Abstract
The replication and transcription activator (RTA) of Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, a homologue of Epstein-Barr virus BRLF1 or Rta, is a strong transactivator and inducer of lytic replication. RTA acting alone can induce lytic replication of KSHV in infected cell lines that originated from primary effusion lymphomas, leading to virus production. During the lytic replication process, RTA activates many kinds of genes, including polyadenylated nuclear RNA, K8, K9 (vIRF), ORF57, and so on. We focused here on the mechanism of how RTA upregulates the K9 (vIRF) promoter and identified two independent cis-acting elements in the K9 (vIRF) promoter that responded to RTA. These elements were finally confined to the sequence 5′-TCTGGGACAGTC-3′ in responsive element (RE) I-2B and the sequence 5′-GTACTTAAAATA-3′ in RE IIC-2, both of which did not share sequence homology. Multiple factors bound specifically with these elements, and their binding was correlated with the RTA-responsive activity. Electrophoretic mobility shift assay with nuclear extract from infected cells and the N-terminal part of RTA expressed in Escherichia coli, however, did not show that RTA interacted directly with these elements, in contrast to the RTA responsive elements in the PAN/K12 promoter region, the ORF57/K8 promoter region. Thus, it was likely that RTA could transactivate several kinds of unique cis elements without directly binding to the responsive elements, probably through cooperation with other DNA-binding factors.
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV8), is found in Kaposi's sarcoma (7). The genomic sequence of KSHV has high similarity to that of Epstein-Barr virus (EBV), another human B-cell-tropic herpesvirus. Both viruses belong to the gammaherpesvirus group and are thought to be oncogenic DNA viruses. For example, EBV is thought to be a causative agent for nasopharyngeal carcinoma (NPC) (21), some types of B-cell lymphomas (21), and gastric tumors (22). KSHV is reported to be tightly linked with primary effusion lymphomas (PEL) (3) and multicentric Castleman's disease (33), as well as Kaposi's sarcoma (7, 31).
KSHV usually resides in the latent form in B cells, as does EBV, but certain stimuli caused by humoral factors such as gamma interferon or other cytokines induce lytic replication of KSHV (5, 28, 29), which leads to virus production followed by virus dissemination. Typically, some chemical agents such as 12-O-tetradecanoylphorbol-13-acetate (TPA), sodium n-butylate, and calcium ionophore strongly induce lytic replication of KSHV in KSHV-infected cell lines, including BCBL1 (29), BC1 (4), and BC3 (2). In these systems, the expression of immediate-early genes seems to be a key event for starting lytic replication. In KSHV replication, the replication and transcription activator (RTA), a homologue of EBV BRLF1/Rta, is reported to play an important role, and its expression is sufficient to induce KSHV lytic replication (14, 25, 34).
EBV BZLF1/Zta, a very important lytic inducer and transactivator of EBV, upregulates downstream genes, including itself and another immediate-early gene, BRLF1/Rta (1). Zta binds its recognition sequence, called ZRE (Zta responsive element), an AP1 recognition sequence-like element (8). On the other hand, BRLF1/Rta has both a DNA-binding and a non-DNA-binding mechanism for activating its target genes (20, 23, 27, 41, 42).
Other gamma-2 herpesviruses, such as murine herpesvirus 68 (MHV68, also called γHV) and herpesvirus saimiri, encode a homologue of RTA, which functions as a key lytic replication inducer (19, 35, 38, 40). A distinct feature of these homologues of animal gamma herpesviruses is that two types of RTA are generated, ORF50a and ORF50b; only the shorter form (ORF50b), which is translated just from the ORF50 open reading frame and is seen in both animal gammaherpesviruses and not in KSHV, is functional (19, 35). This short-form RTA homologue typically directs DNA-binding activity to activate target gene expression (19). The recognition sequence, reported as 5′-CCN9GG, is quite loose and is also recognized by the EBV Rta (15, 16, 17).
Although KSHV RTA is known to activate many kinds of viral genes, including those for the polyadenylated nuclear protein (PAN)/nut-1 (32), K8 (25, 26), K9 (vIRF) (10), ORF57 (26), ORF59 (26), K12 (6), and RTA itself (12, 30), it has remained unclear how RTA regulates their gene expression.
In the case of KSHV, a 60-amino-acid (aa) region at the N terminus that is translated from the first exon of upstream ORF49 is indispensable for the localization of RTA to the nucleus, where it functions (10). Recently, Song et al. reported an RTA responsive element (RRE) sequence in the PAN regulatory region. The sequence is also seen in K12 RRE (6) and is completely different from the CCN9GG consensus recognized by the herpesvirus saimiri, MHV68, and EBV Rta, although the sequence contains CCN8GG. They also reported that RTA binds to the sequence with quite high affinity (32). Lukac et al. showed that RTA bound with the common RRE seen both in the K8 and ORF57 promoter, under limited conditions (24).
Here, we identified two cis elements responding to RTA, termed RRE I-2B and RRE IIC-2 that were specifically activated by KSHV RTA in the K9 (vIRF) regulatory region. They did not share a common sequence and are also different from all of the cis elements reported so far. The responsive elements that we found were activated by RTA through a nondirect DNA-binding mechanism.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
RTA 412, which consists of 412 aa from the amino terminus of the full RTA (691 aa), was amplified by PCR by using the full-length RTA cDNA (10) as a template and the primers RTA-F (5′-CCGAATTCATGGCGCAAGATGACAAG-3′) and RTA-R (SacI; 5′-GGGAGCTCGGGTTGTCGGGAGAATC-3′). The fragment was cloned in pET21a+, termed pET21a+RTA412. The sequence of the fragment was confirmed by sequencing analysis.
pcDNA3.1(−)RTAMycHisB (described as pcDNA-ORF50-cDNA by Chen et al. [10]) was a pcDNA3.1(−)MycHisB (Invitrogen)-based KSHV RTA expression vector for mammalian cells and contained the full RTA coding region (691 aa), as described elsewhere (9, 10).
Reporter plasmids for identification of responsive elements to RTA were essentially based on pE1B Luc, in which a synthetic adenovirus E1B minimal TATA sequence was followed by a firefly luciferase gene in the pSP72 (Promega) backbone. Responsive element I-1 (RE I-1) and RE I-2 were amplified by PCR by using synthetic oligonucleotides: 5′-TAGAGGGGGTGGAAAATTCTC-3′ and 5′-TTCACTCATTTGAAAA-3′ for RE I-1 and 5′-GAAAAGGAAGCTATGTGGTTT-3′ and 5′-TAAGCGGGTTTTTTGCTAAAGCACTT-3′ for RE I-2. The amplified fragments were then treated with 10 U of T4 polymerase (New England Biolabs) and 5 U of T4 polynucleotide kinase (Takara) with four deoxyribonucleotides (dATP, dGTP, dCTP, and dTTP) and ATP, repectively, and inserted into the PvuII site of pE1B as described above. For RE I-3, two synthetic oligonucleotides (5′-GCTTAGGAGTTGGCTATAGGCGGGACCCTGCA-3′ and 5′-TGCAGGGTCCCGCCTATAGCCAACTCCTAAGC-3′) were annealed at room temperature after a heating step at 85°C for 10 min in 0.6 M NaCl.
