Abstract
We report a Sendai virus (SeV) vector system for expression of major histocompatibility complex (MHC) class I/peptide complexes. We cloned the extracellular domain of a human MHC class I heavy chain, HLA-A*2402, and human β-2 microglobulin (β2m) fused with HLA-A*2402-restricted human immunodeficiency virus type 1 (HIV-1) cytotoxic T-lymphocyte (CTL) epitopes (e-β2m) in separate SeV vectors. When we coinfected nonhuman mammalian cells with the SeVs, naturally folded human MHC class I/peptide complexes were secreted in the culture supernatants. Biotin binding peptide sequences on the C terminus of the heavy chain were used to tetramerize the complexes. These tetramers made in the SeV system recognized specific CD8-positive T cells in peripheral blood mononuclear cells of HIV-1-positive patients with a specificity and sensitivity similar to those of MHC class I tetramers made in an Escherichia coli system. Solo infection of e-β2m/SeV produced soluble e-β2m in the culture supernatant, and cells pulsed with the soluble protein were recognized by specific CTLs. Furthermore, when cells were infected with e-β2m/SeV, these cells were recognized by the specific CTLs more efficiently than the protein pulse per se. SeV is nonpathogenic for humans, can transduce foreign genes into nondividing cells, and may be useful for immunotherapy to enhance antigen-specific immune responses. Our system can be used not only to detect but also to stimulate antigen-specific cellular immune responses.
Class I molecules of the major histocompatibility complex (MHC)—human leukocyte antigen (HLA) in humans—play crucial roles in cellular immune responses in humans. An MHC class I/peptide complex has a heterotrimeric structure consisting of a polymorphic glycoprotein called the heavy chain, β-2 microglobulin (β2m), and a short peptide with 8 to 11 amino acids which is a proteolytic fragment of a larger antigen. A unique conformational structure called an epitope results from the bumps created by the peptide embedded in a groove of a heavy chain and a part of the surrounding banks. Cytotoxic T lymphocytes (CTLs) recognize the epitope through their T-cell receptors (TCRs) and remove the exotic cells which present tumors, viral antigens, and so on. When antigen presentation by MHC class I molecules is insufficient, the target cells may escape from immune surveillance by CTLs. Often, invasive tumors partially or totally lose expression of MHC class I/peptide complexes due to a blockage at any step required for synthesis or transport to the cell surface (3). Viruses use different tactics to sneak away from CTLs. Nef in human immunodeficiency virus type 1 (HIV-1), ICP47 in herpes simplex virus type 1, US2, US3, US6, and US11 in cytomegalovirus (CMV), and E19 in adenovirus are examples of viral proteins which disturb the expression of MHC class I/peptide complexes on the surfaces of infected cells (1, 2, 27, 40, 41, 43).
It has been shown that artificially manufactured MHC class I/peptide complexes are useful for studying epitope-specific CTL responses or for enhancing these responses in vitro. Several methods of manufacturing MHC class I/peptide complexes in vitro have been reported. The first is to make the three components independently and mix them (11). The second is to make two molecules, β2m and a heavy chain fused with a peptide sequence at the N terminus via a linker sequence (15). The third is also to make two molecules, a heavy chain and β2m fused with a peptide at the N terminus via a linker sequence (31, 37, 39). Fourth, a fusion protein consisting of a heavy chain and β2m is mixed with a peptide (22, 36). The fifth method is to make a single chain construct of the three components (23). Escherichia coli or baculovirus expression systems have been used to make MHC class I tetramers (4, 39). In this study, we adopted a Sendai virus (SeV) vector system for MHC class I/peptide complex expression.
SeV, a member of the family Paramyxoviridae, has a nonsegmented negative-strand RNA as a genome. It causes severe respiratory disease in mice but is nonpathogenic for humans (16, 17, 24). It has been shown that the SeV vector system is very efficient at production of soluble proteins (44). We would like to show that this system is also very efficient at producing heteromeric molecules in the culture supernatant. Moreover, the same system could be used to express MHC class I/peptide complexes with intended specificities on the cell surface and to stimulate antigen-specific CTLs. Thus, the SeV vector system appears to be not only an efficient but also a versatile system for expression of heteromeric cell surface molecules.
MATERIALS AND METHODS
Cell lines and media.
Cells of the monkey kidney cell lines LLC-MK2 and CV-1 were cultured in minimal essential medium (MEM) (Sigma, St. Louis, Mo.) supplemented with penicillin and streptomycin, each at 100 U/ml (Invitrogen, Carlsbad, Calif.), and 10% heat-inactivated fetal calf serum (FCS) (M10). Epstein-Barr virus-transformed B-lymphoblastoid cell lines (B-LCLs) were established as previously described (26) and maintained in RPMI 1640 (Sigma) supplemented with 100 U of penicillin/ml, 100 U of streptomycin/ml, and 10% heat-inactivated FCS (R10). For CTL culture, R10 supplemented with 10% Lymphocult T (Biotest, Dreieich, Germany) was used.
Construction of recombinant SeV.
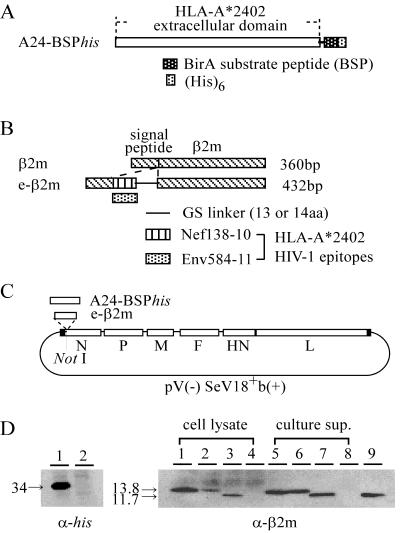
A genetic construct for a soluble HLA class I heavy chain (A24-BSPhis) was generated by a three-step PCR using a cloned HLA-A*2402 cDNA as a template (35). In A24-BSPhis, the external domain of the HLA-A*2402 molecule (from the α1 to the α3 domain of HLA-A*2402) was connected with BirA substrate peptide (BSP), a histidine tag (his), and signals needed for expression in SeV between NotI sites (Fig. 1A).
