Abstract
Dendritic cells (DCs) are among the first cells encountered by human and simian immunodeficiency virus (HIV and SIV) following mucosal infection. Because these cells efficiently capture and transmit virus to T cells, they may play a major role in mediating HIV and SIV infection. Recently, a C-type lectin protein present on DCs, DC-specific ICAM-3-grabbing nonintegrin (DC-SIGN), was shown to efficiently bind and present HIV and SIV to CD4+, coreceptor-positive cells in trans. However, the significance of DC-SIGN for virus transmission and pathogenesis in vivo remains unclear. Because SIV infection of macaques may represent the best model to study the importance of DC-SIGN in HIV infection, we cloned and characterized pig-tailed macaque DC-SIGN and generated monoclonal antibodies (MAbs) against it. We demonstrate that, like human DC-SIGN, pig-tailed macaque DC-SIGN (ptDC-SIGN) is expressed on DCs and macrophages but not on monocytes, T cells, or B cells. Moderate levels of ptDC-SIGN expression were detected on the surface of DCs, and low-level expression was found on macrophages. Additionally, we show that ptDC-SIGN efficiently binds and transmits replication-competent SIVmne variants to CD4+, coreceptor-positive cells. Moreover, transmission of virus between pig-tailed macaque DCs and CD4+ T cells is largely ptDC-SIGN dependent. Interestingly, MAbs directed against ptDC-SIGN vary in the capacity to block transmission of different SIVmne variants. These data demonstrate that ptDC-SIGN plays a central role in transmitting virus from macaque DCs to T cells, and they suggest that SIVmne variants may differ in their interactions with ptDC-SIGN. Thus, SIVmne infection of pig-tailed macaques may provide an opportunity to investigate the significance of DC-SIGN in primate lentiviral infections.
Transmission and dissemination of human immunodeficiency virus (HIV) may depend on its ability to hijack normal trafficking of dendritic cells (DCs) from mucosal sites of infection to draining lymph nodes. Immature DCs found in peripheral tissues efficiently capture antigens (3, 25, 28, 45) and migrate to draining lymph nodes where they elicit T-cell responses. During transit to secondary lymphoid organs, they undergo maturation into competent antigen-presenting cells and acquire a greater ability to attract and activate T cells (1, 3, 22, 47). Interestingly, in situ studies of macaques infected intravaginally with simian immunodeficiency virus (SIV) demonstrate that the infecting virus first encounters DCs, and perhaps macrophages, in the vaginal and cervical epithelium (8, 13, 20, 24, 31, 44). Within a few days, virus is localized to T-cell zones of draining lymph nodes (20, 31, 44). These data suggest that virus infection may be initiated in DCs, but the initial burst of replication and continuous production of virus may occur primarily in CD4+ T cells within lymph nodes.
The ability of DC-T-cell interactions to support HIV and SIV replication has been modeled in vitro using DCs and T cells derived from skin explants as well as blood (6, 21, 34, 38, 39). These studies have shown that contact between T cells and DCs may play a key role in driving HIV and SIV replication. Notably, infection of T cells occurs most efficiently when virus is presented by DCs (15, 38). Indeed, DCs may be well suited for promoting HIV and SIV infection and replication in T cells because they express the appropriate adhesion and stimulatory molecules that facilitate T-cell activation (3), an event that is critical for both viruses to complete their replication cycles (4, 36, 37, 46, 49). Interestingly, DCs can efficiently capture and transmit HIV type 1 (HIV-1) and SIV to CD4+ T cells even without becoming productively infected (6, 34). These data further suggest that both HIV and SIV have evolved the ability to exploit an antigen capture and presentation mechanism in order to initiate and promote their replication in CD4+ T cells.
The mechanism by which DCs capture HIV and transmit it to CD4+ T cells remained unclear until it was recently demonstrated that an HIV-1 envelope (Env) gp120 binding protein (renamed DC-SIGN for DC-specific ICAM-3-grabbing nonintegrin) was primarily expressed in DCs and certain macrophages (10, 16, 42, 43). DC-SIGN is a type II transmembrane protein that contains an external calcium-dependent mannose binding lectin domain. It functions as a cell adhesion molecule, mediating both T-cell activation and DC migration through high-affinity interactions with ICAM-3 and ICAM-2, respectively (14, 16). DC-SIGN has also been recently shown to internalize antigen for presentation to T cells (12). Intriguingly, DC-SIGN efficiently captures and transmits HIV and SIV to CD4+, coreceptor-positive cells (16, 35). However, in contrast to CD4 and coreceptor, it does not mediate viral entry into cells. Interestingly, DC-SIGN is expressed on DCs in the lamina propria of vaginal, intestinal, and rectal mucosal tissues and on DCs present in T-cell zones of lymphoid tissue of humans and macaques (16, 23). Thus, DC-SIGN-positive DCs are well positioned to capture HIV and SIV entering the host through mucosa sites and to transport them to lymph nodes where robust viral replication can be initiated and maintained in T cells. In this respect, DC-SIGN-positive DCs could have a role in both transmission and propagation of HIV and SIV; therefore, blocking the DC-SIGN-virus interaction may help prevent transmission or reduce viral loads in infected individuals.
Because there is no model system to examine early events in HIV transmission and pathogenesis, the SIV macaque model represents the best system to address the significance of DC-SIGN for HIV infection and replication in vivo. Previously, we reported the initial cloning and characterization of pig-tailed macaque DC-SIGN (ptDC-SIGN) (2). Here, we generate monoclonal antibodies (MAbs) against ptDC-SIGN and show that pig-tailed macaque DCs and macrophages express ptDC-SIGN. Furthermore, we demonstrate that SIV transmission from pig-tailed macaque DCs to T cells is enhanced by ptDC-SIGN.
MATERIALS AND METHODS
Cell lines and viruses.
CEMx174 cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated (56°C) fetal bovine serum (FBS), 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (RPMI complete medium). The 293T cell line was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated FBS, 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (DMEM complete medium). The CMMT-CD4-LTR-β-gal (sMAGI) indicator cells were grown in DMEM complete medium plus 200 μg of G418 per ml (Gibco-BRL) and 50 U of hygromycin per ml (CalBiochem, La Jolla, Calif.) (7). THP-1- and THP-1/DC-SIGN- transfected cells were grown in RPMI complete medium with 15% FBS. GHOST cells expressing CD4, or CD4 and CCR5, were grown in DMEM complete plus 500 μg of G418/ml, 100 μg of hygromycin/ml, and 1 μg of puromycin/ml.
To generate virus stocks, 293T cells were transfected with the plasmid proviral clones of SIVmneCL8 (32), SIVmne170 (27), or SIVmne027 (26) by using the FuGene 6 reagent (Roche, Indianapolis, Ind.). Twenty-four hours posttransfection, the cells were washed once with phosphate-buffered saline (PBS) and then cultured for an additional 24 h in fresh DMEM complete medium. Supernatants were harvested, passed through 0.22-μm syringe filters (Corning Inc., Corning, N.Y.), aliquoted, and frozen at −70°C until used for infection experiments. The amount of SIV p27gag antigen was quantitated using a commercial enzyme-linked immunosorbent assay (ELISA; Immunotech-Coulter, Miami, Fla.). The titer of each virus stock was determined using the sMAGI assay as previously described (7).
MAbs to ptDC-SIGN.
