Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) latently infects KS tumors, primary effusion lymphomas (PELs), and PEL cell lines. K12 (T0.7) is the most abundant transcript expressed in latent KSHV infection. We characterize here the K12 transcript from a PEL tumor prior to passage in cell culture. The PEL tumor KSHV K12 transcript contained additional complex nucleotide repeat elements compared to the previously described K12 message of the BCBL-1 PEL cell line. The PEL tumor lacked kaposin B, the predominant BCBL-1 K12 protein product, but encoded kaposins A and C. The K12 transcript was spliced and the splicing event occurred in all KSHV isolates tested. The 5′ end of the K12 transcript was mapped by 5′ RACE (rapid amplification of cDNA ends) and S1 nuclease protection assays and was at the site of an active promoter. This work demonstrates that the K12 transcript contains variable, complex repeat elements, is spliced and is expressed from a novel KSHV promoter.
Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus-8 (HHV-8) is a gamma-2 herpesvirus tightly linked to KS, primary effusion lymphoma (PEL), and multicentric Castleman's disease, an aggressive lymphoproliferative disorder (6, 8, 18, 30). KSHV infection in tumors and PEL cell lines is predominantly latent, and latently infected cells express a limited number of KSHV genes. The latency-associated nuclear antigens 1 and 2 (5, 15, 16, 23, 25), v-cyclin (7, 9, 11), v-FLIP (2, 33), K15 (10, 13, 22, 29), and the T0.7 (27, 35) transcript are expressed in latent infection.
T0.7 was originally described as a 0.7 kb, highly abundant transcript in KS which initiated just upstream of the KSHV K12 open reading frame (ORF) (35). K12 encodes a 60-amino-acid protein termed kaposin A. In situ hybridization analyses by using probe to detect T0.7 sequence demonstrate that it is present in nearly all spindle cells, in all stages of KS, and is also found in PEL cell lines (31, 32). Subsequent work demonstrated that the K12 transcript is almost always much longer than 0.7 kb. Northern blots of KSHV-infected tumors and PEL cell lines probed with T0.7 lacked 0.7-kb transcripts and instead had K12 transcripts which varied between ∼1.5 and ∼2.5 kb, depending on the tumor or cell line (27). In fact, the initial tumor that had the 0.7-kb fragment also had a larger transcript detected by the T0.7 probe (27, 35). These longer transcripts extend further upstream from the initially reported 0.7-kb start site. The upstream sequence contains two GC-rich repeat sequences termed DR2 and DR1. The number of DR2 and DR1 repeat units vary among different KSHV isolates, resulting in variable sizes of the K12 transcripts (27).
Protein expression from the K12 transcript is complex and incompletely understood (17, 27). Investigation of the BCBL-1 (27) PEL cell line K12 transcript showed that expression occurs from more than one reading frame. Kaposin A (frame 1) initiates with an AUG codon but other expressed ORFs initiate from non-AUG codon(s). For instance, the BCBL-1 K12 transcript expresses kaposin B (frame 2) from a CUG codon (27). Kaposin B is the predominant protein expressed by the BCBL-1 K12 transcript, although other polypeptides are also expressed at lower levels. Kaposin B is largely comprised of DR2 (HPRNPARRTPGTRRGAPQEPGAA) and DR1 (PGTWCPPPREPGALLPGNLVPSS) repeat elements (27). Intriguingly, the amino acid sequences of DR2 and DR1 remain the same in all three reading frames. Kaposin C (frame 1) fuses kaposin A with upstream DR2/DR1 repeats and also initiates from a CUG.
Kaposin A has multiple functions in single gene transfer assays (17, 19, 20, 34). It induces focus formation in Rat-3 cells and NIH 3T3 cells and causes anchorage-independent growth and loss of contact inhibition in NIH 3T3 cells. Rat-3 cells expressing kaposin A are tumorigenic in nu/nu mice (19). Kaposin A induces reorganization of cellular F-actin and causes increased adhesion in Jurkat cells (17). The transforming and adhesion effects of kaposin A are mediated through its association with cytohesin-1, a guanine nucleotide exchange factor, which regulates integrin activity (17). In fact, a dominant-negative cytohesin-1-inhibited kaposin A induced focus formation and restored normal actin organization.
We investigated the K12 transcript from a PEL tumor (14) prior to passage in cell culture and found significant differences from the BCBL-1 sequence (27). The PEL tumor K12 sequence contained additional, complex repeat elements upstream of the DR2 and DR1 repeats. Kaposin B, which encodes the predominant BCBL-1 K12 expression product, was absent from the PEL tumor K12 transcript. The K12 transcript was spliced and the splicing event was conserved among KSHV isolates. Novel KSHV promoter activity was identified at the K12 transcription initiation site.
MATERIALS AND METHODS
Cell lines.
BCBL-1 (24), BC-1 (6), BC-2 (6), BC-3 (1), and BCP-1 (3) are KSHV-infected PEL cell lines.
Northern blot analysis.
A 4-mm punch biopsy was obtained with informed consent from a patient with AIDS-related KS according to the human experimental guidelines of the U.S. Department of Health and Human Services and Beth Israel Deaconess Medical Center. Total RNA was isolated from the PEL, PEL cell lines, and the KS biopsy with Trizol Reagent (Gibco) and poly(A) RNA was purified with Messagemaker Reagent Assembly (Gibco). A total of 25 μg of total RNA or 1.5 μg of poly(A) RNA was resolved on a 1% agarose formaldehyde gel, transferred onto a nylon filter, and probed. Primers K12F (GCCCTCGATACGCCTGCTCT) and K12R (TGCCCTCCTCCCTCCTCACT) were used to PCR amplify DNA from within the T0.7 sequence. The amplification product was 32P labeled (T0.7 probe) and used to probe the nylon filter.
cDNA library construction and screening.