FIG. 1.
Construction of SeVs expressing the soluble MHC class I heavy chain or e-β2m. (A) Structure of A24-BSPhis. The extracellular domain of HLA-A*2402 was connected with BSP and the histidine tag via two amino acids (glycine and serine). (B) Structure of e-β2m. An HLA-A*2402-restricted epitope and a 13- or 14-amino-acid glycine-serine linker were inserted between the signal sequence and the coding sequences of the mature β2m protein. In this study, we used two HLA-A*2402-restricted HIV-1 CTL epitopes, Nef138-10 and Env584-11. (C) A24-BSPhis and e-β2m followed by E and S signals of SeV were inserted into the NotI site in the parental pV(−)SeV18+b(+), which generated a full-length SeV V(−) antigenome, as described in Materials and Methods. (D) Expression of A24-BSPhis and e-β2m in SeV-infected cells by Western blot analysis. CV-1 cells were infected with SeVs at an MOI of 3 and were lysed 24 h postinfection, and cell lysates were separated on an SDS-PAGE gel with a 10 to 20% gradient, transferred to a polyvinylidene difluoride membrane, and then detected with an anti-His6 (left) or anti-β2m (right) MAb. In the case of e-β2m, the culture supernatants which were removed from SeV particles by centrifugation were also used. (Left) Lane 1, A24-BSPhis/SeV; lane 2, wild-type SeV (wt/SeV). (Right) Lanes 1 and 5, Nef138-β2m/SeV; lanes 2 and 6, Env584-β2m/SeV; lanes 3 and 7, β2m/SeV; lanes 4 and 8, wt/SeV; lane 9, 20 ng of purified β2m. In lanes 1 to 4, 10 μg of cell lysates was added; in lanes 5 to 8, 10 μl of the culture supernatant was added. The numbers to the left of the gels indicate the sizes (in kilodaltons) of the products.
Initially, the HLA-A*2402 cDNA was amplified using the 5′ primer A24-a (5′-TGCGGCCGCCGTACGAGGATGGCCGTCATGGCGCCCCG-3′), which hybridizes to the 5′ end of the HLA-A*2402 signal sequence and contains a spacer sequence and a NotI site, and the 3′ primer A24-d1 (5′-GTCCCGCAGCTCCATCTTCATTGCCTCAAAGATTCCTCCAAGGGATCCCCATCTCAGGGTGAGGGGCTT-3′), which hybridizes to the 3′ end of the HLA-A*2402 α3 domain and encodes BSP (LGGIFEAMKMELRD) (coding sequence underlined) (29). A24-a was used as the 5′ primer for all three PCR steps. The first PCR product was used as a template for the second PCR and amplified using the 3′ primer A24-d1his (5′-CTACGGCGTACGTCAATGGTGGTGATGGTGGTGGTCCCGCAGCTCCAT-3′), which hybridizes to the 3′ half of BSP and contains a histidine tag (His6), a stop codon, and a spacer sequence. The second PCR product was used as a template for the third PCR and amplified using the 3′ primer A24-d2his (5′-TTGCGGCCGCGATGAACTTTCACCCTAAGTTTTTCTTACTACGGCGTACGTCA-3′), which hybridizes to the stop codon and the spacer sequence and contains the SeV E and S signal sequences and a NotI site (44).
For epitope-fused β2m (e-β2m), human β2m cDNA was cloned by reverse transcription-PCR from mRNA isolated from peripheral blood mononuclear cells (PBMCs) of a healthy donor. The gene was modified to introduce sequences encoding peptides with a binding motif for HLA-A*2402 and a Gly-Ser linker between the C terminus of the signal sequence and the N terminus of the mature β2m protein (Fig. 1B). Two HLA-A*2402-binding peptides were used as epitopes: one was the peptide from HIV-1 Nef corresponding to amino acids 138 to 147 (Nef138-10; RYPLTFGWCF), and the other was the peptide from HIV-1 Env corresponding to amino acids 584 to 594 (Env584-11; RYLRDQQLLGI) (14). The cloned β2m was used as a template for a three-step PCR. Initially, β2m was amplified using the 5′ primer e/b2m-a1 (5′-GGAGGTGGCGGGTCCGGAGGTGGTTCTGGTGGAGGTTCGATCCAGCGTACTCCAAAGATT-3′), which hybridizes to β2m immediately adjacent to the signal sequence and encodes a 13-amino-acid Gly-Ser linker sequence (GGGGSGGGSGGGS), and the 3′ primer b2m-d (5′-TTGCGGCCGCGATGAACTTTCACCCTAAGTTTTTCTTACTACGGCGTACGTTACATGTCTCGATCCCACTT-3′), which hybridizes to the 3′ end of β2m and encodes a spacer sequence, E and S signals, and a NotI site. b2m-d was used as the 3′ primer in all three PCR steps. For Nef138-β2m, e(nef)-a2 (5′-TCTGGCCTGGAGGCTAGATATCCACTGACCTTTGGATGGTGCTTCGGAGGAGGTGGCGGGTCC-3′), which hybridizes to the linker sequence, encodes Nef138-10 (underlined), and contains part of the β2m signal sequence, was used as the 5′ primer for the second PCR. e(env)-a2 (5′-TCTGGCCTGGAGGCTAGATACCTAAGGGATCAACAGCTCCTAGGGATTGGAGGTGGCGGGTCC-3′) was used for Env584-β2m (underlined). Then these PCR products were used as a template for the third PCR and amplified using the 5′ primer e/b2m-a3 (5′-TGCGGCCGCCGTACGGCCGAGATGTCTCGCTCCGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGGCCTGGAGGCT-3′), which contains a β2m signal sequence, a spacer sequence, and a NotI site. We also made a β2m coding fragment by using b2m-a (5′-TGCGGCCGCCGTACGGCCGAGATGTCTCGCTCCGTGGCCTTA-3′), which hybridizes to the 5′ end of the β2m signal sequence and contains the spacer sequence and NotI site, and b2m-d.
