Abstract
Infected-cell protein 27 (ICP27) is an essential herpes simplex virus type 1 (HSV-1) regulatory protein that activates a subset of viral delayed-early and late genes, at least in part through posttranscriptional mechanisms. Previous studies have shown that the amino (N)-terminal half of the protein contains important functional regions, including a leucine-rich nuclear export signal (NES). However, to date, the phenotype of an HSV-1 ICP27 NES mutant has not been reported. In this study, we engineered and characterized dLeu, an HSV-1 deletion mutant that specifically lacks ICP27's NES (amino acids 6 to 19). The phenotype of dLeu was analyzed alongside those of eight other ICP27 N-terminal deletion mutants. We found that in Vero cells, dLeu displays modest defects in viral gene expression and an approximately 100-fold reduction in the production of viral progeny. Unlike wild-type (WT) ICP27, which exhibits a cytoplasmic distribution in addition to its predominant nuclear localization, dLeu ICP27 is highly restricted to the cell nucleus. This strongly suggests that the N-terminal leucine-rich sequence functions as an NES during viral infection. Our analysis of dLeu and the other mutants has enabled us to genetically define the regions in the N-terminal 200 residues of ICP27 which are required for efficient viral growth in Vero cells. Only two regions appear to be important: (i) the leucine-rich NES and (ii) the RGG box RNA-binding domain, encoded by residues 139 to 153. A virus lacking the RGG box-encoding sequence, d4-5, has a phenotype similar to that of dLeu in that it displays modest defects in viral gene expression and grows poorly. Interestingly, deletion of both the NES and RGG box, as well as the sequences in between, is lethal. The resulting virus, d1-5, displays severe defects in viral gene expression and DNA synthesis and is unable to produce significant amounts of infectious progeny. Therefore, the N-terminal portion of ICP27 contains at least two functional domains which collectively are absolutely essential for viral infection.
During its lytic infection, herpes simplex virus type 1 (HSV-1) exerts tight regulatory control over the expression of its genes (for a review of HSV gene regulation, see reference 38). The approximately 80 viral genes are transcribed in the infected-cell nucleus by the host RNA polymerase II and are expressed in three sequential waves. Immediately upon infection, five immediate-early (IE) genes are transcriptionally induced via the action of a virus-encoded virion protein, VP16, working in concert with cellular proteins. Four of the five IE genes encode nuclear proteins (ICP0, ICP4, ICP22, and ICP27) which activate the expression of all later genes. Upon synthesis of the IE proteins, delayed-early (DE) genes are expressed. Many of the DE proteins have enzymatic activities which are involved directly or indirectly in viral DNA replication. Accumulation of these proteins results in the activation of viral DNA replication, which in turn stimulates the expression of the last set of genes, the late (L) genes. These genes predominantly encode viral structural proteins and can be subdivided into leaky-L and true-L classes, depending on whether they are partially or wholly dependent upon viral DNA synthesis for their expression. In addition to sequentially activating its own genes during infection, HSV-1 also uses multiple mechanisms to inhibit the expression of cellular genes.
Of the four IE proteins which regulate viral gene expression, ICP4 and ICP27 are essential for growth under all known conditions, whereas ICP0 and ICP22 are required only under certain conditions. ICP4 is the major transcriptional activator protein of HSV-1, being stringently required for DE/L gene transcription. The role of ICP27 is less well characterized, but it appears to carry out multiple functions. First, studies of viral ICP27 mutants have shown that it plays a critical role in stimulating the expression of certain DE/L gene mRNAs and proteins (21, 22, 24, 33, 35, 40, 50). ICP27 mutants express reduced levels of several DE genes encoding essential DNA replication factors (22, 50). This effect appears to be responsible for the reduced levels of viral DNA replication that are observed in ICP27 mutant-infected cells. Second, ICP27 represses the expression of several viral IE and DE genes at late times after infection (21, 33, 40). Third, ICP27 inhibits the production of host mRNAs and proteins during infection (14, 40, 46, 49).
The molecular mechanisms by which ICP27 exerts its multiple effects on gene expression are only partially understood. However, many of ICP27's effects are mediated at the posttranscriptional level. Consistent with this, ICP27 is an RNA-binding protein (4, 18, 27, 42), although the biological significance of this activity is presently unclear. It is also not known whether ICP27 binds to specific RNA sequences or structures. ICP27's posttranscriptional effects on gene expression are diverse. It inhibits pre-mRNA splicing during the lytic infection (5, 16), an effect which is at least partially responsible for ICP27's inhibitory effects on host gene expression. ICP27 also modulates the efficiency of pre-mRNA polyadenylation (23) and in so doing may stimulate the expression of certain L genes which contain inherently weak polyadenylation sites (22). It has been proposed that ICP27 functions as a transport factor for the nuclear export of viral L mRNAs, which, due to their lack of introns, may not be able to efficiently access the normal cellular mRNA export pathway (20, 42, 46). Consistent with a role in mRNA export, ICP27 has been found to shuttle continuously between the nucleus and cytoplasm (26, 30, 42, 46). Evidence that ICP27 affects nuclear export pathways in infected cells also comes from studies which show that it promotes the cytoplasmic accumulation of unspliced alpha-globin mRNA in HeLa cells (7, 10). ICP27 also affects mRNA stability, in that it posttranscriptionally stabilizes beta interferon mRNA in infected cells (4, 28). In addition to these seemingly diverse posttranscriptional effects, ICP27 may also regulate transcription during infection, as it appears to stimulate the transcription of at least two viral L genes (19) and inhibit the transcription of certain cellular genes (49).
The functional domains of ICP27 have not been fully delineated. Numerous studies, however, indicate that the carboxyl- (C)-terminal half of the protein (from approximately residue 260 to the C terminus at residue 512) is critical (4, 15, 24, 33, 34, 47, 48). Consistent with this, the sequences corresponding to the C-terminal half of ICP27 are conserved in regulatory proteins encoded by all mammalian and avian herpesviruses sequenced to date (39). In contrast, the amino (N)-terminal half of ICP27 also has important functional regions but is not closely conserved in sequence among herpesviruses. In the first 200 amino acid residues of ICP27, at least three functional regions have been described (Fig. 1), each of which can function in the context of heterologous proteins. These are (i) a leucine-rich nuclear export signal (NES), similar to that of the human immunodeficiency virus type 1 Rev protein, mapping to residues 5 to 15 (42); (ii) a nuclear localization signal (NLS), mapping to residues 109 to 137 (25); and (iii) an RGG box RNA-binding domain, mapping to residues 137 to 153 (27). The N-terminal half of ICP27 is also the site of multiple posttranslational modifications, including phosphorylation of serine residues (52), methylation of one or more arginine residues on the RGG box (27), and possibly nucleotidylylation (2, 3).
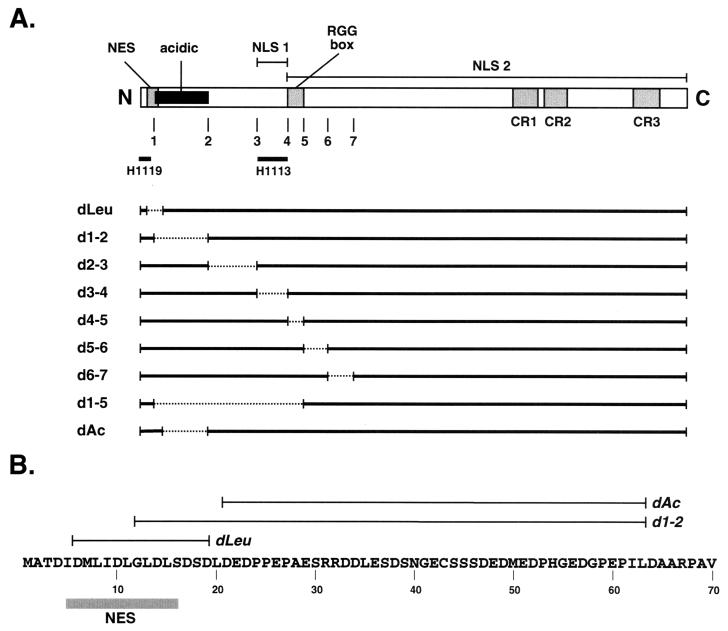
FIG. 1.
HSV-1 ICP27 N-terminal mutants. (A) The bar at the top represents the ICP27 polypeptide, highlighting known motifs and functional sequences. Below the bar, numbers 1 to 7 refer to sites of engineered XhoI restriction sites (34) which were used to construct many of the deletion mutations. The regions of ICP27 that harbor the epitopes for mouse MAbs H1119 and H1113 are indicated. Also indicated are the leucine-rich NES, acidic region, NLS 1, NLS 2, RGG box RNA-binding domain, and regions highly conserved in the ICP27 homologs of other herpesviruses (CR1 to -3). The solid horizontal lines below the bar indicate the ICP27 protein sequences present in the various N-terminal deletion mutants; dashed lines indicate in-frame deletions. (B) Comparison of amino acid sequences deleted in dLeu, d1-2, and dAc. The N-terminal 70 residues (in single-letter amino acid code) of ICP27 from strain KOS1.1 are shown, along with the location of ICP27's N-terminal leucine-rich NES. The extents of the deletions in dLeu, d1-2, and dAc are indicated by lines.