A smaller region of RE I-2 and the RE II region were also generated with synthetic oligonucleotides (Sawady Technology, Tokyo, Japan) for further analyses (Fig. 1). The synthetic oligonucleotides with an XhoI site at the end were as follows: 5′-TCGAGTGAAAAGGAAGCTATGTGGTTTC-3′ and 5′-TCGAGAAACCACATAGCTTCCTTTTCAC-3′ for RE I-2A, 5′-TCGAGTTTCTGGGACAGTCTAAAAAAC-3′ and 5′-TCGAGTTTTTTAGACTGTCCCAGAAAC-3′ for RE I-2B, 5′-TCGAGAAGTGCTTTAGCAAAAAACCCGCTC-3′ and 5′-TCGAGAGCGGGTTTTTTGCTAAAGCACTTC-3′ for RE I-1C, 5′-TCGAGTCTGACATATCTTTTTTGGGTATGGTGGC-3′ and 5′-TCGAGCCACCATACCCAAAAAAGATATGTCAGAC-3′ for RE IIA, 5′-TCGAGTGGGTGGGGGTGGAGGGCGGCAGATTGCCTCAGAC-3′ and 5′-TCGAGTCTGAGGCAATCTGCCGCCCTCCACCCCCACCCAC-3′ for RE IIB, 5′-TCGAGCAGACCCTGCTTTGTATCCCGTACTTAAAATAGAAC-3′ and 5′-TCGAGTTCTATTTTAAGTACGGGATACAAAGCAGGGTCTGC-3′ for RE IIC, 5′-TCGAGCAGACCCTGCTTTGTATCCC-3′ and 5′-TCGAGGGATACAAAGCAGGGTCTGC-3′ for RE IIC-1, and 5′-TCGAGTATCCCGTACTTAAAATAGAAC-3′ and 5′-TCGAGTTCTATTTTAAGTACGGGATAC-3′ for RE IIC-2. Each combination was annealed at room temperature for 30 min after being heated at 85°C for 10 min as described above. The annealed oligonucleotides were precipitated with ethanol, dried, dissolved in distilled water, and then treated with T4 polynucleotide kinase and ATP to add phosphate to the 5′ end as described above. These fragments were then inserted into the XhoI site of pE1B Luc to prepare the RE reporter constructs. RE reporter constructs containing three to five concatenated RE fragments in the antisense orientation were used in the RE reporter gene assay (see below), since one copy of the RE occasionally did not show its responsibility to transactivator (36). Monomer constructs, however, for RE I-2B and RE IIC-2 were also prepared in the same way in order to verify whether such regions were suitable for electrophoretic mobility shift assays (EMSA) (see below).
FIG. 1.
Schematic representation of the RTA-responsive region of the K9 (vIRF) promoter. The positions of sequence fragments used in the present study with the upstream sequence of K9 (vIRF) are indicated. The numbers above the sequence indicate the positions of the nucleotides marked with a dot and are relative to the K9 mRNA start site in the lytic phase (10). Variously shaded bars such as RE I-1, RE I-2, and so on shows the regions described in the present study. The line under the boxes depicts the shorter fragments of each RE. Nucleotides surrounded by a dotted box show putative AP1 and/or SP1 recognition sites, as shown above the line. Previously identified responsive regions. RE I and RE II lie between deletion mutants D4 and D5 and between deletion mutants D8 and D9, respectively, as described elsewhere (10). The RE III region is shown in the hatched box.
The oligonucleotides 5′-TCGAGAATGGGTGGCTAACCTGTCCAAAATATGGGAAG-3′ and 5′-TCGAGCTTCCCATATTTTGGACAGGTTAGCCACCCATTC-3′ for PAN RRE (32) and 5′-TCGAGAGTGTAACAATAATGTTCCCACGGC-3′ and 5′-TCGAGCCGTGGGAACATTATTGTTACACTC-3′ for 57/K8 RRE (24) were synthesized. They were annealed and kinase treated at the 5′ end as described above. They were then cloned in the XhoI site of pE1B Luc as described above. All clones were confirmed by sequencing.
Mutant versions of RE I-2B and RE IIC-2 were also constructed with chemically synthesized oligonucleotides by the same methods given above. The oligonucleotides were as follows: 5′-TCGAGGTTCTCAGGACAGTCTAAAAAC-3′ and 5′-TCGAGTTTTTAGACTGTCCTGAGAACC-3′ for RE I-2Bm1, 5′-TCGAGGTTTCTGAAGGAGTCTAAAAAC-3′ and 5′-TCGAGTTTTTAGACTCCTTCAGAAACC-3′ for RE I-2Bm2, 5′-TCGAGGTTTCTGGGACGACTTAAAAAC-3′ and 5′-TCGAGTTTTTAAGTCGTCCCAGAAACC-3′ for RE I-2Bm3, 5′-TCGAGGTTTCTGGGACAGTCCGGGAAC-3′ and 5′-TCGAGTTCCCGGACTGTCCCAGAAACC-3′ for RE I-2Bm4, 5′-TCGAGTATCCCTGCTTTAAAATAGAAC-3′ and 5′-TCGAGTTCTATTTTAAAGCAGGGATAC-3′ for RE IIC-2m1, 5′-TCGAGTATCCCGTACCCGGAATAGAAC-3′ and 5′-TCGAGTTCTATTCCGGGTACGGGATAC-3′ for RE IIC-2m2, and 5′-TCGAGTATCCCGTACTTAAGGCGGAAC-3′ and 5′-TCGAGTTCCGCCTTAAGTACGGGATAC-3′ for RE IIC-2m3. The underlining indicates the mutated nucleotides, and the locations of all fragments used for the reporter constructs are summarized in Fig. 1.
Cells.
All cell lines used in this experiment were maintained in 5% CO2 in a humidified atmosphere (CO2 incubator). A KSHV- but not EBV-infected cell line, BCBL1 (29), was cultured in RPMI containing 20 μg of gentamicin/ml and 10% fetal bovine serum (FBS). The BJAB cell line, which is infected with neither KSHV nor EBV, was grown in the same medium as BCBL1. Adenovirus-transformed human embryonic kidney fibroblast 293 cells and its derivative, 293L (13), were cultured in Dulbecco modified Eagle medium supplemented with 100 μg of streptomycin/ml, 100 U of penicillin G/ml, and 10% FBS.
For TPA (Sigma) induction of these cells, 2 × 106 cells were induced with 25 ng of TPA/ml.
Expression of ORF50 protein in Escherichia coli.
pET21a+RTA412 was introduced into E. coli strain Origami B DE3 (Novagen). The transformed E. coli was grown in 1 liter of Luria broth at 30°C to an optical density at 600 nm of ca. 0.6 to 0.7. The cells were then harvested by centrifugation at 4,000 × g for 10 min at 4°C and washed twice with and suspended in 50 ml of a resuspension buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 10 mM imidazole). Lysozyme (Nacalai tesque) was added at 1 mg/ml, and the cells were incubated for 30 min on ice. The sample was then sonicated on ice six times at 250 W for 10 s with 10-s intervals. After the lysates were spun at 10,000 × g for 60 min at 4°C to eliminate cell debris, the cleared lysate was collected and passed through a 1-ml nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) column, washed with 5 ml of wash buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 20 mM imidazole) twice. The bound materials were eluted with 3 ml of elution buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 250 mM imidazole), and 0.5-ml fractions were collected. Then, 5 μl of each fraction was subjected to sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with Coomassie brilliant blue (R250; Sigma) and Western blotting analysis with an anti-T7 tag mouse monoclonal antibody (Novagen) or rabbit polyclonal anti-His hexamer antibodies (Santa Cruz Biotechnology, Inc.). This procedure confirmed that the target protein was expressed and that it bound to the Ni-NTA column; the fraction enriched with the target protein was then collected and concentrated with a Centricon 50 filter (Amicon), with the buffer being changed to the resuspension solution.