Each PCR fragment was cloned into the pGEM-T vector (Promega Corp., Madison, Wis.), and we confirmed the absence of PCR errors for each fragment by sequence analysis with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.). Then each fragment was introduced into the NotI site of pV(−)SeV18+b(+), which contains a full-length copy of the positive-sense SeV antigenome (15,384 nucleotides) with an additional 18-nucleotide sequence including a NotI site within the N gene and with two point mutations in the P gene which cause a defect in V mRNA without affecting P protein expression (Fig. 1C) (16, 44), and we obtained A24-BSPhis/pSeV, Nef138-β2m/pSeV, Env584-β2m/pSeV, and β2m/pSeV.
SeV recovery.
Viruses were recovered from each plasmid as previously described (13, 17). Briefly, LLC-MK2 cells were infected with a recombinant vaccinia virus (VV), vTF7-3, expressing T7 polymerase (9) at a multiplicity of infection (MOI) of 2. Then A24-BSPhis/pSeV, Nef138-β2m/pSeV, Env584-β2m/pSeV, or β2m/pSeV, and plasmids pGEM-N, pGEM-P, and pGEM-L, encoding trans-acting proteins, were transfected simultaneously by using the liposomal transfection reagent DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate; Roche Diagnostics, Rotkreuz, Switzerland). Cells were maintained in serum-free MEM in the presence of 40 μg of 1-β-d-arabinofuranosylcytosine (araC)/ml and 100 μg of rifampin/ml to minimize VV cytopathogenecity. Forty-eight hours after transfection, the cells were harvested, washed with phosphate-buffered saline (PBS), and inoculated into the allantoic cavities of 10-day-old embryonated chicken eggs. After 3 days of incubation, the allantoic fluid was harvested. When the presence of viruses was confirmed by hemagglutination test, the allantoic fluid, diluted 107- to 108-fold, was used for the second propagation to remove vTF7-3. We obtained SeV expressing A24-BSPhis (A24-BSPhis/SeV), Nef138-β2m (Nef138-β2m/SeV), Env584-β2m (Env584-β2m/SeV), or β2m (β2m/SeV) at 4.4 × 107, 2.8 × 108, 2.7 × 108, or 3.0 × 108 cell infectious units/ml, respectively.
ELISA for MHC class Ie-β2m complexes or e-β2m.
A sandwich enzyme-linked immunosorbent assay (ELISA) was performed to detect properly folded MHC class I/e-β2m complexes. A 1-μg/ml concentration of the anti-MHC class I monoclonal antibody (MAb) 3F10 (Ancell, Bayport, Minn.), which recognizes only fully assembled MHC class I/peptide complexes, was used as a capture antibody, and a 125-ng/ml concentration of horseradish peroxidase (HRP)-conjugated anti-human β2m (DAKO A/S, Glostrup, Denmark) was used as a detector antibody. For e-β2m detection, samples were directly coated, and the bound e-β2m was detected with 500 ng of HRP-conjugated anti-human β2m/ml. Purified β2m (Biogenesis, Poole, England) was used as a standard protein for measurement. 3,3′,5,5′-Tetramethylbenzidine was used as an HRP substrate in both ELISAs.
Purification of MHC class Ie-β2m complexes.
MHC class I/e-β2m complexes were purified from the culture supernatants of SeV-infected CV-1 cells. The culture supernatants were centrifuged at 40,000 × g to pellet down the SeV particles and were then purified by affinity chromatography on a Hitrap Chelating HP column (Amersham Pharmacia Biotech, Piscataway, N.J.) in 0.02 M NaHPO4 (pH 7.4)-0.5 M NaCl with a 0 to 0.5 M gradient of imidazole.
Preparation of MHC class Ie-β2m tetramers.
Tetramerization of monomeric MHC class I/e-β2m complexes was performed as previously described (4, 7). BSPs of the purified MHC class I/e-β2m complexes were biotinylated by using the BirA enzyme (Avidity, Denver, Colo.). One milligram of MHC class I/e-β2m complexes was incubated with 10 μg of BirA at 25°C for 18 h in a buffer containing 10 mM Tris-Cl (pH 8.0), 50 mM Bicine (pH 8.3), 10 mM ATP, 10 mM magnesium acetate, and 40 μM biotin. Following the reaction, biotinylated MHC class I/e-β2m complexes were purified on a Superdex 200 column (Amersham Pharmacia) in 20 mM Tris-Cl (pH 8.0)-150 mM NaCl. Finally, the buffer was changed to PBS including protease inhibitor cocktail (Roche Diagnostics) and mixed with phycoerythrin (PE)-conjugated streptavidin (Molecular Probe, Eugene, Oreg.) at a 1:1 ratio of biotinylated MHC class I/e-β2m to biotin binding sites.
MHC class I tetramers made in an E. coli system were obtained from the National Institute of Allergy and Infectious Diseases (NIAID) MHC Tetramer Core Facility (Emory University Vaccine Center).
Generation of CTL lines and clones.
A Nef138-10-specific CTL line was induced from PBMCs of HIV-1-infected individuals carrying HLA-A*2402. A total of 3 × 105 PBMCs were cultured in R10 in 96-well round-bottom tissue culture plates. The next day, 105 stimulator cells (autologous phytohemagglutinin [PHA]-stimulated PBMCs irradiated with 3,300 rads and pulsed with Nef138-10 for 1 h at 10 μM) were added and cultured for 2 weeks in the presence of 10% Lymphocult-T, which supplied 100 U of interleukin-2 (IL-2)/ml, and 1 μg of an anti-CD28 MAb (BD Pharmingen, San Diego, Calif.)/ml. The cells were further stimulated with irradiated autologous B-LCLs pulsed with 10 μM concentrations of peptides for another 7 to 10 days. CMV-specific CTL lines were induced in the same way, by using an HLA-A*2402-restricted peptide from CMV pp65 corresponding to amino acids 328 to 337 (CMVpp65/328-9; QYDPVAALF) (19), from PBMCs of HLA-A*2402-positive patients who were also seropositive for both HIV-1 and CMV. For tetramer staining of CTL lines, we used them after the second stimulation. After the fourth stimulation, the cells were cloned by limiting dilution to 0.8 and 8 cells/well in 96-well round-bottom tissue culture plates. Each well contained 105 irradiated autologous B-LCLs and 5 × 104 irradiated allogeneic PBMCs in the presence of 10 μM peptide in the cloning medium (R10 containing 10% lymphocult-T and 5% PHA-blast culture supernatant).