The cDNA of DC-SIGN was cloned from Macaca nemestrina (pig-tailed macaque) monocyte-derived DC total RNA and inserted into the expression vector pcDNA3 (pcDNA-ptDC-SIGN) (Invitrogen, Carlsbad, Calif.) as previously described (2). To generate MAbs against ptDC-SIGN, Jurkat cells were transfected with pcDNA-ptDC-SIGN expression vector and injected into mice. Spleen cell fusions were made with SP2/0 myeloma cells, and individual hybridoma clones were isolated and tested for secretion of anti-DC-SIGN reactive antibodies. Specific binding to ptDC-SIGN was confirmed using 293T cells transiently transfected with pcDNA-ptDC-SIGN. Cross-reactivity with human DC-SIGN (huDC-SIGN) was demonstrated using 293T cells transiently transfected with pcDNA-huDC-SIGN (2). Two hybridoma clones, 8C1 and 11C1, that secrete immunoglobulin G1/κ (IgG1/κ) and IgG2b/κ antibodies against ptDC-SIGN, respectively, were identified and used to generate ascites in mice. MAb DC4, which recognizes the neck domain of DC-SIGN, was kindly provided by R.W. Doms (2).
DC-SIGN deletion constructs.
To construct deletion mutants of ptDC-SIGN, we used a cDNA clone of ptDC-SIGN that was inserted into Bluescript KS(+) (Stratagene, La Jolla, Calif.) (pKS-ptDC-SIGN) (2). Primers for PCR amplification were designed to bind and initiate DNA synthesis of ptDC-SIGN in the reverse direction. The amplified regions of ptDC-SIGN that remained in pKS+ were blunt-end ligated at the primer ends, thereby deleting the specific sequences within ptDC-SIGN. The following primer sets were used to generate the deletion mutants. Primer locations within the ptDC-SIGN sequence are numbered according to the sequence deposited in GenBank (accession no. AF343727). To delete the neck region (ΔNECK78-224), PCR was performed using the primers SIGN-G (5′-GATCGCATCTTGTTTGGATTGTCC-3′; nucleotides [nt] 208 to 231) and SIGN-J (5′-GCAGTGGAACGCCTGTGCCAC-3′; nt 673 to 693). To delete the entire carbohydrate recognition domain (CRD) (ΔCRD232-381), PCR was performed using primers SIGN-H (5′-GTGGCACAGGCGTTCCACTGC-3′; nt 673 to 693) and SIGN-KS (5′-GCGTAGCAGAACTTCACATCAAGC-3′; 1184-pKS+ polylinker sequence). To construct a mutant with a deletion of the carboxyl-terminal 9 amino acids (ΔCRD372-381), we used primers SIGN-I (5′-TTGGGGAGAGCAACCGTTCTTCATC-3′; nt 1090 to 1113) and SIGN-KS. To construct a mutant with a deletion of the carboxyl-terminal 41 amino acids (ΔCRD340-381), PCR amplification was performed with SIGN-L (5′-ATTGCCACTAAATTCCGCACAGTC-3′; nt 994 to 1017) and SIGN-KS. To delete the 90 amino acids from the carboxy-terminal end (ΔCRD291-381), PCR was performed with primers SIGN-M (5′-GAAGCGGTTACTTCTGGAAGACTG-3′; nt 847 to 870) and SIGN-KS. To delete the sequences encoding the first 58 amino acids of the CRD (ΔCRD232-290), PCR was performed using primers SIGN-H and SIGN-O (5′-TTCACCTGGATGGGACTTTCAGAC-3′; nt 868 to 891). Finally, to delete the sequences encoding the central portion of the CRD (ΔCRD291-332), PCR was done using primers SIGN-M and SIGN-N (5′-TGTGCGGAATTTAGTGGCAATGGC-3′; nt 997 to 1020). For each set of primers, PCR amplification was performed using 1 ng of pKS-ptDC-SIGN as a template, a 1 μM concentration of each primer, a 1 mM concentration of each deoxynucleoside triphosphate, and 3 U of Taq Plus long enzyme (Stratagene) per 100-μl reaction mixture. Samples were heated to 94°C for 3 min followed by 35 cycles of amplification. For each primer set, the denaturing step (94°C for 30 s) and extension step (72°C for 10 min) were the same. The annealing temperature was modified for each primer set (53 to 60°C for 30 s) to optimize specific priming. Confirmation of the deletions was made by DNA sequencing. Each clone was excised from pKS+ and introduced into the expression vector, pHM6 (Roche), which adds a hemagglutinin (HA) epitope tag to the amino terminus of the translated protein.
Western blot analysis.
293T cells plated in 6-well dishes were transfected with wild-type or mutant ptDC-SIGN constructs using the FuGene 6 transfection reagent (Roche). Two days posttransfection, cells were lysed in 500 μl of a lysis buffer containing 0.1% sodium dodecyl sulfate, 0.5% deoxycholate, 1.0% Triton X-100, 1 U of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride, and 2 μg of the protease inhibitors pepstatin A, chymostatin, antipain, and leupeptin per ml in PBS. Lysates were clarified by brief centrifugation at 4°C in a microcentrifuge. Proteins were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and transferred to nitrocellulose. HA-tagged ptDC-SIGN proteins were detected using a 1:750 dilution of an anti-HA peroxidase-coupled MAb (Roche) by enhanced chemiluminescence (Amersham Pharmacia, Piscataway, N.J.).
Macaque DC-SIGN capture and transfer assay.
293T cells were transiently transfected with the ptDC-SIGN expression vector pcDNA-ptDC-SIGN or the negative control vector using the FuGene 6 reagent. Twenty-four hours posttransfection, cells were replated in duplicate into wells of U-bottom 96-well plates (105 cells per well in 100 μl). The following day, cells were incubated with virus (1 ng of p27gag antigen) for 2 h at 37°C in a 5% CO2 incubator. Cells were then washed vigorously three times with DMEM complete, resuspended in RPMI complete medium, and added to 2 × 105 CEMx174 cells. At 3- to 4-day intervals over a 14-day period, medium was removed from each culture and replaced with fresh RPMI complete medium. The supernatants were assayed for SIV p27gag antigen by ELISA to monitor virus replication.
To examine the inhibitory properties of the anti-ptDC-SIGN MAbs, cells were treated with the 8C1, 11C1, or control antibody at a final concentration of 25 μg/ml for 20 min at 37°C prior to the addition of virus. Following a 2-h incubation with virus, the cells were washed, resuspended in RPMI complete medium, cocultured with CEMx174 cells, and assayed for virus replication as described above.
Luciferase reporter virus assay for trans-infection by huDC-SIGN.
The HIV-1 env-defective pNL4-3.LucR-E− vector was used to generate pseudotyped viruses containing the luciferase gene (luc) in place of the HIV envelope as reported previously (9). HIV-1 env expression plasmids (ADA, JRFL) and the HIV-1 luc vector were cotransfected into 293T cells by using the FuGene 6 reagent. The vectors were provided by Nathan Landau (Salk Institute for Biological Sciences, La Jolla, Calif.). After 48 h, the supernatants were harvested, frozen in aliquots, and later tested for virus by p24 antigen-capture ELISA. For capture-transfer studies, 100 ng of p24-associated HIV-1 pseudotype virus/ml was incubated with 105 THP-1 or THP-1/DC-SIGN-positive cells for 2 h at 37°C. The cells were washed five times and transferred to 96-well plates (2 × 104 cells/well) containing GHOST cells expressing CD4 and CCR5. For antibody blocking, 50 μl of 11C1 anti-DC-SIGN antibody or medium was incubated with THP-1 or THP-1/DC cells at 50 μg/ml for 30 min at 37°C, and virus was then added for an additional 2 h at 37°C before washing. The cells were followed for 4 days, with the medium changed at day 2. Cells were lysed with 100 μl of GloLysis buffer (Promega), and 50 μl was incubated with 100 μl of Bright Glo substrate (Promega) before analysis with a tube luminometer.