A total of 5 μg of poly(A) RNA was used as a template for each cDNA library. The SUPERSCRIPT Choice System (Gibco) was used to construct cDNA libraries according to the manufacturer's directions except for first-strand cDNA synthesis. In order to enhance synthesis through GC-rich regions, first-strand synthesis was modified. First-strand synthesis for the oligo(dT) primed cDNA library was performed at 50°C for 1 h instead of 37°C. First-strand synthesis for random hexamer-primed cDNA library was performed by first incubation at 37°C for 15 min to allow the primers to anneal to the template RNA and the initial synthesis to occur. An additional 2 μl of Superscript II RT was then added, and the temperature increased to 50°C for 50 min. A total of 150 ng of each cDNA was ligated into Lambda ZAP II (Stratagene) and packaged into Lambda phages by using Gigapack Gold III Packaging extract (Stratagene). A total of 3 × 105 cDNA clones from the oligo(dT) primed library and 3 × 105 clones from the random hexamer primed library were screened with K12 probe. Seven positive clones that contained single inserts were purified from the oligo(dT) primed cDNA library and one positive clone containing a single insert was purified from the random hexamer primed cDNA library.
In vivo excision of cDNAs and sequencing.
In vivo excision of pBluescript containing cDNA inserts from the lambda phage was performed according to the manufacturer's instructions (Stratagene). DNA sequencing was performed at local core facilities. Extensive repeat regions were sequenced by using exonuclease III (Exo-Size Deletion Kit; NEB) to generate unidirectional nested deletions.
RT-PCR and PCR.
Access RT-PCR System (Promega) was used for reverse transcription-PCR (RT-PCR) according to the manufacturer's directions. The DR2/DR1 repeat region was PCR amplified from genomic PEL DNA and RT-PCR amplified from PEL RNA with primers 5′P2 (GATTTACACGTATCGAGGAG; nucleotides [nt] 118703 to 118722) and ZPPA low (CTATCCATGCATTGGGATTG; nt 118094 to 118113 [26]). Primers Splice1 (nt 118698 to 118718) CACCGCTCCTCGATACGTGTA and Splice2 (nt 123630 to 123611) AACCTGACAGAGCACCCTGA amplify a 138-bp product across the K12 mRNA splice site. In the absence of splicing, the PCR product is ∼5.0 kb. PCR enhancer buffer was added to the PCRs which were amplified at 95°C for 3 min, followed by 35 cycles of 95°C for 45 s, 46°C for 30 s, and 68°C for 1.5 min, followed by 68°C for 7 min.
5′ RACE.
5′ RACE System (version 2; Gibco) was used according to the manufacturer's instructions. 5′ RACE (rapid amplification of cDNA ends) was performed on total RNA from PEL tumor cells. The primers were GSP1 (CTCCTCGATACGTGTAAATC), GSP2 (ATCCAAGAGATCCGTCCTC), and GSP3 (CAUCAUCAUCAUCTTGTCTTTATAGCGTTTC). RT-PCR amplified products were cloned into pAMP1 vector.
S1 nuclease protection assay.
PEL DNA between nt 123303 and 123983 was PCR amplified by using primers S1UP (CACCTGCTTTATAAGTAGGA) and S1 DOWN (ATTGTCAGAACAAAGACACA). The PCR product was cloned into PCR2.1 (Invitrogen) and sequenced. S1 nuclease protection was performed with the S1 Assay kit (Ambion) with 5 μg of PEL poly(A) RNA. Probe was generated by linearizing PCR2.1 containing the PCR product 3′ to the insert with HindIII, priming with T7, and transcribing with T7 RNA polymerase with [32P]UTP. After digestion with S1 nuclease, fragments were size separated on an 8 M urea-6% polyacrylamide gel and then visualized by autoradiography.
Plasmids.
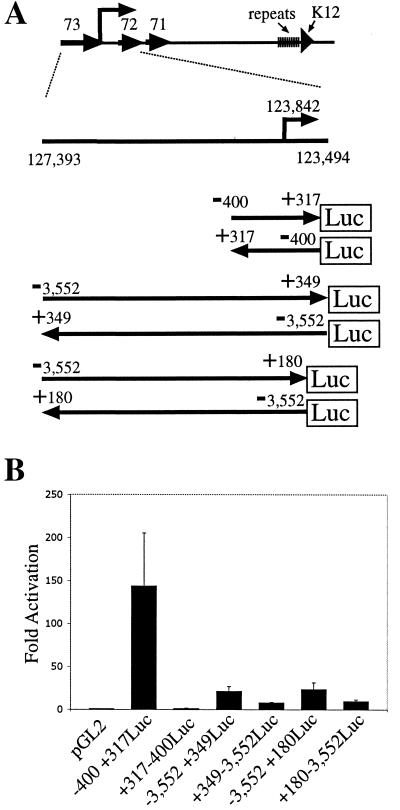
Since the in vivo excision process caused alterations in the K12 repeat region, a reconstructed K12 cDNA clone was made by substituting PCR-amplified, sequence-confirmed repeat elements between the NsiI and BsaAI restriction sites in K12 cDNA. −400+317Luc was generated by inserting the BamHI (nt 123526) to AgeI (nt 124241) fragment into pGL2 Basic (Promega). +317−400Luc was generated by inserting the same BamHI/AgeI fragment in the opposite orientation in GL2 Basic. −3,552+349Luc was constructed by inserting the SspI (nt 123494) to SacI (nt 127393) fragment into pGL2 Basic. +349−3,552Luc was constructed by inserting the SspI/SacI fragment into pGL2 Basic in the opposite orientation. −3,552+180Luc was constructed by inserting the NheI (nt 123663) to SacI (nt 127393) fragment into pGL2 Basic. +180−3,552Luc was constructed by inserting the NheI/SacI fragment into pGL2 Basic in the opposite orientation.
Reporter assays.
Reporter DNA (40 μg) and 5 μg of a vector expressing β-galactosidase from an SV40 promoter were electroporated into 107 BJAB cells at 200 V and 960 μF in 400 μl at room temperature by using a Bio-Rad electroporator. At 48 h after transfection, the cells were lysed in luciferase lysis buffer (Promega), and the luciferase and β-galactosidase activities were determined by using an Optocomp I Luminometer (MGM Instruments). β-Galactosidase activity was used to normalize for transfection efficiency.