Tetramer staining and fluorescence-activated cell sorter analysis.
At first each tetramer was incubated with various dilutions to determine the optimal conditions for staining. Cells were stained with PE-labeled tetramers at 37°C for 15 min at the optimal concentration, washed once with 2% FCS-0.1% NaN3 in PBS, and then stained with anti-CD8-allophycocyanin (BD Pharmingen) at 4°C for 20 min. Cells were washed three times and fixed with 1% paraformaldehyde. Stained cells were analyzed by using a FACSCalibur (Becton Dickinson, Mountain View, Calif.) with CellQuest software (Becton Dickinson) and Flowjo software (Tree Star, San Carlos, Calif.).
51Cr release assay.
Cytotoxicity was measured by a standard 51Cr release assay as previously described (18). Briefly, HLA-A24 matched allogeneic B-LCLs were labeled with 100 μCi of Na251CrO4 for 2 h and washed three times with R10. Labeled target cells (2 × 103) were added to a 96-well round bottom microtiter plate with a corresponding amount of peptide or the culture supernatant of e-β2m/SeV-infected cells. After an hour of incubation, effector cells were added and incubated for 4 h. When SeV-infected LCLs were used as target cells, the cells were infected with SeVs, at an MOI of 10, 20 h before addition of the effector cells. The supernatants were collected and analyzed with a microbeta counter. Spontaneous 51Cr release was determined by measuring counts per minute in the supernatants of wells containing only target cells (cpmspn). Maximum release (cpmmax) was determined by measuring the release of 51Cr from target cells in the presence of 2% Triton X-100. Specific lysis was calculated as (cpmexp − cpmspn)/(cpmmax − cpmspn) ×100, where cpmexp represents the counts per minute in the supernatants of wells containing target and effector cells.
RESULTS
Construction of SeVs expressing soluble MHC class I heavy chain or e-β2m.
We made SeVs expressing soluble MHC class I heavy chain, HLA-A*2402, designated A24-BSPhis, with a BSP sequence and a histidine tag (Fig. 1A). We also made β2m connected with an antigenic peptide (epitope) via a 13- or 14-residue linker at the N terminus and named the construct e-β2m (Fig. 1B). Since it is known that SeV replicates efficiently only when the length of the genome is a multiple of 6 (rule of six) (6), the genome length was adjusted by the linker. In this study the HLA-A*2402-restricted HIV-1 epitope Nef138-10 or Env584-11 was used as the antigenic peptide (14). The constructs carrying A24-BSPhis or e-β2m were inserted into the NotI site of pV(−)SeV18+b(+), which contains the V(−) version of the SeV antigenome (16, 44) (Fig. 1C). Twenty-four hours after infection, A24-BSPhis or e-β2m was expressed in A24-BSPhis/SeV- or e-β2m/SeV-infected CV-1 cells, respectively (Fig. 1D). Considerable amounts of e-β2m were secreted from the infected cells into the culture supernatants (Fig. 1D, right). Because of the epitope and linker peptides, the mobilities of Nef138-β2m and Env584-β2m on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels were slightly slower than that of β2m.
Production of soluble MHC class I/e-β2m complexes by SeV-infected cells.
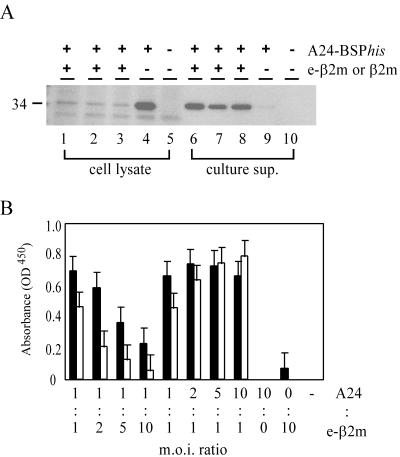
It is known that human MHC class I heavy chain without human β2m fails to be expressed on the cell surface because of its conformational instability (42). When CV-1 cells were infected with A24-BSPhis/SeV alone, A24-BSPhis was detected in the cell lysates but hardly at all in the culture supernatants (Fig. 2A, lanes 4 and 9). When Nef138-β2m/SeV, Env584-β2m/SeV, or β2m/SeV was inoculated together with A24-BSPhis/SeV, large amounts of A24-BSPhis were detected in the culture supernatants (Fig. 2A, lanes 6 to 8) while only small amounts of A24-BSPhis remained in the cell lysates (Fig. 2A, lanes 1 to 3). Thus, the presence of e-β2m stabilized A24-BSPhis by forming stable MHC class I/e-β2m complexes, which could be secreted from the infected cells very efficiently.
FIG. 2.
Secretion of MHC class I/e-β2m complex from SeV-coinfected cells. (A) Detection of A24-BSPhis in SeV-infected cells and culture supernatants. CV-1 cells were infected with A24-BSPhis/SeV with or without e-β2m/SeV at an MOI of 3. Twenty four hours postinfection, cells and culture supernatants were harvested. Cells were lysed, and culture supernatants were centrifuged at 40,000 × g. Ten micrograms of cell lysates (lanes 1 to 5) or 10 μl of supernatants (lanes 6 to 10) was separated on an SDS-PAGE gel with a 10 to 20% gradient. The position of the 34-kDa product is indicated on the left. Lanes 1 and 6, A24BSPhis/SeV plus Nef138-β2m/SeV; lanes 2 and 7, A24BSPhis/SeV plus Env584-β2m/SeV; lanes 3 and 8, A24-BSPhis/SeV plus β2m/SeV; lanes 4 and 9, A24-BSPhis/SeV only; lanes 5 and 10, wild-type SeV. (B) ELISA for MHC class I/e-β2m complex detection. CV-1 cells were infected with A24-BSPhis/SeV and either Nef138-β2m/SeV (solid bars) or Env584-β2m/SeV (open bars) at various MOI ratios. Culture supernatants were harvested 3 days postinfection. After centrifugation at 40,000 × g, culture supernatants were assayed in an ELISA specific for fully assembled MHC class I/peptide complexes (described in Materials and Methods). Averages and standard deviations for three wells are given. Results for one representative experiment out of three are shown.