DC and T-cell isolation and infection.
Peripheral blood was obtained from SIV and simian retrovirus-negative pig-tailed macaques at the Washington Regional Primate Research Center, Seattle, Wash. Peripheral blood mononuclear cells (PBMCs) were isolated as previously described using lymphocyte separation medium (41). To isolate CD14+ monocytes, we used the Miltenyi anti-CD14 microbeads and miniMACS system according to the manufacturer's protocol (Miltenyi Biotec, Auburn, Calif.). Briefly, 108 PBMCs were resuspended in 800 μl of binding buffer (PBS supplemented with 0.5% bovine serum albumin and 2 mM EDTA). MACS CD14 microbeads (200 μl) were added, mixed, and incubated at 4°C for 15 min. The cell-bead mixture was then washed with binding buffer, spun at 200 × g for 10 min, and resuspended in 1 ml of fresh binding buffer. CD14+ cells were positively selected using an MS+/RS+ column tip. Cells eluted from the column were typically greater than 95% CD14+ by fluorescence-activated cell sorter (FACS) analysis. These cells were washed with RPMI complete and cultured with 1,000 U of granulocyte-macrophage colony-stimulating factor (GM-CSF) per ml and 500 U of interleukin-4 (IL-4) per ml (R & D Systems, Minneapolis, Minn.) in RPMI complete for 7 days to generate DCs. At this time, the cells expressed moderately high levels of HLA-DR, moderate levels of CD86, and low levels of CD83 and CD25 by FACS analysis. The CD14− population of cells, which mainly consisted of lymphocytes, was frozen at −70°C until 3 days prior to cocultivation with DCs, at which point they were thawed and stimulated with 10 μg of phytohemagglutinin (PHA) per ml and IL-2 (50 U/ml) in RPMI complete medium. FACS analysis demonstrated that the resulting population of cells consisted of CD3+ CD4+ and CD3+ CD8+ T cells.
To examine transfer of virus from DCs to T cells, DCs (105) were incubated with 1,000 tissue culture infectious doses of SIVmne027 for 3 h at 37°C in a 5% CO2 incubator. DCs were then washed twice with PBS and resuspended in RPMI complete containing IL-2 (50 U/ml), IL-4 (500 U/ml), and GM-CSF (1,000 U/ml). The DCs were then added to cultures of autologous PHA-stimulated peripheral blood lymphocytes (2 × 105). Cultures were maintained in RPMI complete containing IL-2, IL-4, and GM-CSF. At 3- to 4-day intervals, supernatants were harvested, saved at −70°C, and later tested for SIV p27gag antigen by ELISA to monitor virus replication. All infections were performed in triplicate.
To examine the DC-SIGN dependence of virus transmission from DCs to T cells, cultures of DCs were first incubated MAbs 8C1 or 11C1 or control antibodies at a final concentration of 25 μg/ml. Virus was then added to the DC cultures, washed, and incubated with PHA-stimulated peripheral blood lymphocytes as described above. Supernatants were harvested every 3 to 4 days and assayed for SIV p27gag to monitor virus replication.
Flow cytometry.
Transfected 293T cells or primary pig-tailed macaque peripheral blood lymphocytes, monocytes, in vitro-derived macrophages, or DCs were stained in FACS staining buffer (PBS supplemented with 10% heat-inactivated FBS and 0.1% sodium azide) for 30 min on ice with hybridoma supernatants at a final dilution of 1/10, purified antibody at a final concentration of 10 μg per ml, or control antibody at 10 μg per ml. Samples were washed and incubated with a goat anti-mouse Fab fragment conjugated to fluorescein isothiocyanate (FITC) (Dako Corp., Carpinteria, Calif.) on ice for 30 min, washed with PBS, and resuspended in PBS containing 1% paraformaldehyde. For two-color staining of macaque primary PBMCs, phycoerythrin-conjugated antibodies to CD3, CD4, CD14, or CD20 (BD Pharmingen, San Diego, Calif.) at a 1/10 dilution were added after the goat-anti-mouse Fab-FITC staining. Samples were analyzed with a FACScan (Becton Dickinson, San Diego, Calif.) analyzer, and cells were excluded based on forward- and side-scatter characteristics.
To examine ICAM-3 binding to ptDC-SIGN, a recombinant ICAM-3-human Fc chimeric molecule (R & D Systems) was incubated with 293T cells transiently transfected with ptDC-SIGN or control vector (pcDNA3) at a final concentration of 10 μg/ml in FACS staining buffer. After a 30-min incubation on ice, cells were washed once with FACS staining buffer and incubated with a 1/50 dilution of a goat anti-human Fc FITC-conjugated MAb (Biosource International, Camarillo, Calif.) for 30 min on ice. Cells were then washed twice with FACS staining buffer and once with PBS, resuspended in 1% paraformaldehyde in PBS, and analyzed as described above. To assess whether the anti-ptDC-SIGN MAb 8C1 blocked ICAM-3 binding, cells were treated with 8C1 for 30 min prior to incubation with ICAM-3. We could not examine whether the 11C1 antibody blocked ICAM-3 binding to ptDC-SIGN because it cross-reacted with the goat anti-human Fc-FITC secondary antibody. Transfection of 293T cells with ptDC-SIGN typically increased ICAM-3 binding from approximately 0 to 2% to 35 to 40%.
RESULTS
Reactivity of anti-ptDC-SIGN MAbs and expression of ptDC-SIGN.
Previously, we showed that several MAbs raised against huDC-SIGN cross-reacted with ptDC-SIGN. However, none was able to block binding and subsequent transfer of virus to CD4+, coreceptor-positive cells (2). To generate MAbs against ptDC-SIGN, Jurkat cells transfected with a ptDC-SIGN expression vector were used to immunize mice, and spleen cells were harvested for fusions with SP2/0 myeloma cells. Hybridoma supernatants were screened by FACS analysis for reactivity to ptDC-SIGN. MAbs from two hybridomas (8C1 and 11C1) reacted with 293T cells transiently transfected with the ptDC-SIGN expression vector (pcDNA-ptDC-SIGN) but not with a negative control vector (pcDNA3) (Fig. 1). Neither antibody detected ptDC-SIGN by Western blot analysis, suggesting that they recognize conformation-dependent epitopes (data not shown). Furthermore, both antibodies cross-reacted with huDC-SIGN, indicating that they recognize epitopes conserved between the human and pig-tailed macaque molecules.
FIG. 1.
Reactivity of MAbs raised against ptDC-SIGN. 293T cells were transfected with either a control, ptDC-SIGN, or huDC-SIGN expression vector. Two days posttransfection, cells were harvested, bound with either anti-ptDC-SIGN MAb 8C1 or 11C1 (filled curves), or the appropriate isotypic control antibodies (open curves), and a goat-anti-mouse FITC-labeled secondary antibody. Cells were then analyzed by FACS.