The concentrated protein with Ni-NTA agarose was further purified with T7 tag antibody agarose (Novagen) according to the manufacturer's orientation. The finally purified protein as described above was quantified with BCA protein assay reagent (Pierce) according to the manufacturer's protocol.
Transfection and luciferase assay.
For the reporter gene assay, 1 μg of pcDNA3.1(−)MycHisB or pcDNA3.1(−)RTAMycHisB was transfected as the effector plasmid into 3 × 105 293L cells per well (wells were 3 cm in diameter; Iwaki Glass). The cells were prepared 1 day before the transfection, which was performed by using the Superfect transfection reagent (Qiagen), according to the manufacturer's protocol. Each RE E1B Luc reporter plasmid (see Fig. 1) was transfected at 0.1 μg per well. Each transfection assay was done in triplicate, and the mean value and standard deviation were calculated. To normalize the transfection efficiency, 0.1 μg of plasmid pCMVβ (Clontech) per well was also transfected.
For the transfection of BJAB and BCBL1 cells, 5 × 106 cells were washed twice with RPMI without either FBS or antibiotics and transferred to a cuvette with a 0.4-cm polar distance after it was resuspended in 0.4 ml of the same medium. Then, 10 μg of the effector plasmids [pcDNA3.1(−)MycHisB or pcDNA3.1(−)RTAMycHisB] and 1 μg of each RE E1B Luc reporter plasmid were transfected with electroporation (Bio-Rad) at 0.25 kV and 975 μF. One microgram of pCMVβ was cotransfected for normalization. To test the inducibility by TPA, BJAB and BCBL1 cells were transfected with the same amount of reporter constructs under the same conditions. In this experiment, the cells were accurately divided into two wells at 1 day posttransfection, and one of the duplicate wells was treated with TPA (25 ng/ml).
In both cases, cells were harvested 2 days posttransfection and washed with phosphate-buffered saline (PBS). Cells were then lysed in 50 μl of cell lysis buffer (PGC50, Toyo Ink, Tokyo) and spun at 10,000 × g for 10 min at 4°C to obtain cleared lysate. One-fifth of the total cell lysate (∼10 μl) was used to measure luciferase activity. Reaction buffer (50 μl) containing substrate (Picagene LT 7.5 for the 293L cell line and Picagene for the PEL cell line; Toyo Ink) was mixed with the lysate, and the emitted light in relative light units was measured with a Lumat 2000. β-Galactosidase activity was measured colorimetrically with 15 mM chlorophenol red-β-d-galactopyranoside (CPRG; Roche Diagnostics) in Z buffer (0.1 M sodium phosphate, pH 7.5; 10 mM KCl; 1 mM MgSO4; 50 mM 2-mercaptoethanol) by using a Benchmark microplate reader (Bio-Rad) at 570 nm.
The activity of the RE in the presence of RTA is shown as the factor (fold) of the increase or decrease relative to the activity in the absence of RTA with standard deviation.
Western blotting analysis.
BCBL1 cells (2 × 106) were cultured in the presence of TPA (25 ng/ml) and sampled at 0, 2, 4, 8, 12, 24, 48, and 72 h postinduction. Nuclear extract was prepared in 30 μl of buffer C (5 mM HEPES, pH 7.9; 26% glycerol [vol/vol]; 1.5 mM MgCl2; 0.2 mM EDTA; 0.5 mM dithiothreitol [DTT]; 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) (see below). Then, 20 μg of each sample was subjected to SDS-7.5% PAGE, followed by electroblotting (Immun-Blot; Bio-Rad). A mouse monoclonal anti-RTA antibody (α50A) and a horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) were used to detect RTA by using SuperSignal (Pierce) reagents according to the manufacturer's direction.
EMSA.
Nuclear extract (NE) was prepared from BCBL1 cells that were induced with TPA (25 ng/ml) for 24 h or uninduced. After the induction, the cells (total of 108) were harvested and spun at 250 × g at 4°C for 10 min. The medium was withdrawn and the cells were washed with PBS and spun at 250 × g at 4°C for 10 min. The cells were then resuspended in 5 volumes of buffer A (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 0.5 mM DTT; 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). After incubation on ice for 10 min, the cells were spun at 250 × g for 10 min at 4°C and resuspended in 3 volumes of buffer A containing 0.05% Nonidet P-40 (NP-40). The cells were then homogenized on ice with 25 strokes in a Dounce homogenizer. Successful release of the nuclei was confirmed by phase-contrast microscopy, followed by centrifugation at 250 × g at 4°C for 10 min to pellet the nuclei. The pellet was suspended in 1 ml of buffer C (5 mM HEPES, pH 7.9; 26% glycerol [vol/vol]; 1.5 mM MgCl2; 0.2 mM EDTA; 0.5 mM DTT; 0.5 mM PMSF). Sodium chloride was added to 0.3 M accurately, and the sample was mixed well by inversion. The sample was spun at 24,000 × g for 20 min at 4°C after a 30-min incubation on ice. The supernatant was divided into 100-μl aliquots and snap-frozen in dry ice-ethanol. The concentration of the NE was determined by using a Bio-Rad protein assay kit. The NE was stored at −70°C until use.
To prepare 5′ end-labeled probes for EMSA, each RE fragment (I-2B, IIC-2, PAN RRE, and 57/K8 RRE) was subjected to 6% polyacrylamide-bispolyacrylamide (29:1) gel electrophoresis after digestion of the corresponding RE E1B Luc constructs with the restriction enzyme XhoI and a filling-in reaction of the XhoI site with Klenow fragment and substrates (Takara), followed by purification from the acrylamide gel slice. The fragments were treated with shrimp alkaline phosphatase (Roche) according to the manufacturer's protocol. The purified RE fragments, 1 pmol each, were labeled with [γ-32P]ATP (5,000 mCi/mmol; Amersham) and 10 U of T4 polynucleotide kinase (New England Biolabs), followed by spin column gel filtration through 1 ml of Sephadex G25 (Pharmacia) and ethanol precipitation. The total incorporation of the 32P in the purified probes was counted. The probes were then dissolved in distilled water with ca. 105 cpm/μl. Oligonucleotides for the AP1, Oct1, and SP1 consensus sequences were purchased from Promega and labeled in the same way.