To confirm that MHC class I/e-β2m was folded properly like native MHC class I/peptide complexes, culture supernatants of CV-1 cells coinfected with A24-BSPhis/SeV and e-β2m/SeV were assayed by using a MAb which detects only fully assembled MHC class I/peptide complex (see Materials and Methods). Cells were coinfected with various ratios of A24-BSPhis/SeV to Nef138-β2m/SeV or Env584-β2m/SeV, and culture supernatants were assayed on day 3. Considerable amounts of MHC class I/e-β2m complexes were detected in the culture supernatants. The more the ratio of the MOI of e-β2m/SeV to that of A24-BSPhis/SeV was increased, the less A24-BSPhis/e-β2m complex (A24/e-β2m) was secreted when cells were infected with both Nef138-β2m/SeV and Env584-β2m/SeV (Fig. 2B). In contrast, when the MOI ratio was decreased, A24/Nef138-β2m secretion was not affected but A24/Env584-β2m secretion increased. The optimal ratios for A24/e-β2m complex secretion appeared to be dependent on the sequences of the peptides and partly on the ratio of the two vectors (Fig. 2B). These data also confirmed the proper folding of A24-BSPhis.
We purified A24/Nef138-β2m complexes by affinity chromatography using a histidine tag from the culture supernatant of CV-1 cells coinfected with A24-BSPhis/SeV and Nef138-β2m/SeV at an MOI ratio of 1 to 1. About 1 mg of purified A24/Nef138-β2m complex was obtained from 108 CV-1 cells.
Staining of specific CD8 T cells by SeV-made tetramers.
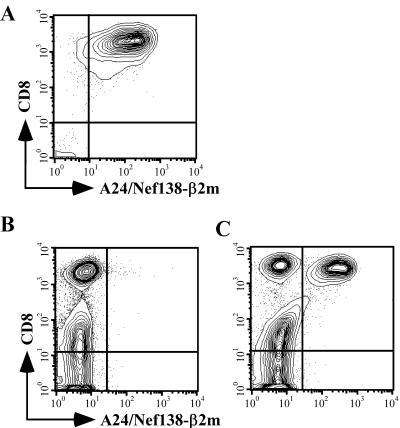
Purified A24/Nef138-β2m complexes were biotinylated at the BSP and multimerized with PE-conjugated streptavidin to form tetramers as previously described (4, 7). A CTL clone specific for Nef138-10 was fully stained with the A24/Nef138-β2m tetramer (Fig. 3A).
FIG. 3.
Staining of CTLs with A24/Nef138-β2m tetramers. (A) A Nef138-10-specific CTL clone was stained with A24/Nef138-β2m tetramers. (B and C) After a second stimulation with cognate peptides, a Nef138-10-specific CTL line (B) and a CMVpp65/328-9-specific CTL line (C) were stained with A24/Nef138-β2m tetramers. A total of 2 × 105 to 5 × 105 cells were stained.
We then tested the specificity of A24/Nef138-β2m tetramers by using CTL lines specific for Nef138-10 or another HLA-A*2402-restricted epitope derived from CMV. Whereas the CMV-specific CTL line was not stained with A24/Nef138-β2m tetramers at all (Fig. 3B), a certain portion of the Nef138-specific CTL lines was stained with A24/Nef138-β2m tetramers (Fig. 3C). These data indicated that tetramers formed by SeV-made MHC class I/e-β2m complexes were fully assembled and recognized by adequate TCRs.
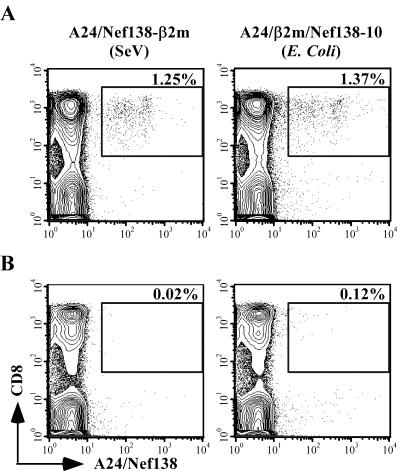
MHC class I/e-β2m complexes in our system have an artificial linker peptide near the epitope which may interfere with recognition by TCRs. In order to test the possibility, we compared the efficiency of our SeV-made tetramers (A24/Nef138-β2m tetramers) with that of E. coli-made tetramers without the linker peptide (A24/β2m/Nef138-10 tetramers). PBMCs from HLA-A*2402-positive uninfected or HIV-1-infected individuals were examined with both tetramers. A total of 1.25 or 1.37% of CD8 T cells among PBMCs from HIV-1-infected individuals and 0.02 or 0.12% of CD8 T cells among PBMCs from non-HIV-1-infected individuals were stained with A24/Nef138-β2m or A24/β2m/Nef138-10 tetramers, respectively (Fig. 4). SeV-made A24/Nef138-β2m tetramers stained Nef138-10-specific CD8 T cells as sensitively as E. coli-made A24/β2m/Nef138-10 tetramers.
FIG. 4.
Staining of PBMCs from HIV-1-infected or uninfected individuals with either the A24/Nef138-β2m (SeV-made) tetramer or the A24/β2m/Nef138-10 (E. coli-made) tetramer. A total of 106 PBMCs from HIV-1-infected individuals (HLA-A2/24, B35/52) (A) or non-HIV-1-infected individuals (HLA-A24, B7/35) (B) were stained with the A24/Nef138-β2m tetramer (left panels) or the A24/β2m/Nef138-10 tetramer (right panels).
Antigenicity of soluble e-β2m and e-β2m/SeV-infected cells.