Nucleotide sequence accession number. The GenBank accession number for the K12 cDNA sequence reported here is AY157025.
RESULTS
Northern blot analysis of the K12 transcript.
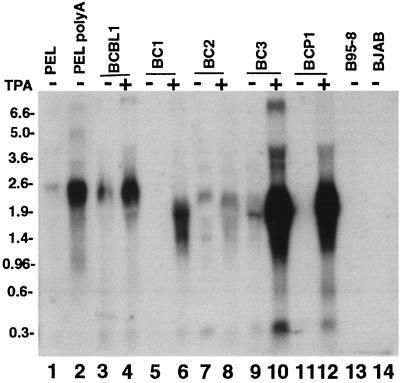
K12 transcripts of a primary PEL tumor prior to passage in cell culture and PEL cell lines were investigated by Northern blotting. K12 transcripts were detected with 32P-radiolabeled T0.7 probe, which contains sequence from the 3′ end of the K12 transcript. As expected, K12 transcript sizes varied since the number of DR2 and DR1 repeats vary among KSHV isolates (27). PEL cell line K12 transcripts were ∼2.5 kb for BCBL-1 (Fig. 1, lanes 3 and 4), ∼1.8 kb for BC-1 (Fig. 1, lane 6), ∼2.3 kb for BC-2 (Fig. 1, lanes 7 and 8), ∼1.8 kb for BC-3 (Fig. 1, lanes 9 and 10), and ∼2.0 kb for BCP-1 (Fig. 1, lane 12). BCBL-1 and BC-3 K12 transcript sizes were similar to other reports (27). Consistent with other observations, the expression of PEL cell line K12 transcripts increased after tetradecanoyl phorbol acetate (TPA) treatment (Fig. 1, lanes 4, 6, 8, 10, and 12) (27). In fact, K12 signal was not detected from BC-1 (Fig. 1, lane 5) and BCP-1 (Fig. 1, lane 11) prior to TPA treatment. K12 probe detected a ∼2.5-kb transcript from both total (Fig. 1, lane 1) and poly(A) (Fig. 1, lane 2) RNA of the PEL tumor. No signal was detected for the B95-8 (EBV-infected, KSHV-uninfected; Fig. 1, lane 13) and BJAB (uninfected; Fig. 1, lane 14) cell lines.
FIG. 1.
Northern blot of PEL cell lines and a PEL tumor for K12. Total RNA (25 μg) was loaded into each lane except for lane 2 in which poly(A) of RNA (1.5 μg) was loaded. RNA was size separated in a 1% agarose-formaldehyde gel and transferred to a nylon membrane, and signal was detected with 32P-labeled T0.7 probe. Cells were harvested directly or after pretreatment with 20 ng/ml of TPA (indicated at top). Total (lane 1) and poly(A) (lane 2) RNA from PEL tumor cells was probed. PEL cell lines BCBL-1 (lanes 3 and 4), BC-1 (lanes 5 and 6), BC-2 (lanes 7 and 8), BC-3 (lanes 9 and 10), and BCP-1 (lanes 11 and 12) are shown. B95-8 (lane 13) is an EBV-infected, KSHV-uninfected cell line. BJAB (lane 14) is an uninfected B lymphoma cell line. Size markers (in kilobases) are indicated at the left.
Generation of a PEL tumor cDNA library and sequencing of K12 cDNA clones.
In order to further investigate the K12 transcript from the PEL tumor, a lambda phage cDNA library was constructed from RNA harvested prior to cell passage, and K12-positive clones were isolated. The release of several K12 cDNA inserts from lambda DNA by EcoRI digestion revealed ∼2.1-kb K12 cDNAs (data not shown). After accounting for polyadenylation, this size is slightly smaller than the ∼2.5-kb K12 migration observed on Northern blotting of the PEL (Fig. 1, lanes 1 and 2), suggesting that the clones were not full length or that the transcript migrates slightly larger than predicted. Prior to sequencing the cDNAs, pBluescript plasmids containing K12 cDNAs were excised from parental lambda phage by in vivo excision after coinfecting bacteria with the lambda phage and helper phage (Stratagene). Sequencing the ends of seven clones from the oligo(dT)-primed cDNA library and one clone from the random hexamer-primed library demonstrated that the 5′ ends of all eight cDNAs were located within 32 nt of each other and the 3′ ends of the oligo(dT)-primed cDNAs were colinear with the previously described K12 (T0.7) 3′ end (27, 34). The proximity of the 5′ ends was consistent with the sequences being close to full length, or with stalling of the reverse transcriptase in this region. DR2 and DR1 repeats and also upstream, complex repeat elements were identified.
It was observed that the in vivo excision process introduced alterations within the central repeat elements since pBluescript cDNAs from single parental lambda phage isolates had variable sequences within these repeat elements. In contrast, the sequence of unique regions at the ends of the cDNAs remained constant. Therefore, the repeat region was further investigated by PCR so as to be independent of phage cloning and in vivo excision. PEL tumor DNA was PCR amplified, PEL tumor RNA was reverse transcribed and PCR amplified, and both products were sequenced. The PCR-amplified genomic DNA sequence was identical to that of the RT-PCR amplified RNA, demonstrating an absence of splicing events in this region. The invariable sequence results were also consistent with an accurate rendering of the repeat sequence. The complete K12 transcript sequence is shown in Fig. 2 with the repeat region from the PCR-amplified sequences.