Binding between the probe and the protein in the NE (20 μg of protein per reaction) was performed in 20 μl of binding buffer [20 mM HEPES, pH 7.9; 4% Ficoll (Ficoll-Paque; Pharmacia); 1 mM MgCl2; 0.5 mM DTT; 2 μg of poly(dI-dC)]. In the case of the purified RTA412, which was derived from pET21a+RTA412, 0.1 μg of protein was used in the reaction. The salt concentration was adjusted by adding KCl to a final concentration of 50 mM. Then, 1 μl of labeled probe was added, and the sample was incubated at room temperature for half an hour. Unlabeled probe was added in the competition analyses at a 50-fold molar excess, unless noted otherwise. In the competition analysis with PAN RRE and RTA412 protein, 100- and 200-fold molar excesses of either cold RE I-2B or RE IIC-2 were added in the reaction. In some cases, 2 μg of specific antibodies against a protein of interest, including RTA, Oct1, and Oct2, was included in the EMSA. Monoclonal anti-Oct1 and anti-Oct2 antibodies were purchased from Santa Cruz Biotechnology, Inc.
RESULTS
RTA activates specific regions of the K9 (vIRF) promoter.
We have already shown that KSHV RTA activates the K9 (vIRF) promoter through specific regions (10). In that report and also as shown in this experiment, we identified two regions that were responsible for RTA activation, bp −479 to −335 between D4 and D5 (RE I) and bp −209 to −121 between D8 and D9 (RE II) upstream of the K9 (vIRF) transcription start site (Fig. 1) (10).
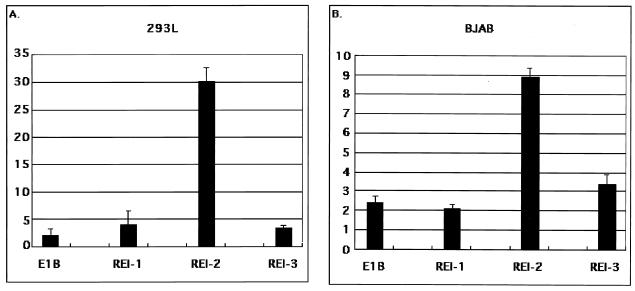
Therefore, we focused on the RE I and the RE II regions for further analysis. We divided the RE I region into three parts: RE I-1 (bp −479 to −424), RE I-2 (bp −426 to −364), and RE I-3 (bp −367 to −335) (Fig. 1). Among these, only RE I-2 was consistently reactive to RTA (Fig. 2A and B) in both 293L cells and BJAB cells. Because there was no difference between the fibroblast cell line, 293L cells, and the B-cell-originated cell line, BJAB, we used 293L cells for further transfection assays.
FIG. 2.
Core RE in RE I. (A) Responsiveness of REs I-1, I-2, and I-3 in 293L cells. (B) Responsiveness of REs I-1, I-2, and I-3 in BJAB cells. The corresponding regions were inserted upstream of the E1B minimal TATA box, which was followed by a firefly luciferase gene. The constructs were transfected into 293L cells (A) and BJAB cells (B), respectively. The emitted light was measured in relative light units 2 days after transfection, which was normalized with β-galactosidase activity in the same reaction lysate. The perpendicular axis gives the fold activity calculated as the normalized activity with RTA divided by that obtained without RTA.
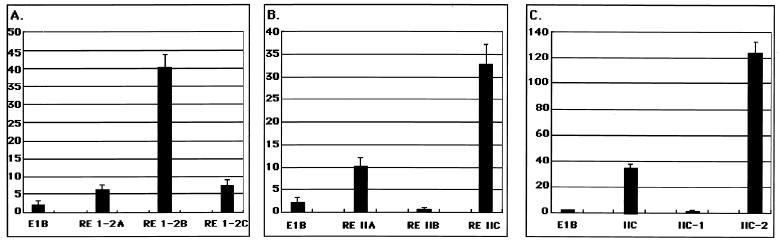
To determine the minimal region of RE I-2 that was required for RTA activation, we fragmented the region into the smaller parts (RE I-2A, RE I-2B, and RE I-2C) listed in Fig. 1 and made new reporter constructs, in which the shorter RE-I fragments were arranged as three to five tandem repeats just upstream of a synthetic E1B minimal TATA box that was followed by a firefly luciferase gene because of the reason mentioned in Materials and Methods. These reporter plasmids were then transfected into 293L cells with the effector plasmid pcDNA3.1(−)RTAMycHisB or its parental vector and with a β-galactosidase expression plasmid, pCMVβ, to normalize the transfection efficiency. The expression of RTA was easily confirmed by Western blotting analysis either with an anti-myc antibody (Invitrogen) or with α50A (data not shown). We assessed the activity of the cis elements by comparing their activity in the presence of pcDNA3.1(−)RTAMycHisB with their activity in its absence. Because the parental reporter plasmid, E1B Luc, was somewhat responsive to RTA expression by a factor of about 2 to 3 and because the basal activity of each reporter plasmid differed to some extent, the responsiveness to RTA was best determined by calculating the fold activity. Using this assessment, RE I-2B significantly responded to RTA in both 293L (Fig. 3A) and BJAB (data not shown) cells. As for the RE II region, it was divided into three parts: RE IIA, IIB, and IIC. Of them, the RE IIC region was constantly reactive to RTA (Fig. 3B). Further analysis showed that RE IIC-2, the latter half of RE IIC, was highly responsive to RTA expression (Fig. 3C), which indicated that there was an RE exclusively in RE IIC-2. Thus, we identified these two elements, RE I-2B and RE IIC-2, as the major responsive elements in the K9 (vIRF) promoter to RTA.
FIG. 3.
Determination of the core responsive segments in RE I-2 and RE II. (A) Responsiveness of REs I-2A, I-2B, and I-C of the RE I-2 region. (B) Responsiveness of RE IIA, RE IIB, and RE IIC of the RE II region. (C) Responsiveness of REs IIC-1 and II-C2 of the RE II-C region. The smaller segments of RE I-2, RE II, and the further RE IIC region (see Fig. 1A) were arranged as three to five tandem repeats upstream of the E1B minimal TATA box, which was followed by a firefly luciferase gene. The assay was performed as described in Materials and Methods. The value of perpendicular axis denotes the fold activity calculated as the normalized activity obtained with RTA divided by that obtained without RTA as in Fig. 2.
RTA upregulates RE I-2B and RE IIC-2 independently of B-cell-specific and other viral factors.
The previous experiment was performed in the absence of TPA, and RTA activated the K9 (vIRF) promoter in cells that were not of B cell origin (293L) as well as in the noninfected B-cell-origin cell line, BJAB, suggesting that the upregulation of RE I-2B and RE IIC-2 by RTA was independent of other viral and B-cell-specific factors. Moreover, none of the potential responsive elements reacted to TPA induction in BJAB or 293L cells, in contrast to the results seen in BCBL1 cells, in which the RE I-2B and RE IIC-2 regions were reactive to TPA, probably owing to the induction of RTA expression (data not shown). We tested here and had tested previously the full promoter region up to −774 bp upstream of the K9 (vIRF) transcription start site, which did not show a response to TPA induction (10). These results proved that the key factor for the upregulation of K9 (vIRF) expression was RTA, not other TPA-responsive cellular factors such as AP1, although there may be cooperation between such factors and RTA, even though they play only a supportive role in the regulation of K9 (vIRF) expression.
The AP1 consensus-like sequence seen in RE III was not functional (Fig. 1), since it did not respond to TPA (data not shown). This was also confirmed by EMSA, in which such elements never competed with the AP1 consensus sequence-like (data not shown). In contrast, SP1 such as motifs seen in RE I-1 and RE IIB were functional, since we observed in an EMSA that the SP1 consensus oligonucleotide did compete with the shifted band when RE I-1 and RE IIB regions were used as probes and an anti-SP1 antibody caused a supershift. A reporter construct containing three to five tandemly arranged AP1 or SP1 consensus sequences did not respond to RTA at all (data not sown).