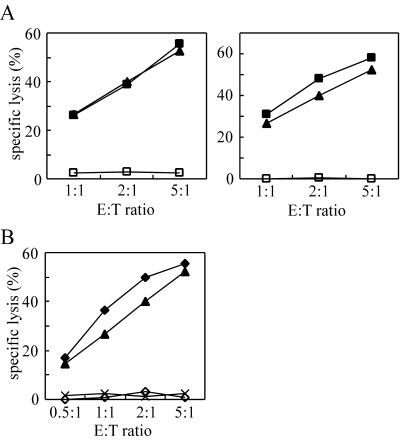
Since it has been reported that e-β2m made in E. coli can stimulate CTLs more stably than the peptide itself (38), we tested this property of SeV-made e-β2m. A CTL killing assay was performed using HLA-A*2402-positive B-LCLs pulsed with either Nef138-β2m, Env584-β2m, or the Nef138-10 peptide. The Nef138-10 peptide was used at 100 nM, a concentration at which the killing activity was saturated (data not shown). CV-1 cells were infected with Nef138-β2m/SeV or Env584-β2m/SeV, and concentrations of Nef138-β2m and Env584-β2m in the culture supernatants were about 300 nM. They were used at a 10-fold dilution, and the final concentrations were about 30 nM. The Nef138-10-specific CTL clone killed the target cells pulsed with Nef138-β2m as efficiently as cognate-free peptides, while they did not kill Env584-β2m-pulsed B-LCLs at all (Fig. 5A, left panel). We also examined culture supernatants of an Nef138-β2m/SeV- or Env584-β2m/SeV-infected human T-cell line, MT-2. Although the concentration of Nef138-β2m in the MT-2 culture supernatant was 10-fold lower than that for CV-1 (10 to 20 nM) and it was also used at a 10-fold dilution, the cells were killed equally (Fig. 5A, right panel). We then tested the possibility of using e-β2m/SeV as a gene therapy tool. For this purpose, we infected B-LCLs with Nef138-β2m/SeV or Env584-β2m/SeV and used the infected cells as targets of a CTL assay 20 h after infection. Only Nef138-β2m/SeV-infected cells, not Env584-β2m/SeV-infected cells, were killed specifically by the Nef138-10-specific CTL clone (Fig. 5B). Although the SeV infectivity of B-LCLs was lower than 30% (data not shown), 55% of target cells were killed at a 5:1 effector-to-target-cell ratio, suggesting that excreted Nef138-β2m bound to the MHC class I molecules on the cell surface. Taken together, these data suggest that Nef138-β2m can be supplied both endogenously and exogenously.
FIG. 5.
Antigenicity of e-β2m. Standard 4-h 51Cr release assays were performed using a Nef138-10-specific CTL clone derived from an HIV-1 infected individual. (A) As target cells, HLA-A24-matched B-LCLs were pulsed with the culture supernatant of Nef138-β2m/SeV (solid squares)- or Env584-β2m/SeV (open squares)-infected CV-1 (left) or MT-2 (right) cells. CV-1 or MT2 cells were infected with SeVs at an MOI of 3, and the culture supernatant was harvested 24 h postinfection and filtered with a 0.22-μm-pore-size membrane. The culture supernatants were used at a 10-fold dilution, and Nef138-β2m and Env584-β2m from CV-1 or MT-2 cells were used at about 30 or 2 nM, respectively. (B) B-LCLs infected with either Nef138-β2m/SeV (solid diamonds), Env584-β2m/SeV (open diamonds), or wild-type SeV (crosses) were also used as target cells. B-LCLs were infected with SeVs at an MOI of 10 and used as target cells 24 h postinfection. As a control, B-LCLs were pulsed with the Nef138-10 peptide at 100 nM (solid triangles).
DISCUSSION
We have described a SeV vector system which can be used to express various forms of MHC class I molecules. First, plenty of MHC class I tetramers which worked as efficiently and specifically as those made in E. coli could be produced with ease. Second, soluble e-β2m could stimulate CTLs as efficiently as cognate peptides when applied to target cells expressing proper MHC class I heavy chains. Third, infection with the viral vector encoding e-β2m could induce expression of proper MHC class I molecules with the intended epitope on the surfaces of target cells expressing proper MHC class I heavy chains.
In the E. coli systems which have been used generally to make tetramers, the heavy chain and β2m must be obtained and purified separately to high purity and then refolded in the presence of the peptide in vitro. The yield of purified MHC class I/peptide complexes is 10 to 15% (11). In our mammalian system, more than 1 mg of MHC class I/e-β2m complexes was produced from 108 CV-1 cells, and almost 100% of purified MHC class I/peptide complexes could be recovered by affinity chromatography using a histidine tag. It has been known that β2m and/or peptide absence leads to misfolding and degradation of the MHC class I heavy chain because of the “quality control” function in the endoplasmic reticulum (ER) (28). Actually, when CV-1 cells were infected with A24-BSPhis/SeV alone, as shown in Fig. 2A, there was little secretion of A24-BSPhis. However, when cells were coinfected with A24-BSPhis/SeV and e-β2m/SeV or β2m/SeV, high levels of A24-BSPhis were secreted. When cells were coinfected with A24-BSPhis/SeV and e-β2m/SeV at the most efficient MOI ratio, A24-BSPhis scarcely remained in the cells 24 h after infection. It is inferred that A24-BSPhis formed stable complexes with e-β2m immediately after its synthesis and was transported to the cell surface and secreted. These results also indicated that the human MHC class I heavy chain, A24-BSPhis, does not form stable complexes with the endogenously expressed monkey β2m in CV-1 cells. This makes it possible to obtain MHC class I/peptide complexes which have a single epitope.
e-β2m display enhanced MHC stabilization and antigenicity compared with those of free peptides because of the adjuvant effect of β2m, especially in the case of peptides with lower affinity with MHC class I (38). Some tetramers are difficult to make, and one of the factors that determines the success of tetramer production is the binding affinity of the peptide for the MHC class I molecule (this information can be found at the NIAID Tetramer Core Facility website, http://www.emory.edu/WHSC/TETRAMER/faq.html). Such MHC class I/peptide complexes, which are impossible to make in the E. coli system, may be produced with this SeV system.