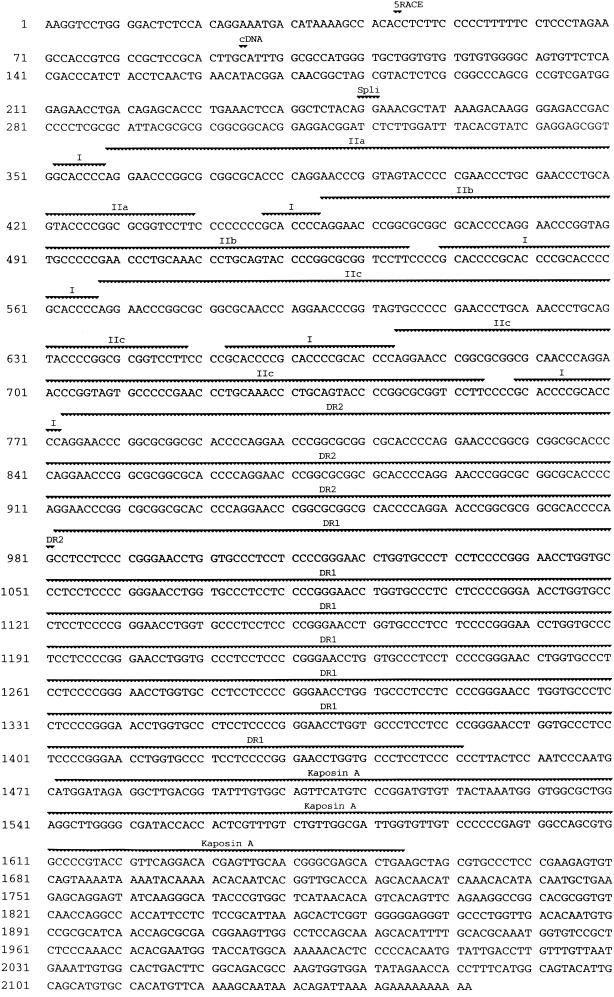
FIG. 2.
Sequence of the PEL K12 transcript. The 5′ ends mapped by cDNA and 5′ RACE analyses are indicated by “cDNA” (nt 95) and “5′RACE” (nt 44), respectively. Repeat elements I, IIa, IIb, IIc, DR2, and DR1 and the kaposin A ORF are indicated. The splice junction is labeled with “Spli” (positions 250 to 251). Sequence analyzed for T0.7 strain differences is from nt 1457 to 2104 (22).
The K12 message is spliced and contains complex GC-rich repeat elements upstream of the DR2 repeats.
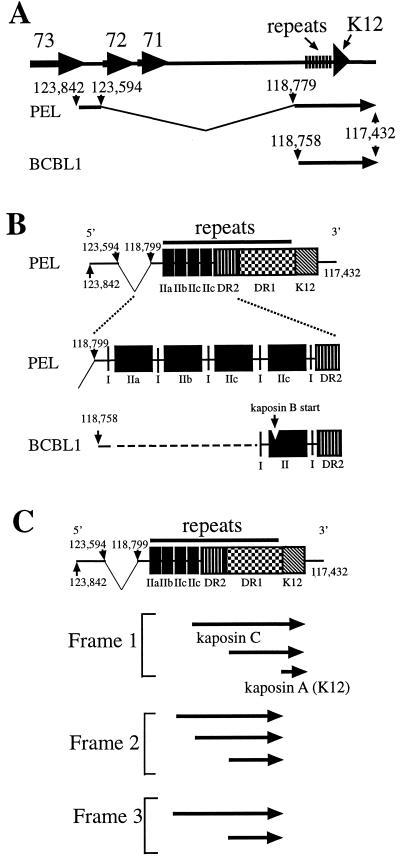
Analysis of the PEL tumor K12 cDNA sequences revealed significant differences from the previously described BCBL-1 sequence (27) (Fig. 3). First, all eight of the PEL tumor cDNA sequences were spliced in contrast to the BCBL-1 K12 transcript, which was unspliced (27). The splice junction was located at nt 118799, 41 nt upstream of the previously reported BCBL-1 transcription start site (nt 118758) (27). nt 118799 was spliced to nt 123594, resulting in introduction of a new exon at the 5′ end of the K12 transcript (Fig. 3A). Canonical splice site donor and acceptor sequences are present at the splice junction.
FIG. 3.
Schematic diagram of the K12 transcript. (A) PEL and BCBL-1 K12 transcripts in relation to the KSHV genome (26). The PEL transcript is spliced between nt 118779 and 123594 and intiates at nt 123842. The BCBL-1 transcript (27) initiates at nt 118758 and is unspliced. The 3′ ends of the PEL and BCBL-1 transcripts are coterminal. ORFs 73, 72, 71, and K12 are indicated. (B) The PEL tumor K12 transcript contains complex repeat elements upstream of the DR repeats. I, IIa, IIb, IIc, DR2, and DR1 repeat elements are indicated. The BCBL-1 type II element contains the CUG kaposin B initiation site which is absent in the PEL K12 transcript. (C) Potential PEL K12 transcript ORFs. Potential ORFs (of at least 30 amino acids) which initiate with CUG or GUG and include the DR1 repeats are indicated by arrows. Kaposin A initiates with ATG. Some arrows represent two potential ORFs with nearby initiation codons. Multiple potential ORFs are present within the DR1 repeats in all reading frames but only the largest such ORF for each reading frame is shown. The 5′ end of the transcript is at nt 123842 and the 3′ end is at nt 117432. Genome coordinates are from BC-1 (26).
A second major difference between the BCBL-1 K12 transcript and that of the PEL tumor was the presence of extensive GC-rich repeat elements upstream of the DR2 and DR1 repeat elements. The first repeat element (termed repeat I GCACCCC) (Fig. 2 and 3B) occurred in multiples of one to four copies of direct repeats and occurred at four locations. In contrast, BCBL-1 has two regions of type I elements (Fig. 3B), with one and two copies, respectively. The second repeat element is 81 nt and occurs in three variations in the PEL K12 transcript termed IIa, IIb, and IIc (Fig. 2 and 3B). Although the BCBL-1 K12 transcript does not contain repeats of type II, it has one copy of a type II element (27) (Fig. 3B). One copy of a type II element is also present in the two other previously reported KSHV genomic sequences of this region, from BC-1 and a KS tumor (21, 26). The BC-1 and BCBL-1 type II sequences are identical and termed II-BC1/BCBL-1, and the KS type II element is termed type II-KS. Table 1 summarizes the nucleotide differences between the type IIa, IIb, IIc, II-BC1/BCBL-1, and II-KS.
