. Albrecht, and E. A. Thompson. 1998. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J. Biol. Chem. 273:22075-22082. [DOI] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. P., W. Wei, and J. M. Sedivy. 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277:831-844. [DOI] [PubMed] [Google Scholar]
- 7.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular nRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campisi, J. 2001. Cellular senescence as a tumor suppressor gene. Trends Cell Biol. 11:S27-S31. [DOI] [PubMed] [Google Scholar]
- 9.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Q., A. Fischer, J. D. Reagan, L. J. Yan, and B. N. Ames. 1995. Oxidative DNA damage and senescence of human diploid fibroblast cells. Proc. Natl. Acad. Sci. USA 92:4337-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Q. M. 2000. Replicative senescence and oxidant-induced premature senescence. Beyond the control of cell cycle checkpoints. Ann. N. Y. Acad. Sci. 908:11-25. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z., E. Knutson, A. Kurosky, and T. Albrecht. 2001. Degradation of p21CIP1 in cells productively infected with human cytomegalovirus. J. Virol. 75:3613-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai, C. Y., and G. H. Enders. 2000. p16INK4a can initiate an autonomous senescence program. Oncogene 19:1613-1622. [DOI] [PubMed] [Google Scholar]
- 14.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubeli, O. Pereira-Smith, M. Peacocke, and J. Campisi. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortunato, E. A., A. K. McElroy, I. Sanchez, and D. H. Spector. 2000. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol. 8:111-119. [DOI] [PubMed] [Google Scholar]
- 18.Fortunato, E. A., and D. H. Spector. 1999. Regulation of human cytomegalovirus gene expression. Adv. Virus Res. 54:61-128. [DOI] [PubMed] [Google Scholar]
- 19.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J.-W. Han, R. J. Lutz, S. Watanabe, E. D. C. McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein, S., E. J. Moerman, J. S. Soeldner, R. E. Gleason, and D. M. Barnett. 1978. Chronologic and physiologic age affect replicative life-span of fibroblasts from diabetic, prediabetic, and normal donors. Science 199:781-782. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein, S., S. Murano, H. Benes, E. J. Moerman, R. A. Jones, R. Thweatt, R. J. Shmookler-Reis, and B. H. Howard. 1989. Studies on the molecular-genetic basis of replicative senescence in Werner syndrome and normal fibroblasts. Exp. Gerontol. 24:461-468. [DOI] [PubMed] [Google Scholar]
- 22.Gribaudo, G., L. Riera, D. Lembo, M. De Andrea, M. Gariglio, T. L. Rudge, L. F. Johnson, and S. Landolfo. 2000. Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J. Virol. 74:4979-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagemeier, C., R. Caswell, G. Hayhurst, J. Sinclair, and T. Kouzarides. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 13:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 26.Hirai, K., and Y. Watanabe. 1976. Induction of α type DNA polymerases in human cytomegalovirus-infected WI-38 cells. Biochim. Biophys. Acta 447:328-339. [DOI] [PubMed] [Google Scholar]
- 27.Ho, A., and S. F. Dowdy. 2002. Regulation of G1 cell-cycle progression by oncogenes and tumor suppressor genes. Curr. Opin. Genet. Dev. 12:47-52. [DOI] [PubMed] [Google Scholar]
- 28.Jault, F. M., J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagawa, S., T. Fujiwara, Y. Kadowaki, T. Fukazawa, R. Sok-Joo, J. A. Roth, and N. Tanaka. 1999. Overexpression of the p21sdi gene induces senescence-like state in human cancer cell: implication for senescence-directed molecular therapy for cancer. Cell Death Differ. 6:765-772. [DOI] [PubMed] [Google Scholar]
- 30.Kalejta, R. F., and T. Shenk. 2002. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 7:D295-D306. [DOI] [PubMed] [Google Scholar]
- 31.Lembo, D., A. Angeretti, M. Gariglio, and S. Landolfo. 1998. Murine cytomegalovirus induces expression and enzyme activity of dihydrofolate reductase in quiescent cells. J. Gen. Virol. 79:2803-2808. [DOI] [PubMed] [Google Scholar]