Mutagenic analyses of RE I-2B and RE IIC.
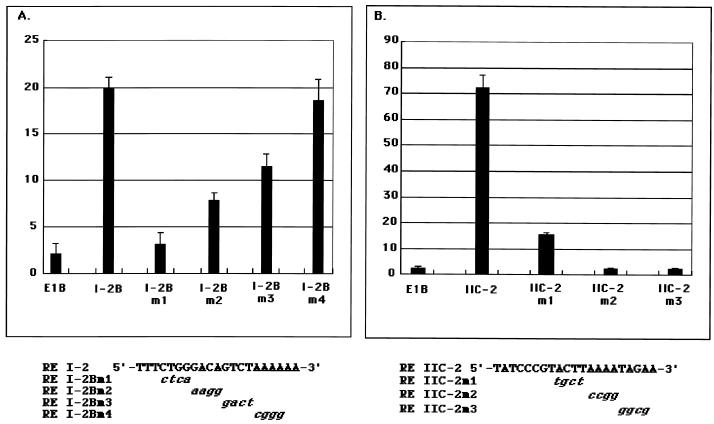
To determine more precisely the region in RE I-2B, we generated four mutants of RE I-2B: RE I-2Bm1, RE I-2Bm2, RE I-2Bm3, and RE I-2Bm4 (Fig. 4A). Of these, RE I-2Bm1 and RE I-2Bm2 were only partially responsive to RTA, at the level of the parental vector (pE1B Luc), and RE I-2Bm2 and RE I-2Bm3 had only partial responsiveness. In contrast, RE I-2Bm4 maintained its activity (Fig. 4A). These observations suggest that the key sequence responsible for RTA in RE I-2B was contained mainly in the RE I-2Bm1 region and partially in the RE I-2Bm2 and RE I-2Bm3 regions. Thus, the first 12 nucleotides of RE I-2B, 5′-TCTGGGACAGTC-3′, seemed most important for the activity.
FIG. 4.
Responsiveness to RTA of the mutants of RE I-2B and RE IIC-2. Results for a mutant series of RE I-2B (A) and RE IIC-2 (B) are shown. Three to five tandem copies of each mutated fragment of RE I-2B and RE IIC-2 were placed upstream of the E1B minimal TATA box, which was followed by a firefly luciferase gene. The assay was done as described above. The value of perpendicular axis denotes the fold activity, calculated as the normalized activity obtained with RTA divided by that obtained without RTA. Mutated nucleotides of each RE are shown under the panel.
We also prepared three kinds of mutants of RE IIC-2: RE IIC-2m1, RE IIC-2m2, and RE IIC-2m3 (Fig. 4B). The reporter analyses showed that the last half of RE IIC-2 contained an important RE for RTA activation because RE IIC-2m2 and RE IIC-2m3 lost their responsiveness to RTA drastically (Fig. 4B). The activity of RE IIC-2m1 was moderate but much lower than in the unmutated RE IIC-2, which suggests that some sequence of this region was also involved in the responsiveness to RTA. The overlapping region between RE IIC-1 and RE IIC-2 was not important, given that RE IIC-1 had no response to RTA (see above). Thus, the main cis element in RE IIC-2 activated by RTA was determined to be 5′-GTACTTAAAATA-3′.
Specific factors bind with the cis-acting sequence.
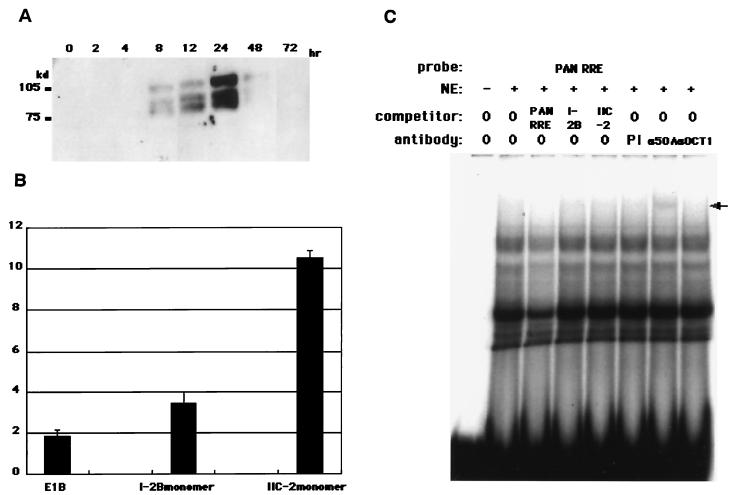
The chronological expression profile of RTA was assessed by SDS-7.5% PAGE, followed by Western blotting analysis with α50A before EMSA was performed. The data showed that RTA expression was first detectable by 4 to 8 h postinduction, peaked by ca. 24 h, and then disappeared (Fig. 5A). Therefore, we used nuclear extract from BCBL1 at 24 h postinduction with TPA for typical EMSA.
FIG. 5.
Chronological expression profile of RTA, responsiveness of monomer RE I-2B and RE IIC-2, and the binding activity of α50A in EMSA. (A) NE was prepared from BCBL1 cells at 0, 2, 4, 8, 12, 24, 48, and 72 h postinduction with TPA. A total of 20 μg of the NE was subjected to Western blotting analyses. RTA was detected with a mouse monoclonal antibody to RTA (α50A) as the first antibody and an anti-mouse IgG Fab fragment conjugated with horseradish peroxidase (see Materials and Methods). (B) Monomer constructs of RE I-2B and RE IIC-2 were transfected into 293L cells with β-galactosidase expression vector (pCMVβ) for normalization of transfection efficiency. The fold activity was calculated as mentioned above (see Materials and Methods). (C) EMSA was performed with PAN RRE as a probe. Cold competitors such as PAN RRE, RE I-2B, and RE IIC-2 were added to the reaction mixture in 50-fold excess for each case. Next, 2 μg of specific antibody to RTA (α50A) and Oct1 (αOct1) was added for supershift and/or binding inhibition analysis. The arrow denotes a supershifted complex with α50A. PI, preimmune serum.
To verify that the monomer of each RE (I-2B and IIC-2) was usable as a probe for EMSA, reporter gene assay was performed with monomer constructs. In each case, the response to RTA was relatively lower than that of the concatenated form but still detectable (Fig. 5B), probably because the concatenated form of each element could amplify its responsiveness to RTA coordinately. α50A was tested as to whether it recognized RTA in EMSA with PAN RRE as a probe (Fig. 5C). In this experiment, α50A caused a weak but still detectable supershift. However, no band disappeared compared to the bands in the assay without the antibody. This suggests that the binding activity of this antibody to RTA-bound complex was relatively weak in the EMSA, that RTA-bound complex itself was not abundant in the case of PAN RRE, and that the bound complex might have overlapped with the other shifted band.