TABLE 1.
KSHV K12 type II element nucleotide variationa
| KSHV K12 element | nt at position:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 12 | 13 | 17 | 20 | 29 | 31 | 38 | 53 | 79 | 80 | 81 | |
| IIa | G | G | C | G | C | C | C | A | G | T | T | C |
| IIb | G | G | C | G | C | C | C | G | A | T | T | C |
| IIc | G | G | C | G | A | C | C | G | A | T | T | C |
| II-BC1/BCBL-1b | T | C | T | A | C | A | C | G | A | |||
| II-KS | T | C | T | A | C | C | A | G | A | |||
Numbers at the top indicate the nt number of the 81 nt type II elements. IIa, IIb, and IIc are present in PEL cells, II-BC1/BCBL-1 is present in BC-1 (26) and BCBL-1 (27) cells, and II-KS is present in a KS tumor (21).
II-BC1/BCBL-1 also has nt A inserted between positions 31 and 32.
In contrast to the type I and II repeats, the DR2 and DR1 repeat sequences were completely conserved between the PEL tumor and BCBL-1 and varied only in the number of copies. The number of DR2 and DR1 copies was initially expected to be similar to that of BCBL-1 since the transcript sizes of the PEL tumor (Fig. 1, lanes 1 and 2) and BCBL-1 (Fig. 1, lanes 3 and 4) were similar. However, the type I and II repeat elements contributed significantly to the transcript size and the PEL tumor contained only 9 DR2 and 20 DR1 elements compared to 17 DR2 and 39 DR1 elements (27) in BCBL-1.
The PEL K12 transcript encodes kaposins A and C but lacks kaposin B.
Analysis of the PEL K12 transcript demonstrates multiple potential ORFs in all three reading frames (Fig. 3C). Frame 1 encodes kaposin A and kaposin C. Frame 1 ORFs also initiate within the DR1 repeats. Frame 2 encodes potential ORFs which initiate within the first and second IIc elements and also within the DR1 repeats (Fig. 3C). Frame 3 of the PEL K12 transcript has an ORF which initiates in the first IIc element and ORFs which initiate within the DR1 repeats.
Comparison of ORFs encoded by the PEL K12 transcript with that of BCBL-1 (27) reveals that frame 1 is similar, but frames 2 and 3 differ significantly. Both the PEL and BCBL-1 transcripts encode frame 1 kaposin A and C and ORFs that initiate within the DR1 repeats. However, the only shared frame 2 feature are the ORFs which initiate within the DR1 repeats. BCBL-1 lacks the frame 2 PEL ORFs that initiate upstream of the DR1 repeats, and the PEL lacks kaposin B, which encodes the predominant protein expressed by the BCBL-1 K12 transcript (27). Kaposin B initiates at a CUG (27) within the type II element of BCBL-1 but this CUG is absent from the PEL K12 transcript due to variant nucleotides at positions 12 and 13 of the type IIa, IIb, and IIc elements in the PEL K12 transcript (Table 1). The PEL and BCBL-1 share frame 3 ORFs which initiate in the DR-1 repeats but BCBL-1 lacks any larger ORF in contrast to the PEL. Therefore, frame 2 and 3 K12 transcript ORFs which initiate upstream of the DR1 repeats differ significantly between the PEL and BCBL-1. However, ORFs which initiate within the DR1 repeats are shared between the PEL and BCBL-1 in all reading frames.
PEL tumor K12 sequence is a KSHV type B subgroup.
T0.7/K12 region sequences from geographically diverse KSHV isolates have been subtyped based on variations at certain nucleotides (between positions 1457 and 2104 in Fig. 2) (22). Based on these variations, the PEL tumor sequence is a type B3 subgroup and is the same as a PEL cell line, VG-1 (4) (Gary Hayward, unpublished data), which was generated from the same subject. Interestingly, four of six cDNAs had a G at position 1583 (Fig. 2), similar to an earlier reported cDNA sequence (35), but different from the A at this position in other reported genomic sequences (21, 26), including VG-1. Two of six cDNAs had an A at this position, suggesting that either RNA editing was occurring or that a mutation occurred at this site after infection of the subject. To further investigate this phenomenon, high-fidelity PCR (Vent polymerase; New England Biolabs) of genomic DNA was performed on this region, amplified DNA ligated into vector, and seven independent clones sequenced. All seven clones had G at nt 1583. Although RNA editing may be occurring, it is more likely that two closely related viruses are present, since the VG-1 cell line derived from the same patient has A at nt 1583 after PCR amplification of genomic DNA. The G at position 1583 changes the kaposin A codon to glycine from serine.
It is possible that the complex repeat elements upstream of the DR2 repeats in the PEL K12 transcript, which are largely absent in BCBL-1 (Fig. 3B), is a strain-related phenomenon. BC-1, a KS tumor, and BCBL-1 have a similar genomic sequence in the region, which differs from the PEL, and are KSHV subtypes A2, A/C, and A3, respectively (22). In contrast, the PEL tumor is KSHV subtype B3.
Identification of the initiation site of the K12 message by using 5′ RACE and S1 analyses.
Experiments were performed to investigate whether the initiation site of the K12 transcript extended upstream of the longest cDNA (Fig. 2, position 95) (BC-1 genomic position 123749 [26]). 5′ RACE was performed on total RNA from PEL tumor cells. Seventeen clones containing single K12 amplified inserts were obtained. Twelve clones had similar sized inserts, one clone had a larger insert and four clones had smaller inserts. Sequence analysis of 2 of the 12 clones with similar size inserts demonstrated 5′ ends corresponding to positions 90 and 95 of Fig. 2. Since the majority of the 5′ RACE clones were of similar size to these and because these 5′ ends were near the positions mapped by the cDNAs, it was likely that reverse transcriptase tended to stall at this region. Sequence analysis of the largest insert extended the 5′ end to position 44 of Fig. 2.