MHC class I molecules which presented glycopeptides were recognized by α/β T cells (30). SeV-made tetramers from mammalian cells may turn out to be useful for analysis of modified peptide antigens.
SeV-made tetramers have artificial linker peptides near the TCR recognition site and may interfere with recognition of MHC class I/peptide complexes by some TCRs. When we compared SeV-made tetramers to E. coli-made tetramers without the linker peptides, the same proportion of cells were stained in PBMCs of HIV-1-infected individuals. This result suggested that the hindrance of epitope recognition by linker peptides was minimal, if there was any. This result may also agree with observations of the crystal structure of soluble TCRs bound to MHC class I ligands (10, 12), which show that room exists for linker peptides attached to the C termini of the antigen peptides (39). Further studies are needed to evaluate possible interference by the linker peptides with TCR-MHC class I interaction. Exogenously pulsed and endogenously expressed Nef138-β2m was recognized by a CTL clone specific for Nef138-10. Supernatants of Nef138-β2m/SeV-infected CV-1 or MT-2 cells were used to pulse target cells. The final concentrations were about 30 or 2 nM, respectively. Cells pulsed with these culture supernatants were killed by the CTLs with a sensitivity similar to that of cells pulsed with Nef138-10 peptides at 100 nM, the concentration at which the target cells could be killed most efficiently (Fig. 5A and data not shown). This result indicated that e-β2m is more stable than free peptides on MHC class I complexes and is efficiently recognized by CTLs even at a low concentration. B-LCLs pulsed with Nef138-β2m derived from MT-2 cells were killed more efficiently than those pulsed with Nef138-β2m derived from CV-1 cells even at a 10-fold-lower concentration. MT-2 is a human CD4-positive T-cell line which is transformed with human T-cell leukemia virus type 1 (HTLV-1). HTLV-1-transformed T cells are known to produce a wide spectrum of lymphokines, including gamma interferon and IL-2, which activate T cells (5, 20, 32). Such cytokines secreted by MT-2 cells may be responsible for the killing activity of the CTL clones.
Gene transduction with SeV enabled e-β2m to be expressed endogenously in human cells. When cells are pulsed with peptides or e-β2m, they are considered to replace the peptides of MHC class I/peptide complexes on the cell surface by chance. But when e-β2m is expressed at high levels in a cell, a certain population of MHC class I heavy chains can associate with e-β2m in the ER and may present the epitopes on the cell surface at a high frequency. Fresh e-β2m is also supplied continuously in the supernatant by the infected cells, so the e-β2m is likely to associate with heavy chains within the cell (in the ER) and on the cell surface. In fact, specific lysis was higher than 50% in spite of a lower percentage of infected cells (less than 30%) (Fig. 5B and data not shown).
In some studies MHC class I genes have been transduced to mammalian cells by DNA transfection to present a specific antigen on the cell surface (15, 23, 36, 37). However, the efficiency of gene transduction by DNA transfection may be too low to apply to clinical trials. In a recent report, target cells which presented peptides covalently linked to β2m delivered by a retroviral vector were recognized and killed by appropriate CTL clones (31). However, with a retroviral vector, efficient gene transduction into nondividing cells, such as dendritic cells (DCs), which are the strongest professional antigen-presenting cells, is difficult, and expression of the protein is considered to be low. On the other hand, in this SeV system, the level of foreign gene expression is very high and the host range is broad (24). Particularly, expression of genes inserted into the V(−) version of the SeV vector is very high (44). The SeV vector can also transduce foreign genes in nondividing cells such as neurons (21). In fact, we confirmed that the green fluorescent protein gene could be transduced into DCs efficiently and expressed at high levels by using an SeV vector expressing green fluorescent protein (unpublished data). Furthermore, SeV replication is independent of nuclear functions and does not transform cells by integrating its genetic information into the cellular genome.
Recently, there have been many studies which used DCs to activate immune responses, particularly in cancer therapy. Pilot DC vaccination studies induced specific anticancer responses, including some clinical responses (8, 25, 33, 34). e-β2m-pulsed or e-β2m/SeV-infected DCs may be useful for immunotherapy against cancers and infection with viruses such as HIV and hepatitis C virus.
Acknowledgments
We thank Y. Ishikawa for providing the HLA-A*2402 gene-containing plasmid. We also thank the NIAID Tetramer Facility and the NIH AIDS Research and Reference Reagent Program for providing the MHC class I tetramer. We acknowledge Hiroko Tomiyama and Masanori Matsui for helpful advice on CTL culture. We are also grateful to Hideki Sakahira and Chikaya Moriya for advice on protein manipulations.
This work was partly supported by grants for AIDS research from the Ministry of Health, Labor and Welfare of Japan; by a Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science (JSPS); and by the Japan Health Sciences Foundation.