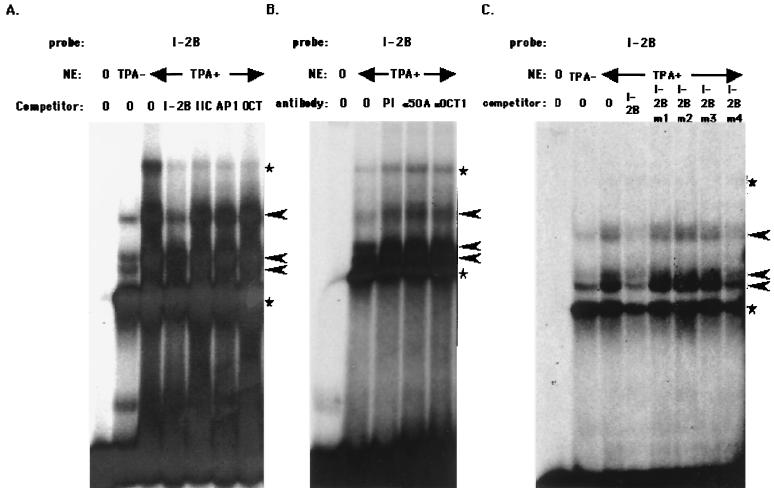
We then sought to determine whether RTA bound with these elements per se or whether there were other factors binding with RTA to regulate the activity. Although the pattern changed somewhat, depending on the conditions of electrophoresis, there were at least three major specific shifted bands when the RE I-2B region was used as a probe (Fig. 6A). All three shifted bands were I-2B specific because they were competed for with I-2 B but not with other fragments, including RE IIC, or the AP1 and Oct consensus sequences (Fig. 6A). The band marked with a star was thought to be nonspecific, since these bands were competed for with other competitors such as RE IIC, AP1, and the octamer-binding protein Oct; otherwise, they were not competed for with the self-competitor. TPA induction seemed to increase the expression of binding factors or enable such factors to bind more efficiently with specific DNA regions (Fig. 6A). Supershift or binding inhibition analyses with a specific antibody against RTA (α50A) or an anti-Oct1 antibody did not affect binding activity (Fig. 6B), indicating that RTA per se did not bind directly with this sequence or at least that RTA did not bind the sequence strongly (see below).
FIG. 6.
EMSA with RE I-2B as a probe. (A) Binding complex of RE I-2B probe and NE. A total of 20 μg of NE from BCBL1 cells induced with TPA or uninduced was incubated with the labeled RE I-2B probe. Each competitor—unlabeled RE I-2B, RE IIC, AP1 (AP1-binding consensus sequence), or Oct (Oct family protein-binding consensus)—shown on the panel was added in a 50-fold molar excess. (B) Supershift analysis with specific antibodies. Next, 20 μg of NE from BCBL1 induced with TPA was incubated with the RE I-2B probe and 2 μg of each of the following antibodies: mouse preimmune serum (PI), α50A (mouse monoclonal anti-RTA antibody), or αOct1 (mouse monoclonal anti-Oct1 antibody). (C) Competition analysis of RE I-2B with its mutants. The labeled RE I-2B probe and each mutant unlabeled probe (in a 50-fold molar excess) were mixed with NE and analyzed. Arrowheads show specifically formed DNA-protein complexes, and the stars indicate nonspecific complexes.
The RE I-2Bm1, RE I-2Bm2, and RE I-2Bm3 fragments, which were not as active as the wild-type fragment, did not compete as well as did the wild-type fragment. In contrast, RE I-2Bm4 did compete well, a finding consistent with its activity (Fig. 4A and 6C). Furthermore, and also consistent with the responsiveness to RTA, when the RE I-2B mutants were used as probes, RE I-2Bm4 bound the factors at the same level as did wild-type RE I-2B (data not shown). On the other hand, RE I-2Bm1 in particular had almost no binding activity as a probe. Therefore, it is likely that these binding factors were involved in the responsiveness to RTA, although the details remain to be clarified.
We analyzed the binding factors for the RE IIC-2 region in the same way (Fig. 7A). In this case, complicated shifted band patterns appeared, although the pattern changed somewhat depending on the conditions of electrophoresis, and the autoradiograph was shown as a long-exposed one to show an Oct1-specific shifted band. Typically, we observed six shifted bands for RE IIC-2 (Fig. 7). All of them, however, were RE IIC-2 specific, because they were all competed for by RE IIC-2 but not by other sequences (Fig. 7A). The slowest-migrating band seemed to show binding to a latency specific factor, because it disappeared or weakened upon TPA induction. The other bands seemed to exist in both the latent and the lytic phases, suggesting that these factors probably bind with the element before RTA is expressed and mediate the transactivation activity of RTA upon its induction.
FIG. 7.
EMSA with RE IIC-2 as a probe. (A) Binding complex of RE IIC-2 probe and NE. A total of 20 μg of NE from BCBL1 cells either induced with TPA or uninduced was incubated with the labeled RE IIC-2 probe. Each competitor—unlabeled RE IIC, RE IIC-1, RE IIC-2, AP1 (AP1-binding consensus sequence), or Oct (Oct family protein-binding consensus)—shown on the panel was added in a 50-fold molar excess. (B) Supershift analyses with specific antibodies. A total of 20 μg of NE from BCBL1 induced with TPA was incubated with the RE IIC-2 probe and 2 μg of each of the following antibodies: mouse preimmune serum (PI), α50A (mouse monoclonal anti-ORF50 antibody), or αOct1 (mouse monoclonal anti-Oct1 antibody). (C) Competition analyses of RE IIC-2 with its mutants. The labeled RE IIC-2 probe and each mutant unlabeled probe (at a 50-fold molar excess) were mixed with NE and analyzed. Arrowheads show specifically formed DNA-protein complexes, and the stars indicate nonspecific complexes.
As with RE I-2B, RTA did not seem to have a strong binding affinity for the RE IIC-2 element. Rather, it is very likely that the multiple DNA-binding factors that appeared in the EMSA mediated RTA's transactivation activity (Fig. 7B; see also below).
The competition analyses suggested that the RE IIC-2m2 and RE IIC-2m3 regions were important for the response to RTA; that is, the RE IIC-2m2 and RE IIC-2m3 regions never competed with the wild-type RE IIC-2 probe (Fig. 7C), which was consistent with the data on responsiveness to RTA (Fig. 5B), and no binding factors were detected when the RE IIC-2m2 and the RE IIC-2m3 fragments were used as probes (data not shown).
One typical competitor was an octamer-binding consensus sequence, which competed with one typical shifted band (Fig. 7A). This result suggested the involvement of octamer-binding factors such as Oct1 and OTA as mediators in RTA transactivation, as shown in the case of the autoregulation of RTA expression (30). This idea was supported by the supershift analyses in EMSA with an anti-Oct1 antibody (Fig. 7B).