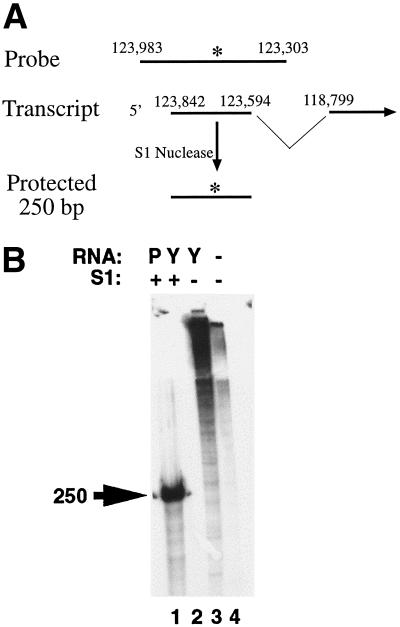
In order to further define the K12 transcript initiation site in a reverse transcriptase-independent manner, S1 nuclease protection experiments were performed. A [32P]UTP-radiolabeled ribonucleotide probe antisense to the K12 transcript and spanning between BC-1 positions 123983 and 123303 (26) (Fig. 4A) was hybridized to RNA and nonannealed, single-stranded regions were then digested with S1 nuclease. PEL tumor RNA (Fig. 4B, lane 1), but not yeast RNA (Fig. 4B, lane 2), protected a ∼250-bp fragment in the presence of S1 nuclease. In the absence of S1 nuclease (Fig. 4B, lanes 3 and 4) probe was not digested and migrated near the gel origin. These results extend the initiation site of the K12 transcript further upstream at or near BC-1 position 123842 (position 1 in Fig. 2).
FIG. 4.
S1 nuclease protection assay to map the 5′ end of the K12 transcript. (A) Schematic diagram of the S1 nuclease protection assay. The single-stranded [32P]UTP-radiolabeled RNA probe spans KSHV (26) nt 123983 to 123303 and extends into the K12 intron (nt 123593 to 118800). After S1 nuclease digestion, a ∼250-bp fragment is protected. nt 123842 is the initiation site of the K12 transcript as determined by S1 nuclease protection. The asterisk indicates the 32P radiolabel. (B) S1 nuclease protection assay. Radiolabeled probe was incubated with PEL tumor poly(A) RNA (lane 1), yeast RNA (lanes 2 and 3), or alone (lane 4) and digested with S1 nuclease (lanes 1 and 2). The protected ∼250-nt fragment in lane 1 is indicated by the arrow.
Splicing of the K12 transcript occurs in other KSHV isolates.
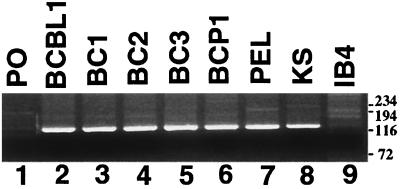
Experiments were performed to investigate whether the splicing event was unique to the PEL tumor or also occurred in other KSHV isolates. RT-PCR with primers flanking the splice sites was performed on RNA from the PEL cell lines BCBL-1, BC-1, BC-2, BC-3, and BCP-1 and a KS tumor. RT-PCR amplified a 135-bp fragment from all PEL cell lines (Fig. 5, lanes 2 to 6), the PEL tumor (Fig. 5, lane 7), and the KS tumor (Fig. 5, lane 8), a finding consistent with the presence of the splice junction. In the absence of splicing, the RT-PCR product was expected to be ∼5 kb. RT-PCR of an EBV-transformed lymphoblastoid cell line which lacks KSHV, IB4, did not amplify a 135-bp fragment (Fig. 5, lane 9). These results indicate that the K12 splicing event is common and perhaps universal among KSHV isolates, including in BCBL-1. However, it does not rule out the possibility that some transcripts may lack the splicing event. The splice extends the transcription initiation site significantly upstream (Fig. 3A).
FIG. 5.
RT-PCR of the K12 transcript splice region. RT-PCR with primers flanking the K12 splice sites amplify a 138-bp product after splicing has occurred but would amplify a ∼5-kb fragment in the absence of splicing. Lanes: 1, PO (primers only); 2 to 6, PEL cell lines BCBL-1, BC-1, BC-2, BC-3, and BCP-1, respectively; 7, PEL tumor; 8, KS tumor; 9, IB4 EBV-infected (KSHV-uninfected) lymphoblastoid cell line. Size markers are shown at the right in base pairs.
A novel KSHV promoter drives K12 transcription.
The region at the K12 transcriptional start site was assayed for promoter activity (Fig. 6). Reporter −400 + 317Luc, which contains 400-bp upstream and 317-bp downstream of the K12 transcription initiation site cloned upstream of the luciferase reporter gene (Fig. 6A), produced ∼150-fold activation in BJAB cells compared to pGL2 vector alone (Fig. 6B). Transfection of BJAB cells with −3,552+349Luc, which contains 3,552 bp upstream and 349 bp downstream of the start site upstream of luciferase (Fig. 6A), produced an activation 21-fold greater than that produced by vector alone (Fig. 6B). Reporter −3,552+180Luc, which contains 3,552 bp upstream and 180 bp downstream of the K12 start site upstream of luciferase (Fig. 6A), produced an activation 23-fold greater than that produced with vector alone (Fig. 6B). Since −3,552+349Luc and −3,552+180Luc had a lower activity than did −400+317, sequence upstream of position −400 appears to dampen promoter activity. The promoter activity of −400+317Luc, −3,552+349Luc, and −3,552+180Luc were orientation specific since +317−400Luc, +349−3,552Luc, and +180−3,552Luc, which contain the promoter in reverse orientation relative to the luciferase gene (Fig. 6A), produced comparatively less or no activity (Fig. 6B).
FIG. 6.