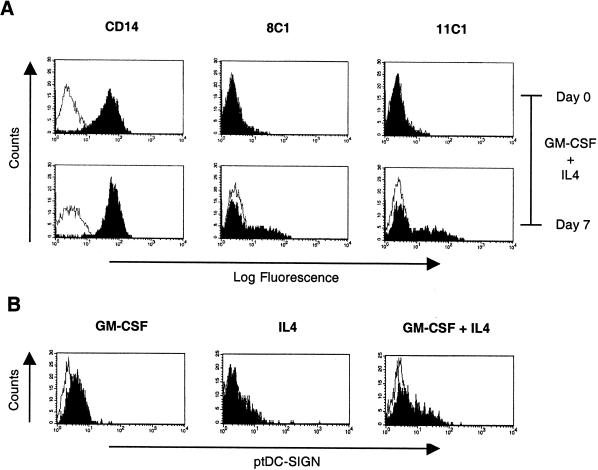
To characterize ptDC-SIGN expression, we stained PBMCs from pig-tailed macaques. Both resting and PHA-stimulated PBMCs were negative for ptDC-SIGN expression, including the CD3+, CD20+, and CD14+ cell populations (data not shown and Fig. 2A). However, moderate levels of ptDC-SIGN expression were detected by 8C1 and 11C1 on 35 to 45% of monocyte-derived DCs generated by culturing CD14+ monocytes with GM-CSF and IL-4 (Fig. 2A). We also examined whether GM-CSF or IL-4 alone could induce DC-SIGN expression from CD14+ monocytes. While both GM-CSF and IL-4 were required to attain moderate surface levels of ptDC-SIGN, GM-CSF alone was capable of inducing low-level expression of ptDC-SIGN as monocytes differentiated into macrophages (Fig. 2B). By contrast, IL-4 did not increase surface expression of ptDC-SIGN. These data demonstrate that macrophages, in addition to DCs, may express ptDC-SIGN in pig-tailed macaques.
FIG. 2.
ptDC-SIGN expression on CD14+ monocytes, macrophages, and dendritic cells. CD14+ monocytes were positively selected from pig-tailed macaque PBMCs and stained for either CD14 or ptDC-SIGN before and after 7 days of culture with GM-CSF and IL-4 (A) or after 7 days of culture with GM-CSF alone, IL-4 alone, or GM-CSF plus IL-4 (B). Cells stained with either anti-CD14 or anti-ptDC-SIGN are shown in the filled curves, and isotype controls are shown in the open curves. Similar results were observed in two independent experiments using cells from two different macaques.
Macaque DC-SIGN and transmission of SIV.
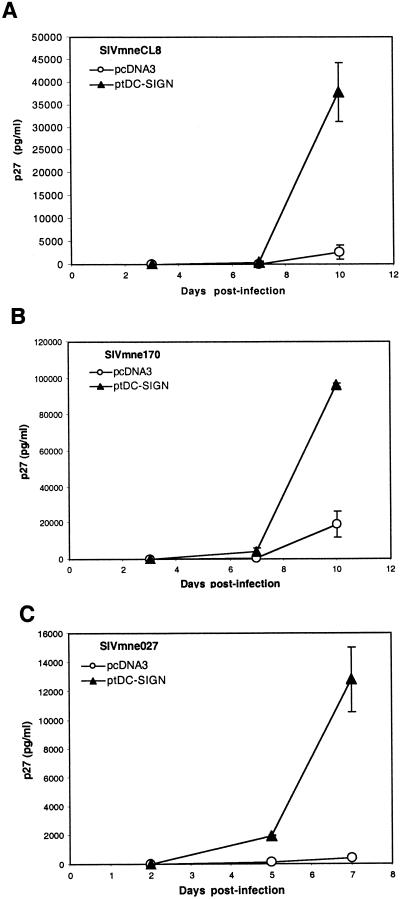
To determine whether ptDC-SIGN could bind and transmit replication-competent SIV, we expressed ptDC-SIGN in 293T cells, incubated the cells with virus for 3 h, removed unbound virus by vigorous washing, and cocultured the cells with CEMx174 cells. Virus replication was measured by assaying culture supernatants every 2 to 3 days for SIV p27gag antigen by ELISA (Fig. 3). Three different variants of the SIVmne strain, SIVmneCL8, SIVmne170, and SIVmne027, were tested. For each virus, p27gag antigen production increased more rapidly in CEMx174 cells cocultured with 293T cells expressing ptDC-SIGN than in CEMx174 cells cocultured with 293T cells that received the control vector (pcDNA3), indicating that ptDC-SIGN could enhance transmission of different SIV variants. Although the variant viruses replicated to different levels, suggesting that transmission of some variants by ptDC-SIGN may be more efficient than others, it was previously shown that these variants replicate at different rates in CEMx174 cells (26, 27). Thus, from these data it is unclear whether there are differences in the efficiency of transmission of variant SIVs by ptDC-SIGN.
FIG. 3.
Enhancement of SIV transmission by ptDC-SIGN. 293T cells were transfected with either a control (pcDNA3) or ptDC-SIGN expression vector (pcDNA-ptDC-SIGN). Cells were incubated with SIVmneCL8 (A), SIVmne170 (B), or SIVmne027 (C), washed, and cultured with CEMx174 cells. Virus production was monitored by measuring p27gag antigen in the culture supernatants over a 7- to 10-day period. Results are representative of at least three independent experiments. The average p27gag antigen values ± the standard error of the mean are shown.
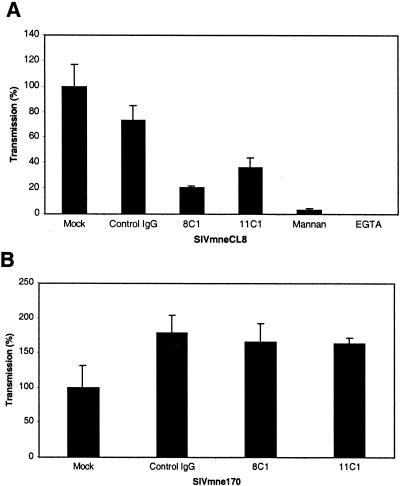
To determine whether the anti-ptDC-SIGN MAbs could inhibit virus binding and subsequent transfer of virus, 293T cells expressing ptDC-SIGN were treated with anti-ptDC-SIGN MAbs or a negative control antibody prior to incubation with virus. Compared to the control antibody, both anti-ptDC-SIGN MAbs, 8C1 and 11C1, reduced binding and transmission of SIVmneCL8 by 79 and 64%, respectively (Fig. 4A). However, the decrease caused by the anti-ptDC-SIGN antibodies was not as great as pretreatment of the cells with either mannan or EGTA, which largely eliminated binding and transmission. Interestingly, neither antibody impaired capture and transfer of the variant SIVmne170 (Fig. 4B). These data suggest that closely related variant viruses may differ in their interactions with ptDC-SIGN.
FIG. 4.
Interference of SIV capture and transmission by anti-ptDC-SIGN MAbs. 293T cells expressing ptDC-SIGN were mock treated or incubated for 20 min with 25 μg of control mouse IgG, 8C1, or 11C1 per ml prior to the addition of SIVmneCL8 (A) or SIVmne170 (B). Cells were also treated with mannan (20 μg/ml) or EGTA (5 mM) prior to the addition of SIVmneCL8. Following a 2-h incubation with virus, cells were washed and cultured with CEMx174 cells. Virus production was measured by assaying supernatants for p27gag antigen by ELISA over a 7-day period. Day 7 p27gag antigen values of the mock-treated cells were normalized to 100%. The amount of p27gag antigen production from each infection is shown relative to the amount produced from the mock-treated cells and represents the average ± the standard error of the mean. Data are representative of at least two independent experiments.