To date, extensive phenotypic characterization of HSV-1 ICP27 N-terminal mutants is limited, but the phenotype of one mutant, d1-2, has been studied in detail (35). This mutant lacks residues 12 to 63, which comprise a highly acidic region. d1-2 is deficient for growth in Vero cells, exhibiting somewhat reduced levels of viral DNA replication and L gene expression. However, d1-2 is not tightly blocked for expression of several L genes, including the true-L gene encoding glycoprotein C (gC). This phenotype differs from those of several C-terminal ICP27 mutants, which are tightly restricted from expressing gC and several other L genes (33, 34, 45, 46, 48).
The deletion in d1-2 impinges on the NES but does not entirely remove it (Fig. 1B). It is therefore unclear to what extent the NES disruption contributes to the phenotype of d1-2. To clarify the role of the NES in ICP27 function, we have engineered a viral mutant, dLeu, which specifically lacks the NES (residues 6 to 19). We have compared its phenotype to those of d1-2 and several other N-terminal ICP27 deletion mutants. These studies have allowed us to genetically define the functional regions in the N-terminal 200 residues of ICP27.
MATERIALS AND METHODS
Cells, viruses, and infections.
Infections were carried out in Vero (African green monkey kidney) cells or in V27 cells, a derivative of Vero cells containing an integrated copy of the ICP27 gene (33). Vero cells were obtained from the American Type Culture Collection. Vero cells were propagated in Dulbecco modified Eagle medium containing 5% heat-inactivated fetal calf serum, 50 U of penicillin per ml, and 50 μg of streptomycin per ml. V27 cells were propagated in the same formulation plus 300 to 600 μg of G418 per ml. All tissue culture reagents were purchased from Life Technologies/Invitrogen (Carlsbad, Calif.).
Strain KOS1.1 (17) was the wild-type (WT) strain of HSV-1 used in this study. Construction of the ICP27 mutants d27-1 (33), d1-2 (35), d2-3 and d6-7 (1), d3-4 and d4-5 (25), and d5-6 and d1-5 (27) has been described previously. The construction of the mutants dLeu and dAc is described below. For all mutants other than d27-1, two genetically independent isolates were obtained, the second of which was designated with the suffix b. All ICP27 mutants were isolated and propagated in V27 cells.
Infections with high-titer stocks were carried out at a multiplicity of infection (MOI) of 1 or 10 PFU per cell in phosphate-buffered saline containing 0.9 mM CaCl2, 0.5 mM MgCl2, 0.1% glucose, and 1% heat-inactivated newborn calf serum. The virus inoculum was allowed to adsorb to the cells for 1 h at 37°C. The inoculum was then replaced with 199 medium containing 2% heat-inactivated newborn calf serum, 50 U of penicillin per ml, and 50 μg of streptomycin per ml, and the infection mixtures were reincubated at 37°C. For viral plaque assays, the medium was the same as for virus infections except that it included 1% heat-inactivated normal pooled human serum (ICN Pharmaceuticals). Plaque assay mixtures were incubated at 37°C for 3 to 4 days to allow plaques to develop. For virus yield experiments (see Fig. 4), infected monolayers were treated at 2 h postinfection (hpi) with glycine-saline solution (pH 3.0) as described previously (35) to lower the level of background due to extracellular virus. The infections were terminated by the addition of 5 ml of sterile milk to each flask, followed by freezing of the flasks at −80°C. Virus was released by three cycles of freeze-thawing, and the amount of infectious virus was determined by plaque assay of the infected cell lysate on V27 cells. For each 10 ml of lysate, 0.5-ml aliquots of 10-fold and higher dilutions were tested for plaque formation; thus, the limit of detection of viral progeny in each infection was 2 × 102 PFU.
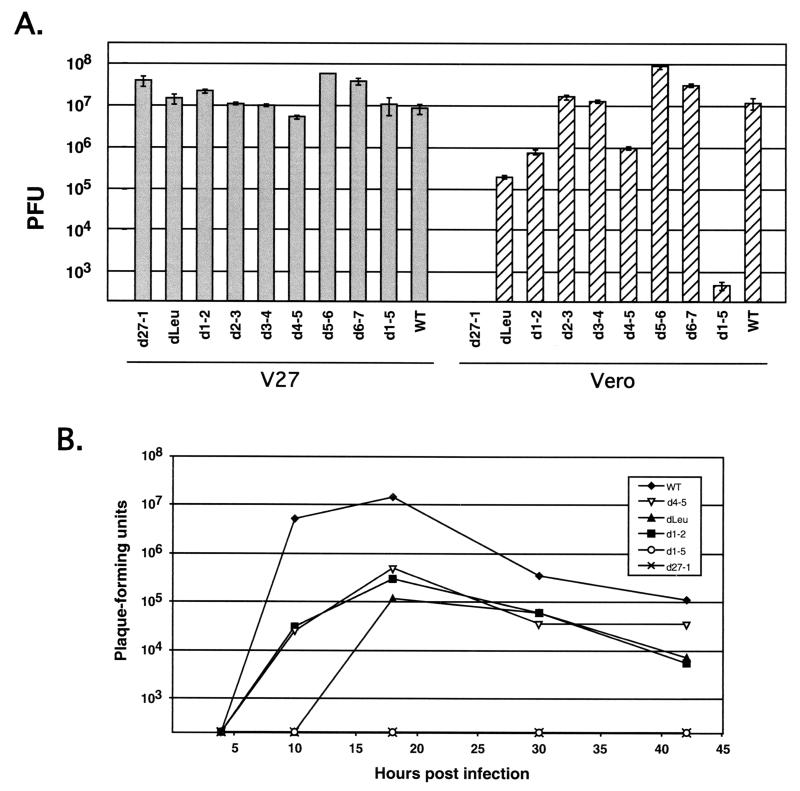
FIG. 4.
Viral yields in infected Vero and V27 cells. (A) Single-cycle yield assays. Replicate cultures of approximately 2 × 106 Vero or V27 cells were infected in duplicate at an MOI of 1 with WT HSV-1 or various ICP27 mutants. After 24 h, the infections were terminated by freezing. Virus was released from the cells by three cycles of freeze-thawing, and the total infectious virus in the lysate was determining by a plaque assay on V27 cells. The gray and hatched bars represent the mean yields in V27 and Vero cells, respectively. Error bars indicate the values of the duplicate samples. (B) Growth curves. Cultures of approximately 2 × 106 Vero cells were infected in duplicate at an MOI of 1 with WT HSV-1 or various ICP27 mutants. Infections were terminated at multiple time points by freezing, and virus yields were determined as for panel A. The values shown represent the mean yields of duplicate infections.
Isolation of dLeu and dAc.
To engineer dLeu, we first constructed a recombinant ICP27 plasmid which has a deletion of the NES sequence. This was accomplished in multiple steps. First, the plasmid pBH27 (32) was altered by oligonucleotide mutagenesis to create plasmids pBH27Jose2 and pBH27Jose3, which have clustered point mutations at codons 5/6 and 19/20. The mutations create KpnI restriction sites. Both engineered plasmids thus have two KpnI restriction sites, one in the ICP27 open reading frame and one in the polylinker region. To construct the deletion mutant plasmid, both plasmids were digested with KpnI and the small fragment of pBH27Jose2 was ligated to the large fragment of pBH27Jose3, which had first been treated with calf intestinal phosphatase. After introduction of the ligated DNA into Escherichia coli, the resulting plasmids were screened to find one which had the small KpnI insert in the proper orientation to give rise to the desired NES deletion. This clone was designated pBH27ΔLeu. The sequence of the mutagenized region was confirmed by DNA sequencing.
To engineer dAc, oligonucleotide mutagenesis was used to engineer an XhoI site at codons 20/21 in pBS27. This plasmid was designated pBSXhoI. To construct the desired deletion mutant, the 3.7-kb XhoI-SstI fragment of pBHXhoI was ligated to the 1.8-kb XhoI-SstI fragment of pBSpm64a, which has an engineered XhoI site at codons 63/64 (35). The resulting plasmid was designated pBSd20-64 and has an in-frame deletion of codons 21 to 63. The sequence of the deleted region was confirmed by DNA sequencing.
For transfer of the dLeu and dAc mutations into HSV-1, the altered ICP27 alleles were engineered into the pPs27pd1 plasmid (32), which contains the ICP27 gene on a 6.0-kb HSV-1 DNA insert. This step was done to increase the extent of HSV-1 sequences flanking the mutation, with the expectation that this would increase the efficiency of in vivo homologous recombination. The new plasmids were called pPs27ΔLeu and pPsd20-64. To introduce the altered genes into recombinant HSV-1 genomes, a marker transfer protocol was used (33). All mutant isolates were plaque purified at least three times.