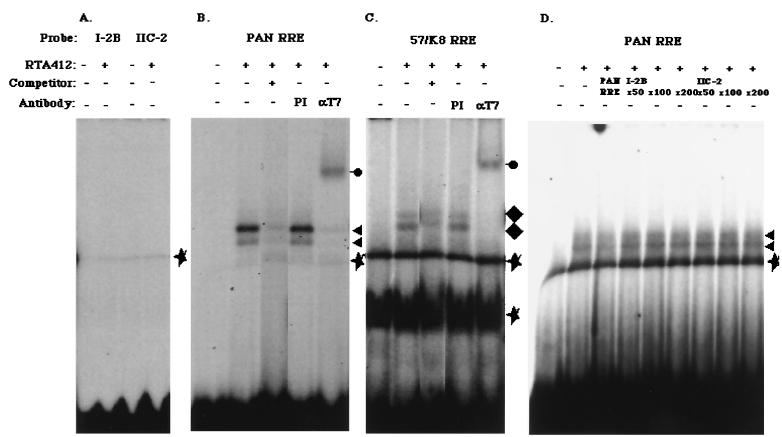
Then, we attempted to determine whether RTA expressed in E. coli bound with the responsive elements, because the N-terminal part of RTA expressed and purified from E. coli was successfully bound with the PAN RRE and the 57/K8 RRE in EMSA (24, 32), and this method would give us direct evidence that RTA bound with the elements. Our construct produced the first 412 aa of RTA, tagged with T7 epitope at the N terminus and with the histidine hexamer at the C terminus, which enabled us to purify the protein doubly with Ni-NTA agarose for His6 and with anti-T7 tag agarose for the T7 epitope. We tested whether the purified protein could bind with RE I-2B and RE IIC-2 to observe the direct evidence as to whether the RTA was bound or not. As reported elsewhere (24, 32), the protein indeed bound with PAN RRE and 57/K8 RRE (Fig. 8B and C). In contrast, RE I-2B and RE IIC-2 did not form a complex in EMSA (Fig. 8A). Furthermore, we tested whether much more excess cold competitor of RE I-2B and RE IIC-2 still competed with PAN RRE binding to RTA. As shown in Fig. 8D, a 200-fold molar excess of the cold competitor could not compete, which confirmed that the responsive elements were principally different ones and could not bind with RTA directly.
FIG. 8.
EMSA with RTA412. E. coli-derived RTA412 protein (0.1 μg) was incubated with RE I-2B and RE IIC-2 (A), PAN RRE (B), and 57/K8 RRE (C) as probes, respectively. PAN RRE and 57/K8 RRE generated specific shifted bands marked by triangles and diamonds, respectively. A mouse monoclonal anti-T7 tag antibody (αT7; Novagen) caused supershift (shown as filled circles with a line). PI refers to mouse preimmune serum, and stars indicate nonspecific complexes. (D) Competition anaysis with RE I-2B and RE II-C2 was shown. In this case, each cold competitor was added by a 50-fold up to a 200-fold molar excess, as indicated in the panel.
DISCUSSION
All gamma-2 herpesviruses, and even EBV, a gamma-1 herpesvirus, encode a homologue of RTA. RTA may have a function in KSHV that is similar to the role of BZLF1/Zta in EBV, which acts centrally in the initiation of the lytic replication of EBV and its own autoregulation. The actual KSHV homologue of BZLF1/Zta is K8α (KSHV bZIP), a bZIP protein that has not been demonstrated to have a transactivation activity and is speculated to be involved solely in lytic replication (39). In contrast, RTA, whose EBV homologue is BRLF1/Rta, has been reported to be a key inducer of lytic replication and to upregulate a variety of KSHV genes, including K9 (vIRF) (10), K8, ORF57 (24, 25, 26), and nut-1/PAN (32), and to autoregulate itself as well (12, 30) as a transcriptional transactivator. Thus, RTA activity leads to efficient lytic replication and finally to the production of mature virus particles.
The mechanism of transactivation by KSHV RTA has not been completely elucidated, although Song et al. reported on the transactivation mechanism of RTA with the nut-1/PAN regulatory element (32). They identified the specific RE for RTA and reported that RTA itself bound with the element quite tightly. Likewise, Lukac et al. reported that the N-terminal part of RTA up to 217 aa also bound with the common RE seen in the ORF57 and the K8 regulatory regions (24, 25, 26). These studies have shown that RTA binding with a specific region is required for its transactivating activity.
Most recently, Chang et al. have shown that RTA binds with a regulatory region of the K12 (Kaposin) gene (6). These authors proved that RTA bound with an element similar to PAN RRE (RTA responsive element) in an EMSA.
We investigated here the mechanism of the activation of the K9 (vIRF) gene by RTA. We reported previously that transcription from the KSHV K9 (vIRF) promoter was upregulated by RTA (10). At that time, SP1-like and/or AP1-like binding motifs had been proposed to be involved in the responsiveness to RTA as common sequences recognized by RTA. Further investigation revealed that such sequences were not responsible for the transactivation by RTA. Instead, we identified two independent cis elements that were upregulated by RTA. We did not consider it likely that the two elements shared common recognition sequences. Reporter gene analyses in cultured cells and competition analyses in EMSA with mutated responsive elements showed that the core responsible sequences were embedded in the 5′-TCTGGGACAGTC-3′ sequence of the RE I-2B element and in the 5′-GTACTTAAAATA-3′ sequence of the RE IIC-2 element. We termed them RRE I-2B and RRE IIC-2 (Fig. 9).
FIG. 9.
Comparison of the identified RTA responsive elements. K9 (vIRF) RRE I-2B and K9 (vIRF) RRE IIC-2 were identified in the present study. Boldface indicates the core sequence for the responsiveness to RTA in K9 (vIRF) RRE I-2B, RRE IIC-2, and Rta-p 2.1 (30). PAN RRE/K12 was described by Song et al. (32) and by Chang et al. (6). 57/K8 RRE was described by Lucac et al. (24). A double underline drawn in K12 RRE shows a core element for its RTA responsiveness (6).
We also investigated whether RTA was itself associated with the complexes seen in the gel shift analyses. Although specific complexes were observed in the analyses and RTA was easily detected in the NE by Western blotting, we could not conclude that RTA was included in the complexes, which suggests that RTA does not bind directly, or at least not tightly, with the responsive elements in these cases, an view confirmed by an experiment with the N-terminal part of RTA expressed in and purified from E. coli. As mentioned above, Song et al. and Lukac et al. showed that E. coli-derived full-length RTA and the N-terminal part of RTA, respectively, bound with the specific regions involved in transactivation by RTA (24, 32). Nonetheless, it was very difficult to identify any common sequences among these elements (Fig. 9). In the present study we tried to use RTA in NE and the N-terminal part of ORF50 protein expressed in E. coli for our binding assay by EMSA. Indeed, the latter bound with the PAN RRE and the ORF57/K8 RRE (24, 32). In contrast, the same experiment was not successful for our case. Therefore, in our case, it is likely that RTA itself does not bind either with RRE I-2B or RRE IIC-2 directly.
EBV Rta was reported to upregulate some gene expression through both direct and indirect mechanisms (20, 23, 27, 42). As mentioned above, RTA binds with the cis elements of the nut1/PAN, ORF57, K8, and K12 promoters (6, 24, 32). Thus, RTA may, like EBV Rta, have two mechanisms to upregulate the expression of target genes: a direct DNA-binding mechanism and a nondirect DNA-binding mechanism. In the latter case, DNA-binding proteins would play an important role in mediating the strong transactivation activity of RTA.
As we reported previously concerning the autoregulation of the RTA promoter by RTA itself (30), Oct1 also seemed to be involved in RE IIC-2, as shown in the competition and supershift analyses in EMSA. The importance of Oct1's involvement in RE IIC-2's responsiveness to RTA is unclear, partly because multiple factors other than Oct1 could be involved since the multiple shifted bands were shown in EMSA. Overexpressed Oct1, however, clearly upregulated the RE IIC-2 activity in response to RTA but did not upregulate the activity of RE I-2B (data not shown), suggesting that Oct1 is likely to play some role in RE IIC-2 transactivation by RTA.