To determine whether ptDC-SIGN plays an important role in the transmission of virus from DC to T cells, we used pig-tailed macaque monocyte-derived DCs as donor cells and autologous PHA-stimulated T cells as recipient cells in a capture and transmission assay. DCs were treated with control or anti-ptDC-SIGN MAbs prior to incubation with variant SIVmne027. Following the incubation with SIVmne027, DCs were vigorously washed and cocultured with the activated T cells. SIVmne027 was used for this experiment because it replicates more efficiently in antigen-presenting cell-T-cell cultures than other SIVmne variants (reference 26 and J. T. Kimata and P. G. Patel, unpublished data). In experiments using cells from two different donors, MAb 8C1 reduced capture and subsequent replication of SIVmne027 by approximately 75% compared to the untreated cells and 68% compared to control antibody treatment (Fig. 5). In contrast, 11C1 had no specific effect on SIVmne027 capture and transmission. The inhibitory activity of 8C1 was confirmed in separate experiments by using donor cells from two additional animals. In these experiments, SIVmne027 capture and transmission was reduced by 73 and 95% compared to the untreated DCs (data not shown). Together, the data demonstrate that transmission of SIV from pig-tailed macaque DCs to T cells is largely dependent on ptDC-SIGN. Finally, because SIVmne027, unlike SIVmneCL8, was sensitive to only 8C1 but not 11C1, the data provide further evidence that variant viruses may interact differently with ptDC-SIGN.
FIG. 5.
Capture and transmission of SIV by macaque DCs. Duplicate cultures of DCs were mock treated or incubated with 25 μg of control IgG, 8C1, or 11C1 per ml for 20 min prior to the addition of SIVmne027. Following a 3-h incubation with virus, the cells were washed and incubated with PHA-stimulated PBMCs and IL-2. Virus production was monitored by p27gag ELISA every 3 to 4 days postinfection. The average p27gag antigen values ± the standard error of the mean are shown. The experiments shown in panels A and B were performed using cells isolated from two different macaques.
Transmission of HIV-1 is inhibited by anti-ptDC-SIGN MAbs.
Because of the high homology between macaque and human DC-SIGN (2, 48), we tested whether the antibodies against ptDC-SIGN could interfere with capture and transfer of HIV-1 by using a previously developed DC-SIGN-virus binding and transmission assay (15). For the experiment, an HIV-luc vector was pseudotyped with two different HIV-1 R5-tropic envelopes (HIV-1JRFL and HIV-1ADA). The luciferase activity in transduced target cells (HOS-CD4-R5 cells) that was observed with THP-1/huDC-SIGN donor cells was normalized to 100% for the virus stocks. huDC-SIGN-negative THP-1 cells were used as a background control. The presence of huDC-SIGN on THP-1 donor cells enhanced transmission of virions pseudotyped with the HIV-1JRFL or HIV-1ADA envelopes (Fig. 6). Preincubation of the donor THP-1/huDC-SIGN cells with MAb 11C1 reduced luciferase activity in the recipient cells to near-background levels (Fig. 6A). In contrast, while MAb 8C1 also had inhibitory activity, it did not lower the level of luciferase activity to that of background, indicating that it may be slightly less effective than 11C1 at blocking HIV-1 binding and transmission (Fig. 6B). These data demonstrate cross-reactive recognition of huDC-SIGN and blocking of HIV-1 capture by the anti-ptDC-SIGN MAbs 8C1 and 11C1.
FIG. 6.
Inhibition of huDC-SIGN-dependent HIV-1 capture and transfer by the anti-ptDC-SIGN MAb. THP-1/huDC-SIGN cells were mock treated or incubated with 50 μg of anti-ptDC-SIGN MAb 11C1 (A) or 8C1 (B) per ml prior to incubation with an HIV-luc reporter virus pseudotyped with either the HIV-1ADA or HIV-1JRFL envelope. Following the incubation with virus, cells were washed and cultured with HOS-CD4-CCR5 cells. Viral infection was determined by measuring luciferase activity in the target cells. THP-1/DC-SIGN− cells were used for background virus capture and transfer. Relative luciferase activity ± the standard error of the mean is shown.
Mapping the binding site for anti-DC-SIGN MAbs.
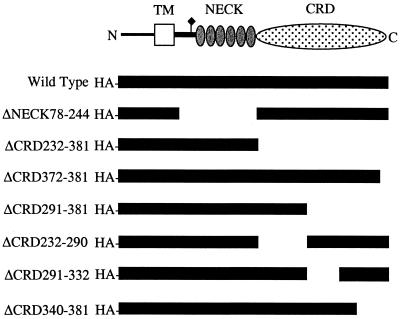
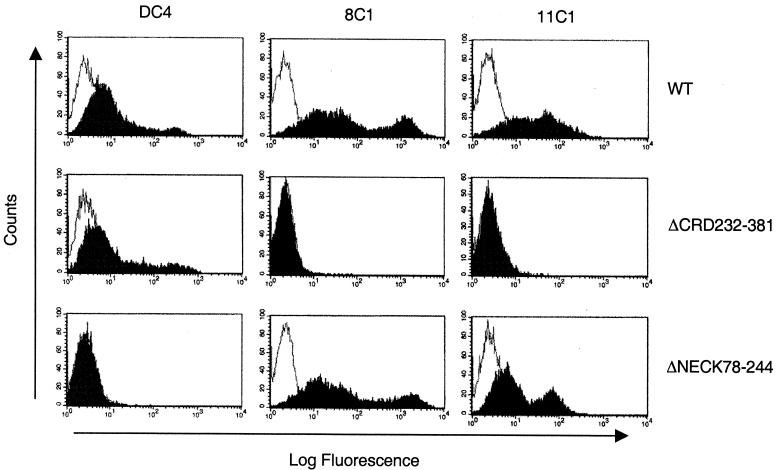
To identify the recognition sites within ptDC-SIGN for MAbs 8C1 and 11C1, we constructed HA epitope-tagged mutants of ptDC-SIGN containing deletions of the neck or CRD (Fig. 7). To show that each deletion mutant expressed the expected-size protein, cell lysates from 293T cells transiently transfected with the constructs were subjected to immunoblot analysis using an anti-HA antibody. A protein of the predicted size was observed for each mutant (data not shown). Surface expression of the wild-type and mutant ptDC-SIGN was examined by FACS analysis using the MAbs 8C1, 11C1, and DC4, which bind the repeat sequence in the neck domain (Table 1 and Fig. 8). Both 8C1 and 11C1 detected the mutant with a deletion of the entire neck domain (ΔNECK78-244) but not a mutant with the entire CRD deleted (ΔCRD232-381). However, MAb DC4, which binds to the neck region of human and macaque DC-SIGN, reacted with ΔCRD232-381, indicating its expression at the cell surface. These data indicate that both 8C1 and 11C1 recognize epitopes in the CRD. Additionally, constructs with deletions of smaller portions of the CRD reacted with the anti-neck antibody DC4 (Table 1). However, with the exception of a mutant with a deletion of the carboxyl-terminal 9 amino acids (ΔCRD372-381), none of these mutants was detected by either 8C1 or 11C1. This provides further evidence that a conformation-dependent epitope may be recognized by both antibodies. Also, one mutant, ΔCRD340-381, was not detected by any of the anti-DC-SIGN MAbs, suggesting that it was not expressed on the surface of cells. Finally, 8C1 did not interfere with recombinant human ICAM-3 binding to ptDC-SIGN (data not shown).
FIG. 7.
Deletion constructs of ptDC-SIGN. The portion of ptDC-SIGN present in each deletion mutant is shown relative to that in the wild-type clone. Also shown is a schematic diagram of the ptDC-SIGN molecule.
TABLE 1.
Binding location of ptDC-SIGN MAbs
| ptDC-SIGN mutant | Anti-DC-SIGN MAba
|
||
|---|---|---|---|
| DC4 | 8C1 | 11C1 | |
| Control | − | − | − |
| Wild type | + | + | + |
| ΔNECK78-224 | − | + | + |
| ΔCRD232-381 | + | − | − |
| ΔCRD372-381 | + | + | + |
| ΔCRD291-381 | + | − | − |
| ΔCRD232-290 | + | − | − |
| ΔCRD291-332 | + | − | − |
| ΔCRD340-381 | − | − | − |
The DC-SIGN MAbs were tested by flow cytometry against 293T cells transfected with the indicated ptDC-SIGN deletion mutant. The control is the pHM6 expression vector without the ptDC-SIGN cDNA insert and the wild type is pHM-ptDC-SIGN.