Analysis of mutant viruses.
With the exception of dAc, Southern blot analysis was performed to confirm the genomic structures of all engineered viruses. Total DNA was isolated from infected cells by the procedure of Gao and Knipe (12). Southern blotting and hybridizations were carried out by standard procedures using ICP27 gene-specific 32P-labeled probes (41). To confirm the genomic structures of the dAc isolates, the N-terminal segments of the dAc and dAc-b ICP27 genes were PCR amplified from infected-cell DNA and subjected to DNA sequence analysis.
Immunoblotting analysis was performed as described previously (37), using anti-ICP27 monoclonal antibodies (MAbs) H1113 and H1119, diluted 1:1,000 and 1:5,400, respectively. H1113 and H1119 were purchased from the Rumbaugh-Goodwin Institute for Cancer Research (Plantation, Fla.). Immunoreactive proteins were detected by enhanced chemiluminescence with a commercially available kit (Amersham). Indirect immunofluorescence was used to assess the intracellular localization of ICP27. Cells were fixed and permeabilized as described previously (31) and stained with H1113 or H1119. For the experiments shown in Fig. 3, H1113 and H1119 were used at a dilution of 1:1,000, and secondary staining was done with a 1:100 dilution of tetramethyl rhodamine isothiocyanate-conjugated donkey anti-mouse immunoglobulin G (Jackson Immunoresearch Laboratories). For the experiment shown in Fig. 8, H1113 was used at a dilution of 1:200, and secondary staining was done with a 1:200 dilution of Cy2-conjugated goat anti-mouse immunoglobulin G (Jackson Immunoresearch Laboratories). Cell staining was visualized with an Olympus BX60 fluorescence microscope linked to a video camera. Images were captured as TIFF by using a CG7 frame grabber (Scion). After the images were imported into the program Photoshop Elements (Adobe), they were adjusted for brightness and contrast. Within a given experiment, all adjustments were done in parallel so that resulting images could be directly compared.
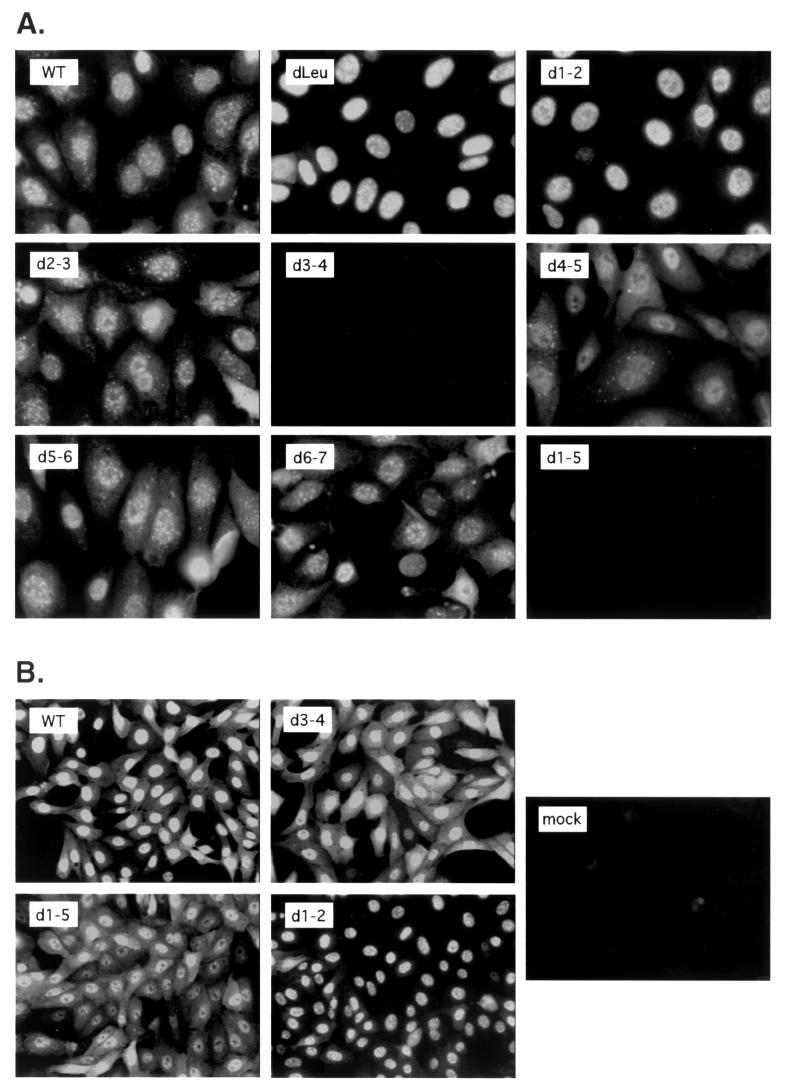
FIG. 3.
Cellular localization of mutant ICP27 polypeptides. Vero cells growing on coverslips were infected at an MOI of 10 with the viruses indicated. At 6 hpi, the cells were fixed and processed for immunofluorescence using anti-ICP27 MAb H1113 (A) or H1119 (B). For each panel, staining, image capture, and image manipulation were done in parallel so that the localization patterns of the different mutant proteins could be directly compared.
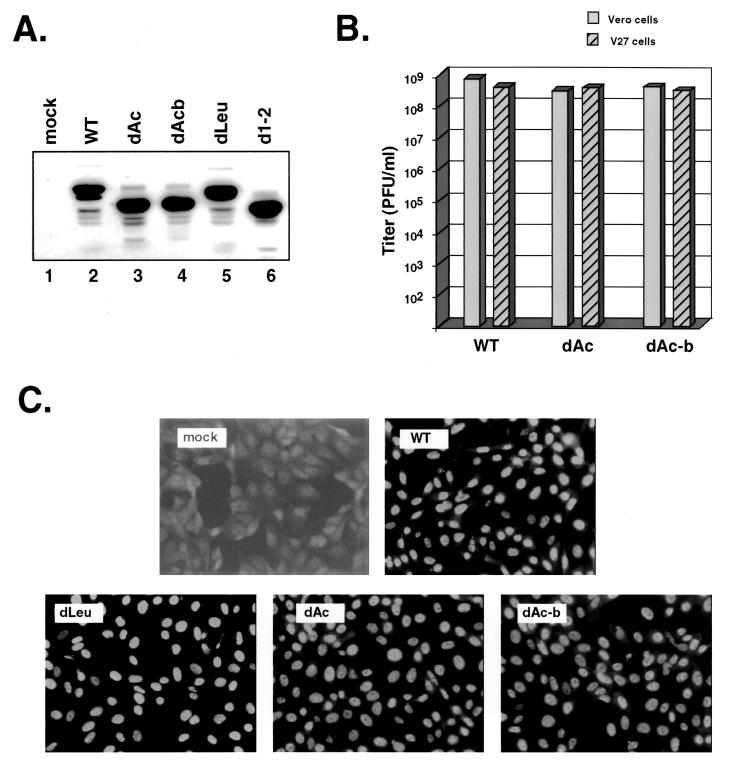
FIG. 8.
Deletion of the acidic region between residues 21 and 63 does not affect viral growth or ICP27 localization. (A) Immunoblot analysis of dAc ICP27. Vero cells were mock infected or infected with the viruses indicated at an MOI of 10. Protein extracts were prepared at 5 hpi, and immunoblotting was carried out with MAb H1113. (B) Plaquing ability of dAc. Stocks of WT HSV-1 or dAc isolates were titrated by plaque assay on Vero cells (gray bars) or V27 cells (hatched bars). (C) Localization of ICP27 in dAc-infected cells. Vero cells were mock infected or infected with the viruses indicated. The cells were fixed at 6 hpi and processed for immunofluorescence with the H1113 MAb.