There is some possibility that Oct1 might be involved in the transactivation of the AAAT sequence of the PAN RRE by RTA because this sequence is seen in K9 (vIRF) RRE IIC-2, Rta-p RRE 2.1 (30), and PAN RRE (32) (Fig. 9). This sequence contains only half of the Oct recognition sequence (5′-ATGCAAAT-3′). Although the consensus is recognized by Oct with the highest affinity, Oct also binds degenerate sequences such as 5′-TAATGRAT-3′, which is seen in the herpes simplex virus type 1 ICP0 promoter (11).
Lukac et al. suggested that an important RTA binding site in the 57/K8 RRE could be 5′-AATGTTCCCAC-3′, the latter half of the inverted repeat structure in the 57/K8 RRE (Fig. 9). If there is an RTA-binding site in PAN RRE, it is probably 5′-GTGGCTAACCTG-3′, whose complementary sequence is somewhat similar to the 57/K8 RRE. Concerning this point, Chang et al. showed that the sequence CTAA in the PAN RRE (32) was a core element for RTA binding and transactivation. In this study, neither of the sequences was present in K9 RRE I-2B, IIC-2, or Rta-p RRE 2.1. We also surveyed whether RRE I-2B and RRE IIC-2 were seen in the regulatory regions that were reported to be upregulated by RTA, such as ORF59 and ORF9 (25). Such elements, however, were not seen, which means that these genes are regulated in a different way. Thus, the mechanisms of gene upregulation by RTA might be quite degenerate.
CBP and HDAC have been reported to interact with RTA to regulate gene expression as coactivators (18). These are general cofactors and not specific DNA-binding proteins. It is, therefore, unlikely that they are important in the specificity of responsive elements for RTA. Rather, it is likely that factors interacting with RTA specify the responsive elements and mediate RTA's strong transactivation activity, even if these factors do not contain activation domains in their sequences. In this regard, Wang et al. identified a protein interacting with RTA (37). This MGC2663 is a member of the Krueppel-associated box-zinc finger proteins and synergizes with RTA to activate the ORF57 and the K8 promoter. However, it remains unclear how much it works because RTA itself binds to their specific regions and strongly activates both of them.
In summary, we identified two independent cis elements in the K9 (vIRF) promoter that are upregulated by RTA. RTA itself did not seem to have a binding activity with these elements. Rather, multiple factors could be involved in their upregulation through binding with the specific elements and an interaction with RTA, the details of which remain to be elucidated.
Acknowledgments
We thank Yun Bao Jing and Misa Kinoshita for their great technical assistance.
This work was funded by grants from the Ministry of Science and Education of Japan (12670282 to K.U. and 09CE2007 to K.Y.) and from the PRESTO Host and Defense Program, Japan Science Technology Corporation (200154023 to K.U.).
REFERENCES
- 1.Adamson, A. L., and S. C. Kenny. 1998. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 product. Virology 251:187-197. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 3.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 5.Chang, J., R. Rolf, D. Dittmer, and D. Ganem. 2000. Inflammatory cytokines and the reactivation of Kaposi's sarcoma-associated herpesvirus lytic replication. Virology 266:17-25. [DOI] [PubMed] [Google Scholar]
- 6.Chang, P.-J., D. Shedd, L. Gradoville, M.-S. Cho, L.-W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y.-N., D. L.-Y. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, C. Parravicini, M. Corbellino, and K. Yamanishi. 2001. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc. Natl. Acad. Sci. USA 98:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary, M. A., and W. Herr. 1995. Mechanism for flexibility in DNA sequence recognition and VP16-induced complex formation by the Oct-1 POU domain. Mol. Cell. Biol. 15:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the RTA gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 13.Foreman, K. E., J. Friborg, W.-P. Kong, C. Woffendin, P. J. Polverini, B. J. Nickoloff, and G. J. Nabel. 1997. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N. Engl. J. Med. 336:163-171. [DOI] [PubMed] [Google Scholar]
- 14.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruffat, H., and A. Sergeant. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 22:1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruffat, H., N. Duran, M. Buisson, F. Wild, R. Buckland, and A. Sergeant. 1992. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J. Virol. 66:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruffat, H., E. Manet, A. Rigolet, and A. Sergeant. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence specific DNA binding protein. Nucleic Acids Res. 18:6835-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1905-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, K. T., A. J. Stevenson, D. J. Goodwin, P. C. Bibson, A. F. Markham, and A. Whitehouse. 1999. The activation domain of herpesvirus saimiri R protein interacts with the TATA-binding protein. J. Virol. 73:9756-9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardwick, J. M., L. Tse., N. Applegren, J. Nicholas, and M. A. Veliuona. 1992. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J. Virol. 66:5500-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 22.Imai, S., S. Koizumi, M. Sugiura, M. Tokunaga, Y. Uemura, N. Yamamoto, S. Tanaka, E. Sato, and T. Osato. 1994. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc. Natl. Acad. Sci. USA 91:9131-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C., N. D. Sista, and J. S. Pagano. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J. Virol. 70:2545-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 27.Manet, E., A. Rigolet, H. Gruffat, J.-F. Giot, and A. Sergeant. 1991. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 19:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monini, P., S. Colombini, M. Sturzl, D. Goletti, A. Cafaro, C. Sgadari, S. Butto, M. Franco, P. Leone, S. Fais, P. Leone, G. Melucci-Vigo, C. Chiozzini, F. Carlini, G. Ascherl, E. Cornali, C. Zietz, E. Ramazzotti, F. Ensoli, M. Andreoni, P. Pezzotti, G. Rezza, R. Yarchoan, R. C. Gallo, and B. Ensoli. 1999. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi's sarcoma. Blood 93:4044-4058. [PubMed] [Google Scholar]
- 29.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 30.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. The octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarid, R., J. S. Olsen, and P. S. Moore. 1999. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv. Virus Res. 52:139-232. [DOI] [PubMed] [Google Scholar]
- 32.Song, M. J., H. J. Brown, T.-T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 34.Sun, R., S.-F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thurau, M., A. Whitehouse, S. Wittmann, D. Meredith, and H. Fickensher. 2000. Distinct transcriptional and functional properties of the R transactivator gene ORF50 of the transforming herpesvirus saimiri strain C488. Virology 268:167-177. [DOI] [PubMed] [Google Scholar]
- 36.Ueda, K., and D. Ganem. 1996. Cellular factors controlling the activity of woodchuck hepatitis virus enhancer II. J. Virol. 70:4714-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, S., A. Liu, M.-H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehouse, A., A. J. Stevenson, M. Cooper, and D. M. Meredith. 1997. Identification of a cis-acting element within the herpesvirus saimiri ORF6 promoter that is responsive to the HVS R transactivator. J. Gen. Virol. 78:1411-1415. [DOI] [PubMed] [Google Scholar]
- 39.Wu, F. Y., J.-H. Ahn, D. J. Alcendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, T.-T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zalani, S., E. H. Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zalani, S., E. A. H. Guthrie, D. E. Gutsch, and S. C. Kenney. 1992. The Epstein-Barr virus immediate-early promoter BRLF1 can be activated by the cellular Sp1 transcription factor. J. Virol. 66:7282-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]