FIG. 8.
Reactivity of anti-DC-SIGN MAbs with deletion mutants of ptDC-SIGN. 293T cells were transfected with vectors expressing the wild-type ptDC-SIGN, a deletion of the complete CRD (ΔCRD232-381), or deletion of the complete neck domain (ΔNECK78-244). Cell expressing each construct were stained with the 8C1, 11C1, or anti-neck antibody DC4 (black curves) or stained with isotypic control antibodies (open curves) and analyzed by FACS.
DISCUSSION
DCs are believed to play a central role in the initiation and establishment of HIV-1 infection in the host. Because they are among the first cells encountered by virus and by virtue of their ability to efficiently capture and transmit HIV-1 and SIV to CD4+ T cells, these cells may act as important vehicles for trafficking virus from the site of infection to draining lymph nodes where robust replication can be initiated in T cells (5, 18, 40). The discovery of DC-SIGN as an attachment factor for primate lentiviruses that can enhance infection of T cells by DCs in trans provides a molecular explanation for how these cells efficiently capture and transmit virus, even in the absence of becoming infected (15, 29). It also raises questions about DC-SIGN′s significance for virus transmission and replication in vivo. Because SIV infection of macaques is the primary model for studying the early events in HIV-1 infection, we examined whether DC-SIGN plays a major role in SIV capture and transfer by macaque DCs. Our data demonstrate that SIV infection of pig-tailed macaques may be a suitable model for studying the significance of DC-SIGN-dependent capture of virus by DCs for virus infection and dissemination.
Using MAbs generated against ptDC-SIGN, we demonstrated that the cell types in pig-tailed macaques which expressed ptDC-SIGN are similar to those in humans (16). CD3+, CD20+, and CD14+ cells in peripheral blood are largely negative for ptDC-SIGN. A small population of cells that do not express these markers are positive for ptDC-SIGN (data not shown). We are currently investigating if these cells represent myeloid DCs, which have been shown to be DC-SIGN positive in human blood (33). As predicted, treatment of CD14+ monocytes with GM-CSF and IL-4 induces ptDC-SIGN expression on DCs. However, these cells express only low and moderate levels of ptDC-SIGN. This contrasts with human monocyte-derived DCs, which express high levels of surface DC-SIGN, and rhesus macaque monocyte-derived DC, which only have low levels of rhesus DC-SIGN (rhDC-SIGN) (15, 16, 48). It is unclear at this time what accounts for the differences in DC-SIGN expression levels between DCs of these macaque species and humans. Although species-specific differences in DC-SIGN expression are possible, an alternative explanation is that both pig-tail and rhesus macaque monocytes are poorly responsive to human GM-CSF and IL-4 compared to human monocytes. Importantly, although the tissue distribution of DC-SIGN-positive cells in rhesus macaques and humans is similar, and rhDC-SIGN can capture and transmit SIV when expressed at high levels (2, 23, 48), if the level of DC-SIGN expression on rhesus macaque DCs in vivo is also too low to capture virus, it would dampen the utility of this species as a model to evaluate the significance of DC-SIGN in virus transmission. It might also cast doubt about the role of DC-SIGN in the pathogenic process. Alternatively, another C-type lectin protein with homology to DC-SIGN may efficiently capture SIV in rhesus macaques. Because pig-tailed macaque DCs express higher levels of DC-SIGN than do rhesus DCs, pig-tailed macaques may prove to be a more useful animal model for such studies than rhesus macaques.
In contrast to human monocyte-derived macrophages, which do not express DC-SIGN (43), pig-tailed macaque macrophages derived from CD14+ monocytes cultured with GM-CSF alone expressed low levels of ptDC-SIGN. Whether or not these cells can also capture and transmit SIV in a DC-SIGN-dependent manner will be important to examine, particularly in light of studies demonstrating that macrophage-resting T-cell interactions support replication of some SIV variants (11, 26). Moreover, it will also be interesting to assess whether DC-SIGN can function as a cis-acting factor that enhances infection of macrophages, because they may have a role in HIV and SIV dissemination and persistence (19). Indeed, the presence of DC-SIGN on CD4+, coreceptor-positive cells significantly enhances infection, and certain types of macrophages express DC-SIGN in vivo (30, 42). However, the level of ptDC-SIGN expression in pig-tailed macaque macrophages may be too low to capture virus. A recent study by Pöhlmann et al. demonstrated that a high level of DC-SIGN expression (60,000 molecules per cell) is required for efficient capture and transmission of HIV-1 (35). On the other hand, if the TH2 cytokine IL-13 can up-regulate ptDC-SIGN expression in macaque macrophages, like it does in human macrophages (43), then ptDC-SIGN may prove to be an important virus attachment and infectivity factor for macaque macrophages.
Previously, it was shown that both rhesus and pig-tailed macaque DC-SIGN capture and transmit virions pseudotyped with either HIV-1 or SIV envelope proteins (2). Those data provided evidence that SIV and HIV interact with macaque DC-SIGN molecules in a manner similar to that with huDC-SIGN. Here, we extend those studies and demonstrate that ptDC-SIGN enhances transmission of different replication-competent variants of SIVmne. Furthermore, MAbs generated against ptDC-SIGN impair virus capture and subsequent transmission to target cells. Importantly, we observed 73 to 95% blocking activity of the 8C1 antibody with DC and T cells from four different macaques, suggesting that ptDC-SIGN is the major factor involved in capture and transmission of SIV by macaque DCs. However, because the transmission of virus was not completely diminished, other factors may participate in the capture of virus by DCs of this macaque species. These data are not inconsistent with those obtained by using human DCs. Capture and transmission of replication-competent HIV-1 by human DCs is also not completely impaired by anti-huDC-SIGN MAbs (15, 29). Moreover, capture and transmission of SIV by rhesus DCs appears to be DC-SIGN independent (48). Thus, factors other than DC-SIGN likely contribute to virus capture by DCs.
The MAbs characterized here are the first to be generated against a macaque DC-SIGN molecule. Both antibodies recognize an epitope(s) conserved between pig-tailed macaque and human DC-SIGN and block the interaction of SIV and HIV with these respective molecules. These data provide further evidence that the specificity and mechanism of SIV and HIV binding to macaque and human DC-SIGN are similar. Additionally, the reactivity of the 8C1 and 11C1 MAbs against ptDC-SIGN deletion mutants indicates that they bind to the CRD of the protein. However, while both antibodies detect a carboxyl-terminal deletion, ruling out the last 9 amino acids of the CRD as part of the epitope for either antibody, we could not further map the binding sites for the antibodies because no other ptDC-SIGN mutant containing a deletion of the CRD was recognized by either antibody. These data suggest that the epitopes may be conformation dependent. This conclusion is supported by the inability of either MAb to detect denatured ptDC-SIGN by immunoblot analysis. Interestingly, while previous studies have shown that anti-huDC-SIGN MAbs prevent ICAM-3 from binding to either huDC-SIGN or rhDC-SIGN (16, 48), at least one of the anti-ptDC-SIGN MAbs does not appear to interfere with ICAM-3 binding to ptDC-SIGN. Because these studies used a recombinant human ICAM-3 molecule to examine binding, it will be necessary to verify these results by testing whether the anti-ptDC-SIGN MAbs also fail to inhibit clustering of pig-tailed macaque DCs and T cells. Taken together, our data indicate that the virus capture function of ptDC-SIGN may be inhibited without interfering with its immune function. Thus, because of their specificity for inhibiting virus-DC-SIGN interactions, the anti-ptDC-SIGN MAbs are attractive inhibitors for testing in vivo against mucosal SIV infection. Mapping the binding site for the MAbs on ptDC-SIGN may help identify the viral envelope binding site. It may also provide insight for the development of novel inhibitors that can specifically prevent virus-DC-SIGN interactions.