Analysis of viral DNA replication levels in HSV-1 infected cells was carried out by [3H]thymidine labeling as previously described (35), except that labeling was performed with 25 μCi of [3H]thymidine (80 Ci/mmol; Amersham) per ml from 7 to 13 hpi. Equal amounts of each purified DNA preparation (2.5 μg) were electrophoresed on a 1% agarose gel, which was then treated with a fluorography enhancing agent (Amplify; Amersham) for 2 h prior to drying and exposure to X-ray film at −80°C. Characterization of viral protein synthesis by metabolic labeling was performed as described previously (34). Analysis of RNA was done as follows. Total RNA was isolated from infected 100-mm-diameter dishes by using the Trizol reagent (Invitrogen). The RNA was resuspended in water, treated with RNase-free DNase (Promega), phenol-chloroform-isoamyl alcohol extracted, and ethanol precipitated. Equal amounts (3.4 μg) of each RNA sample were subjected to electrophoresis through denaturing formaldehyde-agarose gels and transferred to GeneScreen filters (DuPont). Hybridization of 32P-labeled DNA probes was done overnight at 42°C in ULTRAhyb hybridization solution (Ambion). After hybridization, the filters were washed two times for 15 min each in 2× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) and then two times for 15 min each in 0.1× SSC-0.1% SDS. Linearized plasmids were used as the hybridization probes and were labeled by using a random primer labeling kit (Invitrogen). The plasmids used for probes were pE/3583 (13) (ICP8 probe), pEcoRI-BamHI-I-I (11) (gC probe), and pUL42 (UL42 gene probe). The pUL42 plasmid was constructed in the following manner. A 599-bp fragment corresponding to the N-terminal coding region of the HSV-1 UL42 gene was PCR amplified from a preparation of HSV-1 (strain KOS1.1) DNA by using the primers GATCGAATTCTTCCCCTGGCGGTGTGGCCC and GATCGAATTCCGAACGTGGTGGGCGTGGCA. After the PCR product was cut with EcoRI, the fragment was ligated into the EcoRI site of pUC19. This plasmid was designated pUL42.
RESULTS
Isolation of an ICP27 NES deletion mutant.
Previous work has indicated that ICP27 possesses a leucine-rich NES mapping to residues 5 to 15 (42, 47). To characterize the phenotype of a viral mutant lacking the NES, we engineered the mutant dLeu, having a deletion of ICP27 codons 6 to 19 (Fig. 1 and Table 1). dLeu was isolated and propagated in V27 cells, a derivative of Vero cells which possess a stably transfected ICP27 gene and therefore complement growth defects of viral ICP27 mutants (33). Two genetically independent isolates of dLeu were obtained, the second of which was designated with the suffix b. We wished to characterize two independent isolates to ensure that any mutant phenotype could be attributed to the engineered deletion rather than to any possible secondary mutations.
TABLE 1.
HSV-1 ICP27 mutants used in this study
| Mutant | Residues deleted | No. of residues deleted | Amino acid alteration(s) |
|---|---|---|---|
| dLeu | 6-19 | 14 | Residue 5, I to G; residue 20, L to T |
| d1-2 | 12-63 | 52 | Residue 64, D to E |
| d2-3 | 64-108 | 45 | Residue 109, G to E |
| d3-4 | 109-138 | 30 | Residue 139, G to E |
| d4-5 | 139-153 | 15 | Residue 138, R to L; residue 154, G to E |
| d5-6 | 154-173 | 20 | Residue 153, P to L; residue 174, G to E |
| d6-7 | 174-200 | 27 | Residue 173, P to L |
| d1-5 | 12-153 | 142 | Residue 154, G to E |
| dAc | 21-63 | 43 | Residue 64, D to E |
Our laboratory has previously isolated several other HSV-1 mutants which have small in-frame deletions in the N-terminal portion of the ICP27 gene. These are d1-2, d2-3, d3-4, d4-5, d5-6, d6-7, and d1-5 (Fig. 1A; Table 1) (1, 25, 27, 35). Although we previously characterized the ability of these mutants to replicate in Vero cells, only d1-2 has been analyzed in detail with respect to its phenotype (35). We therefore decided to study the phenotype of dLeu alongside those of these previously isolated N-terminal mutants.
ICP27 deletion mutant proteins are stable and localize to the nucleus.
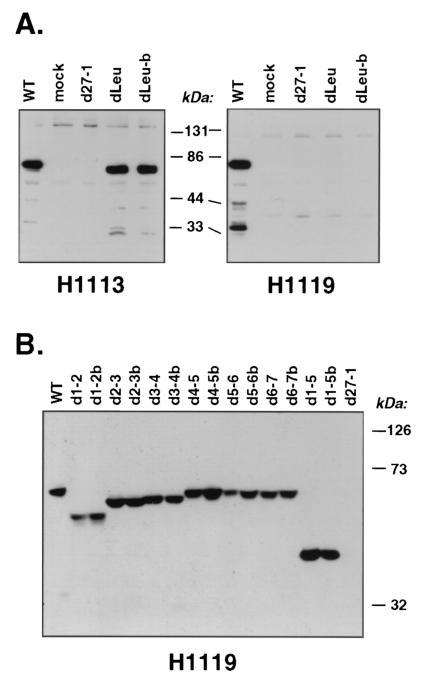
To see if dLeu and the other deletion mutants express stable ICP27 polypeptides, we carried out a set of immunoblotting experiments. Total proteins from infected Vero cells were probed with either of two MAbs specific for ICP27. MAb H1119 binds to residues 1 to 11, whereas MAb H1113 binds to residues 109 to 137 (25). The first experiment focused on dLeu (Fig. 2A); a second experiment focused on the remaining N-terminal mutants (Fig. 2B). Several conclusions can be drawn from the results of these two experiments. First, based on the intensities of the signals, all mutant ICP27 polypeptides are expressed at levels comparable to that of WT ICP27. Thus, the deletions do not appear to adversely affect protein expression or stability. Second, the electrophoretic mobilities of the mutant proteins are consistent with the engineered deletions. The d1-2, d2-3, d3-4, and d1-5 proteins migrated the most rapidly, consistent with their relatively large deletions (≥30 residues), whereas the remaining mutants migrated only slightly faster than WT ICP27, consistent with their smaller deletions. Third, the immunoreactivities of the mutant polypeptides were as expected. The dLeu ICP27 protein was readily detectable with H1113, but not with H1119 (Fig. 2A). This was not surprising, as the engineered deletion removes much of the H1119 epitope. As expected, H1119 reacts with all of the other N-terminal deletion mutant proteins (Fig. 2B). Additional immunoblotting experiments indicated that the d3-4 and d1-5 ICP27 polypeptides do not react with the H1113 MAb (data not shown), as expected since this epitope is missing in these mutants.
FIG. 2.
Immunoblot analysis of N-terminal deletion mutants. (A) Analysis of dLeu. Vero cells were mock infected or infected with the indicated viruses at an MOI of 10. Protein extracts were prepared at 5 hpi, and immunoblotting was carried out using MAbs H1113 and H1119, as indicated. The migration of marker proteins is shown. (B) Analysis of additional N-terminal mutants. The experiment was performed as for panel A, except that infections were carried out at an MOI of 1 and protein extracts were prepared at 16 hpi. Immunoblotting was performed with the H1119 MAb.
NES mutations restrict ICP27 to the cell nucleus.
Several of the deletions affect ICP27's leucine-rich NES and its major NLS (Fig. 1). It was therefore of interest to examine the nucleocytoplasmic distribution of the ICP27 molecules expressed by dLeu and the other mutants. Among the mutants which have been previously described, only d3-4 and d4-5 have been characterized with respect to the intracellular localization of ICP27 (25). In the first experiment, Vero cells were infected with WT or mutant viruses and the cells were fixed at 6 hpi. Immunofluorescence was carried out using the H1113 MAb (Fig. 3A). WT ICP27 exhibited a predominantly nuclear localization but showed some cytoplasmic distribution as well, a pattern consistent with that seen in previous studies (9, 33, 48). The d3-4 and d1-5 ICP27 molecules failed to react with H1113, as expected since the H1113 epitope is lacking in these molecules. To assess the localization of these proteins, a second immunofluorescence analysis was performed using H1119 (Fig. 3B). Together, the results of both experiments and several repeat experiments allow us to make several conclusions about the nucleocytoplasmic distribution of the mutant ICP27 molecules. First, the localizations of dLeu- and d1-2-encoded ICP27s differ significantly from those of WT ICP27 and the other mutants in that they are much more restricted to the nucleus. This is consistent with a defect in the nuclear export of ICP27. Second, all of the N-terminally deleted mutant proteins, even ones missing the major NLS (d3-4 and d1-5), show a significant degree of nuclear localization. This is consistent with past results for d3-4 and with the fact that ICP27 possesses a second C-terminal NLS, corresponding to residues 152 to 512 (Fig. 1A) (25). However, the d3-4 and d1-5 proteins show increased cytoplasmic accumulation compared to WT ICP27, a result which is likely attributable to a decreased rate of nuclear import due to the deletion of the major NLS. Third, the d4-5 and d1-5 proteins, and to some extent the d3-4 protein, fail to localize efficiently to nucleoli. This has been previously reported for d4-5 and is consistent with our previous conclusion that the RGG box forms part of a nucleolar localization signal for ICP27 (25).
Growth of N-terminal ICP27 mutants in Vero cells.