Reporter assays of the K12 transcript promoter region. BJAB (uninfected) cells were co transfected with a luciferase reporter and a beta galactosidase expression vector. Luciferase values were normalized by beta galactosidase activity. (A) Schematic diagram of the K12 promoter region. The genome spanning between the SacI (nt 127393) and SspI (nt 123494) is shown in relation to the KSHV genome. K12 transcription initiates at nt 123842. Numbers indicate genomic positions (26). K12 promoter reporter constructs are shown. Promoter regions were cloned upstream (−400+317Luc, −3,552+349Luc, and −3,552+180Luc) or in reverse orientation (+317−400Luc, +349−3,552Luc, and +180−3,552Luc) of the luciferase (Luc) reporter gene in the pGL2 vector. (B) Fold activation of luciferase after transfection of pGL2 vector, −400+317Luc, +317−400Luc, −3,552+349Luc, +349−3,552Luc, −3,552+180Luc, and +180−3,552Luc into BJAB cells. The data shown are from four experiments. The standard deviation is shown with error bars.
DISCUSSION
We investigated the KSHV K12 transcript from a PEL tumor prior to passage in cell culture. In contrast to the BCBL-1 PEL cell line sequence (27), the K12 transcript contained additional highly complex repeat elements (types I and II) (Fig. 2 and 3B) upstream of the DR2 repeats. It is possible that this difference is due to KSHV strain differences as evidenced by their absence in non-subtype B KSHVs BCBL-1, BC-1, and a KS tumor (21, 22), although further investigation is needed to confirm whether this finding is a strain-related phenomenon. The PEL K12 transcript was spliced, and the splicing event was highly conserved among KSHV subtypes, including BCBL-1, as demonstrated by RT-PCR with primers flanking the splice site (Fig. 5). Since the previously reported BCBL-1 transcript (27) was unspliced, it is possible that there are two species of BCBL-1 K12 mRNAs, one of which is unspliced, or that there was incomplete 5′ mapping of the BCBL-1 transcript. Consistent with the finding of this splice junction in multiple isolates, the canonical splice donor and acceptor sequences are preserved in all reported sequences of this region.
The absence of K12 signal on Northern blotting of the BC-1 and BCP-1 PEL cell lines and the relatively low K12 signal of other PEL cell lines prior to TPA treatment (Fig. 1) raises the possibility that passage in cell culture alters expression of the K12 transcript. KSHV latently infects the vast majority of cells in KSHV-infected tumors or PEL cell lines, and only a small percentage of cells undergo lytic KSHV infection. TPA treatment of PEL cell lines dramatically increases the percentage of cells undergoing lytic infection. The absent or low levels of K12 signal on Northern blots and the high level of induction after TPA treatment are consistent with K12 transcript expression during lytic infection in PEL cell lines. In contrast, in primary KS tumors, in situ hybridization for K12 (T0.7) message demonstrates signal in nearly all spindle cells (31, 32) and Northern blotting of KS tumors detects K12 signal (27) consistent with latent infection. Interestingly, Northern blot analysis of the VG-1 PEL cell line, which was derived from the same tumor as the primary PEL cells used in this work, had barely detectable K12(T0.7) message. However, incubation of VG-1 cells with TPA upregulated the VG-1 K12 message to a level similar to that of the PEL tumor (data not shown). LANA1 expression was present in the majority of PEL tumor cells (14), a finding consistent with latent infection, so it is unlikely that the K12(T0.7) message in the PEL tumor cells resulted from lytic infection.
Despite the sequence variation upstream of the DR2 repeats, complete DR2 and DR1 sequence identity was maintained between the PEL tumor and BCBL-1. In fact, all reported sequences of these repeat elements are identical. DR2 and DR1 each encode different repetitive 23-amino-acid sequences that each remain the same in all reading frames (27). Since nucleotide sequence alterations can easily disrupt the amino acid sequence in at least one reading frame, the nucleotide sequence identity is essential to maintaining amino acid sequence identity in all reading frames. Therefore, DR2 and DR1 sequence identity among KSHV subtypes is consistent with an underlying importance for the virus's ability to express these proteins from all reading frames. This finding combined with the low conservation of sequence upstream of the DR2 elements, including the absence of the BCBL-1 kaposin B initiation codon in the PEL tumor, suggest that expression of the DR2 and DR1 repeat elements is of primary importance regardless of which initiation codons are used to express them. In fact, kaposin B, the BCBL-1 predominant K12 message expression product, is almost completely comprised of DR2 and DR1 repeats (27). Interestingly, although the number of DR2 and DR1 repeat elements vary significantly between KSHV subtypes, the ratio of DR2 to DR1 elements tends to remain at about 1 to 2. BC-1 contains 7 DR2 and 13 DR1 elements, BCBL-1 contains 17 DR2 and 39 DR1, a KS sequence contains 11 DR2 and 17 DR1 elements, and the PEL tumor contains 9 DR2 and 20 DR1 elements (Fig. 2) (21, 26, 27). Maintenance of this ratio of DR2 to DR1 repeats may be important for proper folding or structure of the proteins.
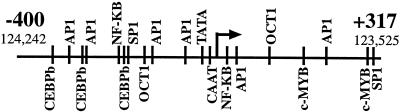
Reporter assays revealed potent promoter activity in the region surrounding the K12 transcriptional start site. This start site is located in the 3′ end of ORF73 (LANA1, LNA, and LNA1) and just upstream of ORF72 (v-cyclin) (Fig. 3A). Of note, despite the proximity to the K12 promoter, ORF72 is expressed from a promoter upstream of LANA1 as part of a multicistronic transcript (12, 28). A TATA box is located ∼35 nt upstream of the K12 start site and a CAAT element at ∼25 nt upstream of the start site (Fig. 7). The −400+317Luc reporter exerted robust activity in BJAB cells (Fig. 6B). Consistent with the strong promoter activity of −400+317Luc, there are multiple potential transcription factor binding sites located both upstream and downstream of the transcription initiation site, including sites for SP1, NF-κB, Oct1, and AP-1 (Fig. 7). The multiple AP1 sites are consistent with the promoter's responsiveness to TPA (Fig. 1) (27). Regulation of the promoter activity also appears to be exerted by sequences more than 400 bp upstream of the start site since inclusion of such sequence in −3,552+349Luc and −3,552+180Luc resulted in reduced reporter activity (Fig. 6B).