An unexpected result of this study is that the SIVmne variants differ in their sensitivities to the blocking effect of the anti-ptDC-SIGN antibodies in virus capture-transfer assays. There are a number of amino acid differences in the envelope proteins of these three variant viruses, particularly in the V1 variable region (26, 27). While we have not directly shown that envelope alone is important for the enhanced transmission of virus by ptDC-SIGN, we hypothesize that structural changes in envelope conferred by mutations account for the differences in sensitivity of the variant viruses to the anti-ptDC-SIGN MAbs and, therefore, the binding capacity to ptDC-SIGN. Defining the envelope mutations that affect sensitivity to the antibodies may provide insight into the determinants involved in DC-SIGN binding. It is intriguing that SIVmneCL8, which has the fewest glycosylation sites of the three variants, is the most sensitive to the anti-ptDC-SIGN antibodies, suggesting that perhaps glycosylation is an important determinant of ptDC-SIGN binding. However, a recent finding has shown that glycosylation may not be important for HIV-1 envelope binding to DC-SIGN (17). Thus, while carbohydrates may not be the main binding determinants in the viral envelope protein, they may alter the affinity of the interaction. Further studies using these SIVmne envelope variants and mutants may help elucidate the importance of carbohydrates and other determinants for envelope-ptDC-SIGN interactions. Finally, it will also be important to examine whether our anti-ptDC-SIGN MAbs can impair DC capture and transmission of viruses displaying SIVmneCL8 or SIVmne170 envelopes, since the context of ptDC-SIGN expression may influence virus and antibody binding. We did not assess whether ptDC-SIGN was critical for transfer of SIVmneCL8 or SIVmne170 with macaque DCs in this study because neither virus is efficiently transmitted from DCs to T cells in vitro (J. T. Kimata and P. G. Patel, unpublished data). Envelope chimeric viruses using SIVmne027 as a vector will be necessary to test the sensitivity of viruses expressing these envelopes to anti-ptDC-SIGN MAbs in DC-T-cell capture-transmission assays.
In summary, primary pig-tailed macaque cells express ptDC-SIGN in similar cell types as human cells. Moreover, pig-tailed macaque DCs capture and transmit SIVmne in a largely DC-SIGN-dependent manner. Additionally, the MAbs against ptDC-SIGN interfere with SIV and HIV capture but not ICAM-3 binding. Thus, pig-tailed macaques and the anti-ptDC-SIGN MAbs may be useful for testing the relevance of DC-SIGN-positive DCs in SIV transmission in vivo. However, an intriguing result is that the SIVmne variants have differing sensitivities to the blocking effects of the anti-ptDC-SIGN MAbs in virus capture-transmission assays. These data raise important questions, should DC-SIGN prove to be a critical factor for virus transmission. First, any novel microbicide or vaccine that interferes with virus attachment to DC-SIGN will have to be successful against different virus variants. Secondly, viruses may have the capacity to evolve resistance to DC-SIGN inhibitors.
Acknowledgments
We thank Stephanie Fargo for assistance with the FACS analysis. We gratefully acknowledge the Washington Regional Primate Research Center, University of Washington, Seattle, for providing pig-tailed macaque blood. GHOST cells were obtained from the AIDS Reagent and Reference Program, Division of AIDS, NIAID, NIH (from D. R. Littman and V. N. KewalRamani).
J.E.B. was supported by an NIH training grant in viral pathogenesis (AI07522). This work was supported by NIH grants AI47725 and AI49803 to J.T.K.
REFERENCES
- 1.Adema, G. J., F. Hartgers, R. Verstraten, E. de Vries, G. Marland, S. Menon, J. Foster, Y. Xu, P. Nooyen, T. McClanahan, K. B. Bacon, and C. G. Figdor. 1997. A dendritic-cell-derived C-C chemokine that preferentially attracts naive T cells. Nature 387:713-717. [DOI] [PubMed] [Google Scholar]
- 2.Baribaud, F., S. Pohlmann, T. Sparwasser, M. T. Kimata, Y. K. Choi, B. S. Haggarty, N. Ahmad, T. Macfarlan, T. G. Edwards, G. J. Leslie, J. Arnason, T. A. Reinhart, J. T. Kimata, D. R. Littman, J. A. Hoxie, and R. W. Doms. 2001. Functional and antigenic characterization of human, rhesus macaque, pig-tailed macaque, and murine DC-SIGN. J. Virol. 75:10281-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, D., J. W. Young, and J. Banchereau. 1999. Dendritic cells. Adv. Immunol. 72:255-324. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinsky, M. I., T. L. Stanwick, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron, P., M. Pope, A. Granelli-Piperno, and R. M. Steinman. 1996. Dendritic cells and the replication of HIV-1. J. Leukoc. Biol. 59:158-171. [DOI] [PubMed] [Google Scholar]
- 6.Cameron, P. U., P. S. Freudenthal, J. M. Barker, S. Gezelter, K. Inaba, and R. M. Steinman. 1992. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 257:383-387. [DOI] [PubMed] [Google Scholar]
- 7.Chackerian, B., N. L. Haigwood, and J. Overbaugh. 1995. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology 213:386-394. [DOI] [PubMed] [Google Scholar]
- 8.Cimarelli, A., G. Zambruno, A. Marconi, G. Girolomoni, U. Bertazzoni, and A. Giannetti. 1994. Quantitation by competitive PCR of HIV-1 proviral DNA in epidermal Langerhans cells of HIV-infected patients. J. Acquir. Immune Defic. Syndr. 7:230-235. [PubMed] [Google Scholar]
- 9.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 10.Curtis, B. M., S. Scharnowske, and A. J. Watson. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:8356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, Z., S. M. Lang, V. G. Sasseville, A. A. Lackner, P. O. Ilyinskii, M. D. Daniel, J. U. Jung, and R. C. Desrosiers. 1995. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell 82:665-674. [DOI] [PubMed] [Google Scholar]
- 12.Engering, A., T. B. Geijtenbeek, S. J. van Vliet, M. Wijers, E. van Liempt, N. Demaurex, A. Lanzavecchia, J. Fransen, C. G. Figdor, V. Piguet, and Y. van Kooyk. 2002. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J. Immunol. 168:2118-2126. [DOI] [PubMed] [Google Scholar]
- 13.Frankel, S. S., B. M. Wenig, A. P. Burke, P. Mannan, L. D. Thompson, S. L. Abbondanzo, A. M. Nelson, M. Pope, and R. M. Steinman. 1996. Replication of HIV-1 in dendritic cell-derived syncytia at the mucosal surface of the adenoid. Science 272:115-117. [DOI] [PubMed] [Google Scholar]
- 14.Geijtenbeek, T. B., D. J. Krooshoop, D. A. Bleijs, S. J. van Vliet, G. C. van Duijnhoven, V. Grabovsky, R. Alon, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 1:353-357. [DOI] [PubMed] [Google Scholar]
- 15.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 16.Geijtenbeek, T. B., R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, G. J. Adema, Y. van Kooyk, and C. G. Figdor. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100:575-585. [DOI] [PubMed] [Google Scholar]
- 17.Geijtenbeek, T. B., G. C. van Duijnhoven, S. J. van Vliet, E. Krieger, G. Vriend, C. G. Figdor, and Y. van Kooyk. 2002. Identification of different binding sites in the dendritic cell-specific receptor DC-SIGN for ICAM-3 and HIV-1. J. Biol. Chem. 277:11314-11320. [DOI] [PubMed] [Google Scholar]