The capacity of dLeu and the other mutants to replicate in Vero cells was next studied. In the first set of experiments, titers of viral stocks were determined on Vero or V27 cells (Table 2). For a given mutant, the ratio of its titer on Vero cells to that on the complementing V27 cells is a measure of how efficiently it is able to form plaques on Vero cells. The results indicate that the mutants fall into three classes in this respect. The first consists of mutants which are replication competent and includes mutants d2-3, d3-4, d5-6, and d6-7. As seen in Table 2, experiment 1, these mutants display Vero/V27 plaquing ratios which are ≥1 or above, similar to WT strain KOS1.1. In some experiments (experiment 1 and data not shown), it was noted that the d3-4 plaques on Vero cells were slightly smaller than WT plaques, suggesting a modest growth defect that has been previously noted (25). The second class of mutants consists of those which are replication deficient, i.e., mutants which are able to form plaques to some extent on Vero cells but not as efficiently as the WT virus. This class includes dLeu, d1-2, and d4-5. These mutants display Vero/V27 plaquing ratios that are significantly less than 1. Moreover, the Vero cell plaques formed by these mutants are significantly smaller than WT HSV-1 plaques. When tested side by side, d1-2 and dLeu generally showed approximately the same plaquing defect in Vero cells, being reduced 5- to 30-fold compared to WT HSV-1. d4-5 formed plaques somewhat more efficiently but still showed a 3- to 10-fold plaquing deficit. It is noteworthy that the plaquing ability of all three mutants in Vero cells varied somewhat from experiment to experiment. In some experiments, d1-2 and dLeu formed plaques that were very difficult to count accurately (data not shown). These observations suggest that some characteristic of the Vero cell cultures, such as cell density, passage number, or serum composition, may influence the ability of these ICP27 mutants to replicate or spread between cells. The third class of mutants are those which are replication incompetent and consists of a single mutant, d1-5. Like d27-1, d1-5 was unable to form plaques in Vero cells, within the limits of detection of the assay (103 PFU/ml). Lower dilutions of either viral stock could not be tested for plaque formation, because they led to cytopathic effects which completely destroyed the cell monolayer.
TABLE 2.
Plaque formation by HSV-1 ICP27 mutants
| Expt | Virus stock | Titer (PFU/ml) in cell linea:
|
Vero titer/V27 titer | |
|---|---|---|---|---|
| Vero | V27 | |||
| 1 | d27-1 | <1 × 103 | 5 × 108 | ≤0.000002 |
| d1-2 | 4 × 107b | 2 × 108 | 0.2 | |
| d1-2b | 2 × 107b | 2 × 108 | 0.1 | |
| d2-3 | 7 × 108 | 6 × 108 | 1.2 | |
| d2-3b | 6 × 108 | 5 × 108 | 1.2 | |
| d3-4 | 1 × 108c | 8 × 107 | 1.3 | |
| d3-4b | 4 × 108c | 4 × 108 | 1.0 | |
| d4-5 | 1 × 108b | 3 × 108 | 0.33 | |
| d4-5b | 2 × 108b | 4 × 108 | 0.33 | |
| d5-6 | 1 × 109 | 1 × 109 | 1.0 | |
| d5-6b | 1 × 109 | 1 × 109 | 1.0 | |
| d6-7 | 7 × 108 | 5 × 108 | 1.4 | |
| d6-7b | 7 × 108 | 6 × 108 | 1.2 | |
| KOS1.1 | 3 × 109 | 2 × 109 | 1.5 | |
| 2 | d1-2a | 5 × 107b | 3 × 108 | 0.16 |
| dLeu | 2 × 107 | 9 × 107 | 0.22 | |
| dLeub | 2 × 107 | 1 × 108 | 0.2 | |
| KOS1.1 | 4 × 108 | 2 × 108 | 2.0 | |
| 3 | d27-1 | <1 × 103 | 3 × 108 | ≤0.0000033 |
| d1-2 | 8 × 106b | 2 × 108 | 0.04 | |
| d1-2b | 6 × 106b | 2 × 108 | 0.03 | |
| d4-5 | 1 × 107b | 1 × 108 | 0.1 | |
| d4-5b | 2 × 107b | 1 × 108 | 0.2 | |
| d1-5 | <1 × 103 | 1 × 108 | ≤0.00001 | |
| d1-5b | <1 × 103 | 8 × 107 | ≤0.000013 | |
| dLeu | 3 × 106b | 8 × 107 | 0.038 | |
| dLeub | 4 × 106b | 9 × 107 | 0.044 | |
| KOS1.1 | 1 × 109 | 5 × 108 | 2.0 | |
Titers of virus stocks were determined in parallel on monolayers of Vero or V27 cells. Plaques were stained on day 4 and counted, and the numbers were used to determine viral titers. In the case of d27-1 and d1-5, 103 PFU/ml is the limit of detection, since virus stock dilutions lower than 1,000-fold led to cytopathic effects which destroyed the Vero cell monolayers.
Very small plaques.
Small plaques.
We further characterized the replication abilities of the N-terminal mutants by carrying out single-cycle viral yield assays. Replicate cultures of Vero or V27 cells were infected in duplicate at an MOI of 1 with WT HSV-1, the ICP27 null mutant d27-1, or the various N-terminal deletion mutants. After 24 h, the infected cultures were harvested, and the yields of virus in the infected-cell lysates were determined by titration on permissive V27 cells (Fig. 4A). As expected, when the infections were carried out in V27 cells, both WT HSV-1 and the ICP27 mutants grew productively. However, the yields varied greatly when the infections were carried out in Vero cells. These results were consistent with the results of the plaquing assay, in that the mutants again fell into the same three classes. The mutants d2-3, d3-4, d5-6, and d6-7 were all replication competent, growing comparably to the WT virus and replicating as well in Vero cells as in V27 cells. In some repeat experiments, the mutants d2-3 and d3-4 showed a small reduction in virus yield in Vero cells compared to V27 cells (data not shown). The second category consisted of mutants dLeu, d1-2, and d4-5. These mutants were replication deficient, growing significantly less efficiently, by 1 to 2 log units, than the WT virus but more efficiently than d27-1. Finally, the mutant d1-5 was severely replication defective, being similar to d27-1 in its inability to yield significant viral progeny in Vero cells.
It is possible that the viruses which failed to replicate as efficiently as the WT in the experiment described above (dLeu, d1-2, 4-5, and d1-5) are delayed for growth and might have yielded higher levels of progeny if the infections were allowed to proceed for longer than 24 h. To investigate this possibility, a viral growth curve experiment was done, in which Vero cells were infected at an MOI of 1 and viral yields were determined at various times postinfection (Fig. 4B). The results indicated that dLeu, d1-2, and d4-5 replicate 1 to 2 log units less efficiently than WT virus even if the infections are carried out for as long as 44 h. In this experiment, d1-5 was indistinguishable from d27-1 in its severe replication defect, as at no time point did either virus produce any progeny above the limits of detection of the experiment.
DNA replication by mutant viruses.
Although it is not absolutely required for HSV-1 DNA replication, ICP27 significantly enhances the levels of viral DNA synthesis (21, 33). ICP27's ability to transactivate the expression of viral DE genes encoding DNA replication factors appears to be responsible for this effect (22, 50). We assessed the ability of various N-terminal mutants to replicate their DNA. Included in this experiment were the C-terminal mutant n504R, which has previously been shown to be able to efficiently stimulate viral DNA synthesis (33, 50), and the null mutant d27-1. Replicate cultures of Vero cells were mock infected or infected at an MOI of 10, and the cells were labeled with [3H]thymidine from 7 to 13 hpi. After purification of total infected-cell DNA, equal amounts of DNA (2.5 μg) were digested with BamHI. Fluorographic analysis of the dried gel (Fig. 5) indicated that several of the mutants replicated their DNA, as newly synthesized viral BamHI fragments were clearly visible. A comparison of viral DNA synthesis in the WT-infected cells (lane 1) and the d27-infected cells (lane 3) confirms the stimulatory effect of ICP27 on viral DNA synthesis. Of the N-terminal mutants analyzed, only d1-5 (lane 4) was incapable of stimulating viral DNA synthesis above the level seen in the d27-1 infection. All of the other N-terminal mutants, including dLeu, showed significant amounts of viral DNA replication.
FIG. 5.
Viral DNA replication by ICP27 mutants. Replicate cultures of Vero cells were mock infected or infected with WT HSV-1 or ICP27 mutants at an MOI of 10. The cells were labeled with [3H]thymidine from 7 to 13 hpi. After purification of total cellular DNA, equal amounts (2.5 μg) were digested with BamHI and electrophoresed on a 1% agarose gel. The gel was treated with a fluorography enhancing agent, dried, and exposed to X-ray film. An autoradiograph is shown. The lines to the right indicate the migration of viral BamHI restriction fragments.
Expression of viral mRNAs by the ICP27 deletion mutants.
We next examined the abilities of the ICP27 mutants to express some specific DE and L mRNAs which are known to be regulated by ICP27. For this series of experiments, we analyzed the N-terminal mutants which showed significant growth defects, i.e., dLeu, d1-2, d4-5, and d1-5. In addition, the null mutant d27-1 and the C-terminal mutant n504R were analyzed.