- 32.Lembo, D., G. Gribaudo, R. Cavallo, L. Riera, A. Angeretti, L. Hertel, and S. Landolfo. 1998. Human cytomegalovirus stimulates cellular dihydrofolate reductase activity in quiescent cells. Intervirology 42:30-36. [DOI] [PubMed] [Google Scholar]
- 33.Lembo, D., G. Gribaudo, A. Hofer, L. Riera, M. Cornaglia, A. Mondo, A. Angeretti, M. Gariglio, L. Thelander, and S. Landolfo. 2000. Expression of an altered ribonucleotide reductase activity associated with the replication of murine cytomegalovirus in quiescent fibroblasts. J. Virol. 74:11557-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundberg, A. S., W. C. Hahn, P. Gupta, and R. A. Weinberg. 2000. Genes involved in senescence and immortalization. Curr. Opin. Cell Biol. 12:705-709. [DOI] [PubMed] [Google Scholar]
- 37.Lundberg, A. S., and R. A. Wienberg. 1999. Control of cell cycle and apoptosis. Eur. J. Cancer 35:1886-1894. [DOI] [PubMed] [Google Scholar]
- 38.Margolis, M. J., S. Pajovic, E. L. Wong, M. Wade, R. Jupp, J. A. Nelson, and J. C. Azizkhan. 1995. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J. Virol. 69:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McConnell, B. B., M. Starborg, S. Brookes, and G. Peters. 1998. Inhibitors of cyclin-dependent kinases induces features of replicative senescence in early passage human diploid fibroblasts. Curr. Biol. 8:351-354. [DOI] [PubMed] [Google Scholar]
- 40.McElroy, A. K., R. S. Dwarakanath, and D. H. Spector. 2000. Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J. Virol. 74:4192-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mocarski, E. S., Jr., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin., B. Roizman, and S. E. Strauss (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 42.Muganda, P., R. Carrasco, and Q. Qian. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 68:8028-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 45.Planas-Silva, M. D., and R. A. Weinberg. 1997. The restriction point and control of cell proliferation. Curr. Opin. Cell Biol. 9:768-772. [DOI] [PubMed] [Google Scholar]
- 46.Rolle, S., M. De Andrea, D. Gioia, D. Lembo, L. Hertel, S. Landolfo, and M. Gariglio. 2001. The interferon-inducible 204 gene is transcriptionally activated by mouse cytomegalovirus and is required for its replication. Virology 286:249-255. [DOI] [PubMed] [Google Scholar]
- 47.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle disregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. L. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 49.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 50.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, J. R., and O. M. Pereira-Smith. 1996. Replicative senescence: implications for in vivo aging and tumor suppression. Science 273:63-67. [DOI] [PubMed] [Google Scholar]
- 53.Speir, E., R. Modali, E.-S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 54.Stein, G. H., L. F. Drullinger, A. Soulard, and V. Dulic. 1999. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol. Cell. Biol. 19:2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tahara, H., K. Kamada, E. Sato, N. Tsuyama, J.-K. Kim, H. Hara, K. Oda, and T. Ide. 1995. Increase in expression levels of interferon-inducible genes in senescent human diploid fibroblasts and in SV40-transformed human fibroblasts with extended lifespan. Oncogene 11:1125-1132. [PubMed] [Google Scholar]
- 56.Tapon, N., K. H. Moberg, and I. K. Hariharan. 2001. The coupling of cell growth to the cell cycle. Curr. Opin. Cell Biol. 13:731-737. [DOI] [PubMed] [Google Scholar]
- 57.Tsugu, A., K. Sakai, P. B. Dirks, S. Jung, R. Weksberg, Y. L. Fei, S. Mondal, S. Ivanchuk, C. Ackerley, P. A. Hamel, and J. T. Rutka. 2000. Expression of p57KIP2 potently blocks the growth of human astrocytomas and induces cell senescence. Am. J. Pathol. 157:919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyms, A. S., J. M. Davis, J. R. Clarke, and D. J. Jeffries. 1987. Synthesis of cytomegalovirus DNA is an antiviral target late in virus growth. J. Gen. Virol. 68:1563-1573. [DOI] [PubMed] [Google Scholar]
- 59.Vogt, P. K., and S. I. Reed (ed.). 1998. Cyclin-dependent kinase (CDK) inhibitors. Curr. Top. Microbiol. Immunol. 227:1-165.9479823 [Google Scholar]
- 60.Wiebush, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]