FIG. 7.
Schematic diagram of the K12 promoter region from −400 to +317. The arrow indicates the transcription initiation site. Potential transcription factor binding sites are indicated. The BC-1 genomic coordinates are also given (26).
The K12 locus is clearly complex. It encodes many potential ORFs, but the relative importance of the different ORFs is not clear. In fact, although kaposin A has multiple functions (17, 19, 20, 34), the role(s) of the DR2/DR1-containing proteins remain to be defined. The conservation of DR2 and DR1 repeat sequences among KSHV subtypes is consistent with important function(s) exerted by these proteins, which future work should elucidate.
Acknowledgments
H.L. and T.K. contributed equally to this work.
We thank Elliott Kieff, Mary Ballestas, Mark Birkenbach, and Gary Hayward for helpful discussions and Jo Ann Proper for assistance with biopsy material. Mary Ballestas harvested PEL DNA and RNA.
This work was supported by grants CA82036 (K.M.K.) and T32 CA09031 (T.K.) from the National Cancer Institute.
REFERENCES
- 1.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. SAid, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 2.Bertin, J., R. C. Armstrong, S. Ottilie, D. A. Martin, Y. Wang, S. Banks, G. H. Wang, T. G. Senkevich, E. S. Alnemri, B. Moss, M. J. Lenardo, K. J. Tomaselli, and J. I. Cohen. 1997. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc. Natl. Acad. Sci. USA 94:1172-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff, C., S. J. Gao, L. E. Healy, S. Matthews, A. J. Thomas, L. Coignet, R. A. Warnke, J. A. Strauchen, E. Matutes, O. W. Kamel, P. S. Moore, R. A. Weiss, and Y. Chang. 1998. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood 91:1671-1679. [PubMed] [Google Scholar]
- 4.Brander, C., T. Suscovich, Y. Lee, P. T. Nguyen, P. O'Connor, J. Seebach, N. G. Jones, M. van Gorder, B. D. Walker, and D. T. Scadden. 2000. Impaired CTL recognition of cells latently infected with Kaposi's sarcoma-associated herpes virus. J. Immunol. 165:2077-2083. [DOI] [PubMed] [Google Scholar]
- 5.Burysek, L., and P. M. Pitha. 2001. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J. Virol. 75:2345-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 7.Cesarman, E., R. G. Nador, F. Bai, R. A. Bohenzky, J. J. Russo, P. S. Moore, Y. Chang, and D. M. Knowles. 1996. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J. Virol. 70:8218-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382:410. [DOI] [PubMed] [Google Scholar]
- 10.Choi, J. K., B. S. Lee, S. N. Shim, M. Li, and J. U. Jung. 2000. Identification of the novel K15 gene at the rightmost end of the Kaposi's sarcoma-associated herpesvirus genome. J. Virol. 74:436-446. [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, M. A., M. A. Sturzl, C. Blasig, A. Schreier, H. G. Guo, M. Reitz, S. R. Opalenik, and P. J. Browning. 1997. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 89:1868-1874. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glenn, M., L. Rainbow, F. Aurad, A. Davison, and T. F. Schulz. 1999. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J. Virol. 73:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, D., M. E. Ballestas, K. M. Kaye, J. M. Gulizia, G. L. Winters, J. Fletcher, D. T. Scadden, and J. C. Aster. 1998. Primary-effusion lymphoma and Kaposi's sarcoma in a cardiac-transplant recipient. N. Engl. J. Med. 339:444-449. [DOI] [PubMed] [Google Scholar]
- 15.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 17.Kliche, S., W. Nagel, E. Kremmer, C. Atzler, A. Ege, T. Knorr, U. Koszinowski, W. Kolanus, and J. Haas. 2001. Signaling by human herpesvirus 8 kaposin A through direct membrane recruitment of cytohesin-1. Mol. Cell 7:833-843. [DOI] [PubMed] [Google Scholar]
- 18.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with or without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 19.Muralidhar, S., A. M. Pumfery, M. Hassani, M. R. Sadaie, N. Azumi, M. Kishishita, J. N. Brady, J. Doniger, P. Medveczky, and L. J. Rosenthal. 1998. Identification of Kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J. Virol. 72:4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muralidhar, S., G. Veytsmann, B. Chandran, D. Ablashi, J. Doniger, and L. J. Rosenthal. 2000. Characterization of the human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) oncogene, kaposin (ORF K12). J. Clin. Virol. 16:203-213. [DOI] [PubMed] [Google Scholar]
- 21.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole, L. J., J. C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. G. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus is encoded by ORF73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renne, R., W. Zhong, B. Herdier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 25.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Eman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp, T. V., H. W. Wang, A. Koumi, D. Hollyman, Y. Endo, H. Ye, M. Q. Du, and C. Boshoff. 2002. K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J. Virol. 76:802-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M.-F. d'Agay, J.-P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 31.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle tumor) cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturzl, M., C. Blasig, A. Schreier, F. Neipel, C. Hohenadl, E. Cornali, G. Ascherl, S. Esser, N. H. Brockmeyer, M. Ekman, E. E. Kaaya, E. Tschachler, and P. Biberfeld. 1997. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi's sarcoma. Int. J. Cancer 72:68-71. [DOI] [PubMed] [Google Scholar]
- 33.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meini, F. Neipel, C. Mattrmann, K. Burns, J.-L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. I. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins(FLIPs) prevent apoptosis induced by death receptors. Nature. 386:517-521. [DOI] [PubMed] [Google Scholar]
- 34.Tomkowicz, B., S. P. Singh, M. Cartas, and A. Srinivasan. 2002. Human herpesvirus-8 encoded Kaposin: subcellular localization using immunofluorescence and biochemical approaches. DNA Cell Biol. 21:151-162. [DOI] [PubMed] [Google Scholar]
- 35.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]