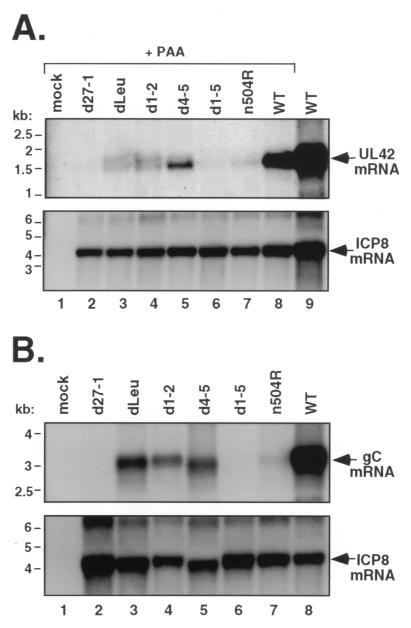
In the first experiment, expression of the DE UL42 gene, which encodes an essential DNA replication factor, was analyzed. Expression of UL42 mRNA is highly dependent on ICP27 (50). To control for differences in viral DNA replication among the mutant infections, which are known to affect UL42 levels (50), all infections except one with a duplicate WT sample were carried out in the presence of phosphonoacetic acid (PAA), a specific inhibitor of HSV-1 DNA replication. Total RNA was harvested from the infected cells at 6 hpi, and equal amounts were subjected to Northern blot analysis with a UL42 probe. The results (Fig. 6A) indicated that the mutants vary considerably in their ability to express the 1.7-kb UL42 transcript. Two mutants, d27-1 and d1-5, did not express any detectable UL42 mRNA at this time point. The four other mutants expressed detectable levels of UL42 mRNA but at levels that were much reduced compared to that for WT virus (lane 8). Among these mutants, d4-5 expressed the most UL42 mRNA, d1-2 and dLeu expressed lesser amounts, and n504 expressed the least. These results correlate with those of the DNA replication experiment (Fig. 5), in that all mutants expressing detectable UL42 mRNA in this assay (dLeu, d1-2, d4-5, and n504R) were competent for viral DNA replication, whereas those that were unable to express UL42 mRNA in this assay (d27-1 and d1-5) were deficient in DNA replication. To ensure that all of the cultures had been successfully infected, the Northern blot filter was stripped and reprobed for ICP8 mRNA. Like UL42, ICP8 is a DE gene encoding an essential DNA replication factor. However, its expression is not dependent upon ICP27 (34, 35, 50). The results (Fig. 6A) indicated that all of the cultures had been efficiently infected, since all of the ICP27 mutants expressed ICP8 mRNA at levels comparable to that for the WT virus.
FIG. 6.
Expression of UL42 and gC mRNAs in mutant-infected Vero cells. (A) Expression of UL42 mRNA. Replicate cultures of Vero cells were mock infected or infected at an MOI of 10 with the viruses indicated. Where indicated, PAA was included in the culture medium at a concentration of 400 μg/ml to inhibit viral DNA replication. At 6 hpi, total RNA was isolated, and equal amounts were analyzed by Northern blotting with a UL42 probe (top panel). The blot was then stripped and reprobed with an ICP8 mRNA-specific probe (bottom panel). (B) Expression of gC mRNA. Replicate cultures of Vero cells were mock infected or infected with the viruses indicated at an MOI of 10. At 9.5 hpi, total RNA was isolated and equal amounts were analyzed by Northern blotting with a gC gene-specific probe (top panel). After stripping of the blot, a second hybridization was performed using an ICP8-specific probe (bottom panel).
We next looked at the abilities of the mutants to stimulate the expression of an ICP27-dependent L gene. For this analysis, we chose the true-L gene encoding gC, which is highly dependent upon ICP27 (33, 40, 45). Since gC mRNA expression also requires viral DNA replication, infections were carried out in the absence of a DNA synthesis inhibitor. Total infected-cell RNA was isolated at 9.5 hpi, and gC mRNA levels were assessed by Northern blotting (Fig. 6B, top panel). The results indicate that, with the exception of d27-1 and d1-5, which were completely unable to express gC mRNA, the N-terminal mutants examined (dLeu, d1-2, and d4-5) were relatively proficient at producing gC message (lanes 3 to 5). In comparison, the n504R mutant, which replicates its DNA efficiently (Fig. 5), is much more restricted for gC mRNA expression (lane 7). Reprobing this same blot with an ICP8 probe indicated that all of the infections expressed abundant ICP8 mRNA, although the amounts varied slightly among the mutants. As a side note, it appeared that both the gC and ICP8 mRNAs expressed by the d4-5 mutant (lane 5) migrated slightly more rapidly than the corresponding mRNAs produced in the other infections. This effect was not observed when infections were carried out in the presence of PAA (Fig. 6A). The molecular basis of this apparent mRNA size difference is under investigation.
Protein synthesis by the mutant viruses.
To more globally assess viral gene expression by the various N-terminal mutants, we examined total viral and cellular protein synthesis at a late time after infection. Vero cells were mock infected or infected at an MOI of 10 with WT HSV-1, d27-1, n504R, or the various N-terminal mutants. The cells were metabolically pulse-labeled with [35S]methionine-cysteine from 9 to 9.5 hpi. Total cell proteins were separated by SDS-polyacrylamide gel electrophoresis, and the patterns of protein synthesis of the various infections were assessed by autoradiography (Fig. 7). As seen previously (33-35), the pattern of protein synthesis in d27-1-infected cells (lane 2) at late times was dramatically different from that of the WT virus (lane 12). The ICP27 null mutant overexpressed certain DE proteins, including ICP6 and ICP8, and dramatically underexpressed several viral L proteins, including ICP5 and ICP25. Several infection-specific proteins were undetectable or barely detectable in d27-1-infected cells. The remaining ICP27 mutants varied in their patterns of protein synthesis. The mutant that was the most clearly defective was d1-5 (lane 10). This mutant exhibited a protein synthesis pattern that was quite similar to that of d27-1, although it appeared to express slightly higher levels of ICP5 and some other infection-specific proteins. The C-terminal mutant n504R (lane 11) also showed severe defects in viral protein synthesis, as has been previously noted (33). In contrast, the other N-terminal mutants (dLeu, d1-2, d2-3, d4-5, d5-6, and d6-7) were much more like the WT virus than the null mutant in their patterns of protein synthesis. Notably, these mutants were able to induce expression of several infection-specific proteins that were not expressed to a significant extent in the d27-1, n504R, or the d1-5 infections (e.g., the proteins indicated by one and three asterisks in Fig. 7). However, several significant differences from the WT pattern were apparent. First, dLeu and d1-2 (lanes 3 and 4, respectively) showed reduced expression of several infection-specific proteins, such as ICP5, compared to the WT. Second, the mutant d4-5 expressed abundant ICP5 but appeared to be somewhat deficient in expressing some other infection-specific proteins, in particular the proteins indicated by one and two asterisks in Fig. 7. Third, several mutants (d2-3, d3-4, and d4-5) appeared to overexpress some infection-specific proteins, most notably ICP6. Two of the mutants, d5-6 and d6-7 (lanes 8 and 9, respectively), could not be distinguished from the WT virus in their patterns of protein synthesis.
FIG. 7.
Protein synthesis in ICP27 mutant-infected Vero cells. Replicate cultures of Vero cells were mock infected or infected at an MOI of 10 by the viruses indicated. At 9 to 9.5 hpi, the cultures were labeled with [35S]methionine-cysteine and total cell protein samples were prepared. The proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed by autoradiography. The positions at which molecular mass markers migrated are indicated at the left, and the positions of known viral proteins are indicated at the right. The positions of unidentified infection-specific proteins discussed in the text are indicated at the right by asterisks.
The acidic region of ICP27, between residues 21 and 63, is not required for viral growth or normal nucleocytoplasmic distribution.
In all of the experiments described above, the phenotype of d1-2 was found to be very similar to that of NES mutant, dLeu. The deletion in d1-2 was engineered prior to the discovery of NES motifs and was designed to remove the sequences encoding an acidic region of ICP27 between residues 12 and 63 (35). However, the deletion extends into the NES, removing several leucine residues (Fig. 1B). The common phenotype of d1-2 and dLeu suggests the possibility that the phenotype of d1-2 is due entirely to disruption of the NES rather than to deletion of the acidic residues. To more directly address the function of the acidic residues in this region of ICP27, we engineered a mutant, designated dAc, in which most of the N-terminal acidic residues (amino acids 21 to 63) were deleted but in which the NES was left intact (Fig. 1). As with the other mutants, two genetically independent viral isolates were obtained. Immunoblot analysis indicated that ICP27 polypeptides expressed by the dAc isolates were stable and of the expected size (Fig. 8A). To determine whether the deletion affects viral growth, we compared the abilities of the viral stocks to form plaques on Vero and V27 cells. This analysis (Fig. 8B) demonstrated that the dAc mutant has a WT plaquing phenotype, as it forms plaques on Vero cells as efficiently as it does on V27 cells. Furthermore, the size and morphology of dAc plaques on Vero cells are equivalent to those of WT HSV-1 (data not shown).