- 18.Grouard, G., and E. A. Clark. 1997. Role of dendritic and follicular dendritic cells in HIV infection and pathogenesis. Curr. Opin. Immunol. 9:563-567. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch, V. M., M. E. Sharkey, C. R. Brown, B. Brichacek, S. Goldstein, J. Wakefield, R. Byrum, W. R. Elkins, B. H. Hahn, J. D. Lifson, and M. Stevenson. 1998. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat. Med. 4:1401-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, J., M. B. Gardner, and C. J. Miller. 2000. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J. Virol. 74:6087-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ignatius, R., F. Isdell, U. O'Doherty, and M. Pope. 1998. Dendritic cells from skin and blood of macaques both promote SIV replication with T cells from different anatomical sites. J. Med. Primatol. 27:121-128. [DOI] [PubMed] [Google Scholar]
- 22.Inaba, K., N. Romani, and R. M. Steinman. 1989. An antigen-independent contact mechanism as an early step in T cell-proliferative responses to dendritic cells. J. Exp. Med. 170:527-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jameson, B., F. Baribaud, S. Pohlmann, D. Ghavimi, F. Mortari, R. W. Doms, and A. Iwasaki. 2002. Expression of DC-SIGN by dendritic cells of intestinal and genital mucosae in humans and rhesus macaques. J. Virol. 76:1866-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanitakis, J., S. Escaich, C. Trepo, and J. Thivolet. 1991. Detection of human immunodeficiency virus-DNA and RNA in the skin of HIV-infected patients using the polymerase chain reaction. J. Investig. Dermatol. 97:91-96. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, G., A. Nusrat, M. D. Witmer, I. Nath, and Z. A. Cohn. 1987. Distribution and turnover of Langerhans cells during delayed immune responses in human skin. J. Exp. Med. 165:763-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimata, J. T., A. Mozaffarian, and J. Overbaugh. 1998. A lymph node-derived cytopathic simian immunodeficiency virus Mne variant replicates in nonstimulated peripheral blood mononuclear cells. J. Virol. 72:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimata, J. T., and J. Overbaugh. 1997. The cytopathicity of a simian immunodeficiency virus Mne variant is determined by mutations in Gag and Env. J. Virol. 71:7629-7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight, S. C. 1996. Bone-marrow-derived dendritic cells and the pathogenesis of AIDS. AIDS 10:807-817. [DOI] [PubMed] [Google Scholar]
- 29.Kwon, D. S., G. Gregorio, N. Bitton, W. A. Hendrickson, and D. R. Littman. 2002. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 16:135-144. [DOI] [PubMed] [Google Scholar]
- 30.Lee, B., G. Leslie, E. Soilleux, U. O'Doherty, S. Baik, E. Levroney, K. Flummerfelt, W. Swiggard, N. Coleman, M. Malim, and R. W. Doms. 2001. cis expression of DC-SIGN allows for more efficient entry of human and simian immunodeficiency viruses via CD4 and a coreceptor. J. Virol. 75:12028-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, C. J., P. Vogel, N. J. Alexander, S. Sutjipto, A. G. Hendrickx, and P. A. Marx. 1992. Localization of SIV in the genital tract of chronically infected female rhesus macaques. Am. J. Pathol. 141:655-660. [PMC free article] [PubMed] [Google Scholar]
- 32.Overbaugh, J., L. M. Rudensey, M. D. Papenhausen, R. E. Benveniste, and W. R. Morton. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson, S., A. Rae, N. Hockey, J. Gilmour, and F. Gotch. 2001. Plasmacytoid dendritic cells are highly susceptible to human immunodeficiency virus type 1 infection and release infectious virus. J. Virol. 75:6710-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinchuk, L. M., P. S. Polacino, M. B. Agy, S. J. Klaus, and E. A. Clark. 1994. The role of CD40 and CD80 accessory cell molecules in dendritic cell-dependent HIV-1 infection. Immunity 1:317-325. [DOI] [PubMed] [Google Scholar]
- 35.Pohlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 75:4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polacino, P. S., H. A. Liang, and E. A. Clark. 1995. Formation of simian immunodeficiency virus long terminal repeat circles in resting T cells requires both T cell receptor- and IL-2-dependent activation. J. Exp. Med. 182:617-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polacino, P. S., H. A. Liang, E. J. Firpo, and E. A. Clark. 1993. T-cell activation influences initial DNA synthesis of simian immunodeficiency virus in resting T lymphocytes from macaques. J. Virol. 67:7008-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pope, M., M. G. H. Betjes, N. Romani, H. Hirmand, P. U. Cameron, L. Hoffman, S. Gezelter, G. Schuler, and R. M. Steinman. 1994. Conjugates of dendritic cells and memory T lymphocytes from skin facilitate productive infection with HIV-1. Cell 78:389-398. [DOI] [PubMed] [Google Scholar]
- 39.Pope, M., D. Elmore, D. Ho, and P. Marx. 1997. Dendrite cell-T cell mixtures, isolated from the skin and mucosae of macaques, support the replication of SIV. AIDS Res. Hum. Retrovir. 13:819-827. [DOI] [PubMed] [Google Scholar]
- 40.Rowland-Jones, S. L. 1999. HIV: the deadly passenger in dendritic cells. Curr. Biol. 9:R248-R250. [DOI] [PubMed] [Google Scholar]
- 41.Rudensey, L. M., J. T. Kimata, R. E. Benveniste, and J. Overbaugh. 1995. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology 207:528-542. [DOI] [PubMed] [Google Scholar]
- 42.Soilleux, E. J., L. S. Morris, B. Lee, S. Pohlmann, J. Trowsdale, R. W. Doms, and N. Coleman. 2001. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. J. Pathol. 195:586-592. [DOI] [PubMed] [Google Scholar]
- 43.Soilleux, E. J., L. S. Morris, G. Leslie, J. Chehimi, Q. Luo, E. Levroney, J. Trowsdale, L. J. Montaner, R. W. Doms, D. Weissman, N. Coleman, and B. Lee. 2002. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J. Leukoc. Biol. 71:445-457. [PubMed] [Google Scholar]
- 44.Spira, A. I., P. A. Marx, B. K. Patterson, J. Mahoney, R. A. Koup, S. M. Wolinsky, and D. D. Ho. 1996. Cellular targets of infection and route of viral dissemination after an intravaginal inoculation of simian immunodeficiency virus into rhesus macaques. J. Exp. Med. 183:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinman, R. M. 1991. The dendritic cell system and its role in immunogenicity. Annu. Rev. Immunol. 9:271-296. [DOI] [PubMed] [Google Scholar]
- 46.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, H. L., and J. G. Cyster. 1999. Chemokine up-regulation and activated T cell attraction by maturing dendritic cells. Science 284:819-822. [DOI] [PubMed] [Google Scholar]
- 48.Wu, L., A. A. Bashirova, T. D. Martin, L. Villamide, E. Mehlhop, A. O. Chertov, D. Unutmaz, M. Pope, M. Carrington, and V. N. KewalRamani. 2002. Rhesus macaque dendritic cells efficiently transmit primate lentiviruses independently of DC-SIGN. Proc. Natl. Acad. Sci. USA 99:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Y. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]