The region deleted in dAc has been reported to contain a sequence, termed the export control sequence, which negatively regulates the nuclear export of ICP27 (48). Thus, it is conceivable that the deletion of residues 21 to 63 might affect the nucleocytoplasmic distribution of ICP27. To test this, we carried out an immunofluorescence experiment. Localization of the dAc-encoded protein at 6 hpi was predominantly nuclear, with significant cytoplasmic distribution (Fig. 8C). This pattern was very similar to that of WT ICP27. In contrast, dLeu-encoded ICP27 was highly restricted to the cell nucleus, as observed in earlier experiments. We conclude that deletion of residues 21 to 63 does not significantly affect the growth or nucleocytoplasmic distribution of ICP27 in infected Vero cells.
DISCUSSION
The function of ICP27's N-terminal NES as defined by a viral deletion mutant.
Although there is evidence that ICP27 carries out multiple functions in infected cells, recent evidence suggests that a primary function of the protein is to stimulate the export of certain viral mRNAs to the cytoplasm (10, 20, 42, 47). The biological and biochemical properties of ICP27 support such a model in that the protein binds to RNA via an RGG box domain (27, 42) and possibly KH domain-like regions (47) and shuttles continuously between the nucleus and cytoplasm (26, 30, 42, 46). Consistent with its ability to exit the nucleus, ICP27 has a short leucine-rich sequence which is similar to the NES of human immunodeficiency virus type 1 Rev and many other cellular and viral proteins that interact with the CRM-1 nuclear export factor (8, 42). Furthermore, evidence has been presented that ICP27's leucine-rich motif functions as a bona fide NES in the context of viral infection (42). On the other hand, the biological importance of the leucine-rich NES has been called into question by a recent study which showed that the REF/TAP nuclear export system, not CRM-1, is responsible for ICP27-dependent HSV-1 mRNA export in Xenopus oocytes (20).
Given the above background, we wished to characterize the phenotype of an HSV-1 mutant that expresses an ICP27 molecule lacking its NES. Although no such mutant has been previously isolated, two studies have addressed the question of whether ICP27's NES is essential for viral infection. First, Zhi and Sandri-Goldin used a plasmid transfection-virus infection protocol to ask whether an NES-negative ICP27 expressed from a transfected plasmid can complement the growth of a viral ICP27 deletion mutant in rabbit skin cells (42, 52). No complementation above background levels was observed, suggesting that the NES is essential for viral growth. Second, Soliman and Silverstein engineered a mutant ICP27 gene having leucine-to-alanine coding substitutions in the NES sequence at codons 13 and 15 (47). This allele failed to generate viable recombinants in a marker rescue assay, again suggesting that the NES is essential for growth.
In the present study, we have directly addressed whether the NES is essential by isolating dLeu, an HSV-1 ICP27 NES deletion mutant. Somewhat surprisingly, dLeu is not completely replication defective. In a single-cycle infection, dLeu generates approximately 1,000-fold more progeny than an ICP27 null mutant (Fig. 4). It is also able to form plaques on Vero cells, although these plaques are smaller than WT HSV-1 plaques and arise with reduced efficiency. Thus, we conclude that the N-terminal leucine-rich region is not absolutely essential for viral growth in Vero cells. This is in contrast to certain sequences in the C-terminal half of ICP27 which completely eliminate virus growth when deleted or altered (24, 33, 34, 48).
On the other hand, our results also demonstrate that the NES is an important functional region of ICP27, since dLeu shows an approximately 100-fold reduction compared to the WT virus in Vero cells. Similar levels of growth attenuation are seen for dLeu when its growth is assayed in HeLa, HEp-2, or HEL human cell lines (J. Lengyel and S. A. Rice, unpublished data). The phenotype of dLeu in Vero cells can be used to infer the function of the amino-terminal leucine-rich region. First, loss of this sequence leads to a striking change in the nucleocytoplasmic distribution of ICP27. Whereas the WT protein shows substantial cytoplasmic localization in addition to its predominant nuclear distribution, the dLeu ICP27 polypeptide is highly restricted to the nucleus. This strongly suggests that the N-terminal leucine-rich region functions as an NES during viral infection, an interpretation consistent with the earlier conclusions of others (42). Second, loss of the NES leads to specific defects in gene expression. When examined by assaying late viral protein synthesis, these defects are not especially severe compared to the striking defects seen in ICP27 null mutant-infected cells (Fig. 7). However, several viral proteins are specifically reduced or lacking in dLeu-infected cells. Moreover, we found that the mRNA levels for two ICP27-dependent genes, UL42 and gC, are reduced in dLeu-infected cells.
The fact that gene expression defects of dLeu are not dramatic indicates that an ICP27 molecule lacking an NES can still efficiently activate many ICP27-dependent genes. If the activation of these genes requires that ICP27 exit the nucleus (e.g., if it functions as a mediator of mRNA export), then ICP27 may have a secondary NES or be able to associate with another NES-bearing cellular or viral protein. Alternately, ICP27 may activate viral genes by a mechanism which does not require nuclear export. In this regard, at least two other mechanisms have been proposed to explain how ICP27 might activate L genes. First, ICP27 may activate the expression of some L genes, which have inherently weak polyadenylation signals, by increasing the cleavage and polyadenylation of their transcripts (22). Second, ICP27 may directly stimulate the rates of transcription of some viral L genes (19). Presumably, neither of these mechanisms would require that ICP27 exit the nucleus.
The phenotype of dLeu in Vero cells appears to be identical to that of d1-2, an N-terminal deletion mutant that we have previously characterized (35). The d1-2 mutant has a deletion of residues 12 to 63, removing several of the leucine residues that comprise the NES as well as a number of acidic residues that are C terminal to them. Previously, we found that d1-2 was partially defective in ICP27's ability to stimulate viral DNA synthesis. In the present study, we found that both d1-2 and dLeu carried out relatively efficient DNA replication. We are unsure why the results for d1-2 differ quantitatively between the present and past studies; this may reflect, at least in part, the different times of [3H]thymidine labeling that were used in the two experiments. In any case, it appears from both studies that d1-2- and dLeu-encoded ICP27 molecules can stimulate substantial amounts of viral DNA synthesis in the absence of a functional NES.
Identification of two important functional regions in the N-terminal 200 residues of ICP27.
The phenotype of dLeu was characterized alongside those of eight other N-terminal deletion mutants. Together, the nine deletions extend over all portions of the N-terminal 200 residues, with the exception of the N-terminal four residues. It is very unlikely that unintended, secondary mutations contribute to the phenotype of any of these mutants, since two genetically independent isolates of each virus were found to be phenotypically identical. In addition, all mutant stocks were isolated and amplified in complementing V27 cells, thus avoiding any selection for suppressor mutations. Thus, our work allows for a relatively unbiased and systematic identification of the essential sequences in the N-terminal portion of ICP27. The results suggest that only two relatively small regions are required for efficient growth. The first is the leucine-rich NES, discussed above. The second is an RGG box RNA-binding domain, mapping to residues 139 to 153 (27). As with the deletion of the NES, deletion of the RGG box attenuates viral growth, but does not completely eliminate it. Like dLeu, the RGG box mutant d4-5 exhibits modest deficiencies in viral gene expression and grows poorly.
If one assumes that ICP27's RNA-binding activity is critical to its function, then it is somewhat surprising that d4-5 is only slightly attenuated. The RGG box is the only well-defined RNA-binding region in ICP27 (27), and it appears to be required for ICP27's interaction with RNA in vivo (43). However, some recent data suggest that ICP27 may also interact with RNA via C-terminal KH domain-like regions (47). It is also possible that direct RNA binding is not an essential activity of ICP27. ICP27 has been shown to physically interact with a number of cellular proteins involved in mRNA metabolism, including pre-mRNA splicing factors (5, 6, 44), hnRNP K (51), and the REF family of nuclear export factors (20). It is thus possible that ICP27 could function in posttranscriptional gene regulation primarily via protein-protein interactions. Perhaps the RGG box of ICP27 has evolved as a secondary interaction motif to strengthen ICP27's contacts in RNA-protein complexes.
It is significant that a relatively large N-terminal deletion which removes both the NES and the RGG box leads to a much more severe phenotype than deletion of either region alone. The virus bearing this deletion, d1-5, expresses a stable ICP27 polypeptide which shows a significant degree of nuclear localization. However, the protein is almost completely nonfunctional, as d1-5-infected cells exhibit severe defects in viral gene expression and DNA synthesis and are unable to produce significant amounts of viral progeny. Although several C-terminal ICP27 mutants are similarly defective for plaque formation (21, 33, 36, 47), d1-5 is the first example of an N-terminal mutant which is severely replication defective. The replication defect of d1-5 may be due to a synergistic negative effect of deleting both the NES and RGG box. It is also possible that the additional deletion of the sequences between the NES and RGG box (i.e., residues 20 to 138) plays a role in the lethal phenotype of the d1-5 mutation. This possibility is discussed further below.
Nonessential regions in the N-terminal portion of ICP27.
Our mutational analysis indicates that several regions in the N-terminal region of ICP27 (residues 21 to 63, 64 to 108, 109 to 138, 154 to 173, and 174 to 200) can be removed with very little or no effect on viral replication. Thus, these regions have no essential role in ICP27 function, at least in the context of the productive infection of Vero cells. As discussed below, evidence that at least two of these regions play a role in ICP27 function has previously been presented.
First, the acidic-region deletion in dAc has been reported to contain an export control sequence (ECS) at residues 31 to 34 which negatively regulates the activity of the NES (48). Moreover, Murata et al. found that an ICP27 gene alteration at codon 50 confers viral resistance to leptomycin B, an inhibitor of CRM-1-dependent nuclear export (29). Those authors suggested that the resistance mutation at codon 50 may negatively affect the function of the ECS and thus promote nuclear export. Our results are not readily consistent with the existence of an ECS in ICP27. However, it is important to point out that our immunofluorescence experiments show only steady-state protein localization and are therefore not a true assay for nuclear export. More direct assays for ICP27 nuclear export, such as interspecies heterokaryon assays (26), will have to be used to further test the existence of the ECS.
Second, it is interesting that the d3-4 viral mutant is replication competent. Koffa et al. recently showed that ICP27 directly interacts with the cellular REF nuclear export adaptors and demonstrated that this interaction is critical for ICP27-mediated mRNA export in Xenopus oocytes (20). The REF-interacting region of ICP27 was mapped to residues 10 to 138. Moreover, those investigators demonstrated that d3-4-encoded ICP27, lacking residues 109 to 138, does not detectably interact with REF in HeLa cells. Our demonstration that d3-4 virus replicates efficiently in Vero cells suggests that the REF interaction is not critical for ICP27 function during viral infection. Our results could be reconciled with those of Koffa et al., however, by hypothesizing that ICP27 can function by accessing either of two nuclear export systems: the CRM-1 system via the leucine-rich NES or the REF pathway via an interaction involving residues 109 to 138. With this in mind, it is interesting that the d1-5 mutation, which presumably eliminates contacts with both export pathways, leads to a severe block to viral growth. However, as pointed out above, the d1-5 protein also lacks the RGG box sequence, which clearly plays a role in ICP27 function. Our hypothesis could be tested by constructing a mutant in which the encoded ICP27 molecule retains its RGG box but lacks both the leucine-rich NES and the REF interaction region. The construction of such a mutant is under way in our laboratory.
Acknowledgments
We thank the other members of our laboratory, as well as Leslie Schiff and the members of her laboratory, for their cheerful help and thoughtful suggestions. Many thanks are also due to Keith Perkins and Anna Strain for critical reviews of the manuscript.
This research was supported by a grant from the National Institutes of Health (RO1-AI42737) to S.A.R.
REFERENCES
- 1.Aubert, M., S. A. Rice, and J. A. Blaho. 2001. Accumulation of herpes simplex virus type 1 early and leaky-late proteins correlates with apoptosis prevention in infected human HEp-2 cells. J. Virol. 75:1013-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 269:17401-17410. [PubMed] [Google Scholar]
- 3.Blaho, J. A., C. Mitchell, and B. Roizman. 1993. Guanylylation and adenylylation of the alpha regulatory proteins of herpes simplex virus require a viral beta or gamma function. J. Virol. 67:3891-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, C. R., M. S. Nakamura, J. D. Mosca, G. S. Hayward, S. E. Straus, and L. P. Perera. 1995. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J. Virol. 69:7187-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, H. E., D. A. Matthews, S. Wadd, J. E. Scott, J. Kean, S. Graham, W. C. Russell, and J. B. Clements. 2000. Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. J. Virol. 74:11322-11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant, H. E., S. E. Wadd, A. I. Lamond, S. J. Silverstein, and J. B. Clements. 2001. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 75:4376-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, P., K. S. Ellison, R. Verity, and J. R. Smiley. 2000. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated alpha-globin pre-mRNA in infected HeLa cells. J. Virol. 74:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 1998. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology 249:203-210. [DOI] [PubMed] [Google Scholar]
- 9.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252:162-178. [DOI] [PubMed] [Google Scholar]
- 10.Ellison, K. S., S. A. Rice, R. Verity, and J. R. Smiley. 2000. Processing of alpha-globin and ICP0 mRNA in cells infected with herpes simplex virus type 1 ICP27 mutants. J. Virol. 74:7307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frink, R. J., R. Eisenberg, G. Cohen, and E. K. Wagner. 1983. Detailed analysis of the portion of the herpes simplex virus type 1 genome encoding glycoprotein C. J. Virol. 45:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, M., and D. M. Knipe. 1989. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J. Virol. 63:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, M., and D. M. Knipe. 1993. Intragenic complementation of herpes simplex virus ICP8 DNA-binding protein mutants. J. Virol. 67:876-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardwicke, M. A., and R. M. Sandri-Goldin. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwicke, M. A., P. J. Vaughan, R. E. Sekulovich, R. O'Conner, and R. M. Sandri-Goldin. 1989. The regions important for the activator and repressor functions of herpes simplex virus type 1 alpha protein ICP27 map to the C-terminal half of the molecule. J. Virol. 63:4590-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes, R. G., Jr., and W. H. Munyon. 1975. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J. Virol. 16:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram, A., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J. Gen. Virol. 77:1847-1851. [DOI] [PubMed] [Google Scholar]
- 19.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 icp27 is required for transcription of two viral late (gamma2) genes in infected cells. Virology 283:273-284. [DOI] [PubMed] [Google Scholar]
- 20.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGregor, F., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J. Virol. 70:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLauchlan, J., A. Phelan, C. Loney, R. M. Sandri-Goldin, and J. B. Clements. 1992. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J. Virol. 66:6939-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahan, L., and P. A. Schaffer. 1990. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J. Virol. 64:3471-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mears, W. E., V. Lam, and S. A. Rice. 1995. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol 69:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128-137. [DOI] [PubMed] [Google Scholar]
- 27.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosca, J. D., P. M. Pitha, and G. S. Hayward. 1992. Herpes simplex virus infection selectively stimulates accumulation of beta interferon reporter gene mRNA by a posttranscriptional mechanism. J. Virol. 66:3811-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murata, T., F. Goshima, T. Koshizuka, H. Takakuwa, and Y. Nishiyama. 2001. A single amino acid substitution in the ICP27 protein of herpes simplex virus type 1 is responsible for its resistance to leptomycin B. J. Virol. 75:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phelan, A., and J. B. Clements. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 78:3327-3331. [DOI] [PubMed] [Google Scholar]
- 31.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 32.Rice, S. A., and D. M. Knipe. 1988. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J. Virol. 62:3814-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, S. A., and V. Lam. 1994. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J. Virol. 68:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, S. A., V. Lam, and D. M. Knipe. 1993. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J. Virol. 67:1778-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68:988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, S. A., L. S. Su, and D. M. Knipe. 1989. Herpes simplex virus alpha protein ICP27 possesses separable positive and negative regulatory activities. J. Virol. 63:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roizman, B., and D. M. Knipe. 2001. Herpes simplex virus and their replication, p. 2399-2460. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 39.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2398. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 40.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandri-Goldin, R. M. 1998. Interactions between a herpes simplex virus regulatory protein and cellular mRNA processing pathways. Methods 16:95-104. [DOI] [PubMed] [Google Scholar]
- 44.Sandri-Goldin, R. M., and M. K. Hibbard. 1996. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J. Virol. 70:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, I. L., M. A. Hardwicke, and R. M. Sandri-Goldin. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 186:74-86. [DOI] [PubMed] [Google Scholar]
- 46.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soliman, T. M., and S. J. Silverstein. 2000. Herpesvirus mRNAs are sorted for export via Crm1-dependent and -independent pathways. J. Virol. 74:2814-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soliman, T. M., and S. J. Silverstein. 2000. Identification of an export control sequence and a requirement for the KH domains in ICP27 from herpes simplex virus type 1. J. Virol. 74:7600-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer, C. A., M. E. Dahmus, and S. A. Rice. 1997. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J. Virol. 71:2031-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 70:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wadd, S., H. Bryant, O. Filhol, J. E. Scott, T. Y. Hsieh, R. D. Everett, and J. B. Clements. 1999. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 274:28991-28998. [DOI] [PubMed] [Google Scholar]
- 52.Zhi, Y., and R. M. Sandri-Goldin. 1999. Analysis of the phosphorylation sites of herpes simplex virus type 1 regulatory protein ICP27. J. Virol. 73:3246-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]