REFERENCES
- 1.Ahn, K., A. Gruhler, B. Galocha, T. R. Jones, E. J. Wiertz, H. L. Ploegh, P. A. Peterson, Y. Yang, and K. Fruh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, K., T. H. Meyer, S. Uebel, P. Sempe, H. Djaballah, Y. Yang, P. A. Peterson, K. Fruh, and R. Tampe. 1996. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. 15:3247-3255. [PMC free article] [PubMed] [Google Scholar]
- 3.Algarra, I., T. Cabrera, and F. Garrido. 2000. The HLA crossroad in tumor immunology. Hum. Immunol. 61:65-73. [DOI] [PubMed] [Google Scholar]
- 4.Altman, J. D., P. A. Moss, P. J. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D. A., F. B. Nelson, E. L. Reinherz, and D. J. Diamond. 1991. The human interferon-gamma gene contains an inducible promoter that can be transactivated by tax I and II. Eur. J. Immunol. 21:1879-1885. [DOI] [PubMed] [Google Scholar]
- 6.Calain, P., and L. Roux. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective-interfering RNA. J. Virol. 67:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford, F., H. Kozono, J. White, P. Marrack, and J. Kappler. 1998. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity 8:675-682. [DOI] [PubMed] [Google Scholar]
- 8.Fong, L., and E. G. Engleman. 2000. Dendritic cells in cancer immunotherapy. Annu. Rev. Immunol. 18:245-273. [DOI] [PubMed] [Google Scholar]
- 9.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garboczi, D. N., P. Ghosh, U. Utz, Q. R. Fan, W. E. Biddison, and D. C. Wiley. 1996. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384:134-141. [DOI] [PubMed] [Google Scholar]
- 11.Garboczi, D. N., D. T. Hung, and D. C. Wiley. 1992. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA 89:3429-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, K. C., M. Degano, L. R. Pease, M. Huang, P. A. Peterson, L. Teyton, and I. A. Wilson. 1998. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science 279:1166-1172. [DOI] [PubMed] [Google Scholar]
- 13.Hasan, M. K., A. Kato, T. Shioda, Y. Sakai, D. Yu, and Y. Nagai. 1997. Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3′ proximal first locus. J. Gen. Virol. 78:2813-2820. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda-Moore, Y., H. Tomiyama, K. Miwa, S. Oka, A. Iwamoto, Y. Kaneko, and M. Takiguchi. 1997. Identification and characterization of multiple HLA-A24-restricted HIV-1 CTL epitopes: strong epitopes are derived from V regions of HIV-1. J. Immunol. 159:6242-6252. [PubMed] [Google Scholar]
- 15.Kang, X., P. F. Robbins, E. B. Fitzgerald, R. Wang, S. A. Rosenberg, and Y. Kawakami. 1997. Induction of melanoma reactive T cells by stimulator cells expressing melanoma epitope-major histocompatibility complex class I fusion proteins. Cancer Res. 57:202-205. [PubMed] [Google Scholar]
- 16.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, A., Y. Sakai, T. Shioda, T. Kondo, M. Nakanishi, and Y. Nagai. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1:569-579. [DOI] [PubMed] [Google Scholar]
- 18.Kawana, A., H. Tomiyama, M. Takiguchi, T. Shioda, T. Nakamura, and A. Iwamoto. 1999. Accumulation of specific amino acid substitutions in HLA-B35-restricted human immunodeficiency virus type 1 cytotoxic T lymphocyte epitopes. AIDS Res. Hum. Retrovir. 15:1099-1107. [DOI] [PubMed] [Google Scholar]
- 19.Kuzushima, K., N. Hayashi, H. Kimura, and T. Tsurumi. 2001. Efficient identification of HLA-A∗2402-restricted cytomegalovirus-specific CD8+ T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 98:1872-1881. [DOI] [PubMed] [Google Scholar]
- 20.Lal, R. B., and D. L. Rudolph. 1991. Constitutive production of interleukin-6 and tumor necrosis factor-alpha from spontaneously proliferating T cells in patients with human T-cell lymphotropic virus type-I/II. Blood 78:571-574. [PubMed] [Google Scholar]
- 21.Li, H. O., Y. F. Zhu, M. Asakawa, H. Kuma, T. Hirata, Y. Ueda, Y. S. Lee, M. Fukumura, A. Iida, A. Kato, Y. Nagai, and M. Hasegawa. 2000. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 74:6564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mottez, E., C. Jaulin, F. Godeau, J. Choppin, J. P. Levy, and P. Kourilsky. 1991. A single-chain murine class I major transplantation antigen. Eur. J. Immunol. 21:467-471. [DOI] [PubMed] [Google Scholar]
- 23.Mottez, E., P. Langlade-Demoyen, H. Gournier, F. Martinon, J. Maryanski, P. Kourilsky, and J. P. Abastado. 1995. Cells expressing a major histocompatibility complex class I molecule with a single covalently bound peptide are highly immunogenic. J. Exp. Med. 181:493-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai, Y. 1999. Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding. Rev. Med. Virol. 9:83-99. [DOI] [PubMed] [Google Scholar]
- 25.Nestle, F. O., S. Alijagic, M. Gilliet, Y. Sun, S. Grabbe, R. Dummer, G. Burg, and D. Schadendorf. 1998. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat. Med. 4:328-332. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson, K., and G. Klein. 1982. Phenotypic and cytogenetic characteristics of human B-lymphoid cell lines and their relevance for the etiology of Burkitt's lymphoma. Adv. Cancer Res. 37:319-380. [DOI] [PubMed] [Google Scholar]
- 27.Paabo, S., L. Severinsson, M. Andersson, I. Martens, T. Nilsson, and P. A. Peterson. 1989. Adenovirus proteins and MHC expression. Adv. Cancer Res. 52:151-163. [DOI] [PubMed] [Google Scholar]
- 28.Pamer, E., and P. Cresswell. 1998. Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol. 16:323-358. [DOI] [PubMed] [Google Scholar]
- 29.Schatz, P. J. 1993. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Bio/Technology 11:1138-1143. [DOI] [PubMed] [Google Scholar]
- 30.Speir, J. A., U. M. Abdel-Motal, M. Jondal, and I. A. Wilson. 1999. Crystal structure of an MHC class I presented glycopeptide that generates carbohydrate-specific CTL. Immunity 10:51-61. [DOI] [PubMed] [Google Scholar]
- 31.Tafuro, S., U. C. Meier, P. R. Dunbar, E. Y. Jones, G. T. Layton, M. G. Hunter, J. I. Bell, and A. J. McMichael. 2001. Reconstitution of antigen presentation in HLA class I-negative cancer cells with peptide-β2m fusion molecules. Eur. J. Immunol. 31:440-449. [DOI] [PubMed] [Google Scholar]
- 32.Tendler, C. L., S. J. Greenberg, W. A. Blattner, A. Manns, E. Murphy, T. Fleisher, B. Hanchard, O. Morgan, J. D. Burton, D. L. Nelson, et al. 1990. Transactivation of interleukin 2 and its receptor induces immune activation in human T-cell lymphotropic virus type I-associated myelopathy: pathogenic implications and a rationale for immunotherapy. Proc. Natl. Acad. Sci. USA 87:5218-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurner, B., I. Haendle, C. Roder, D. Dieckmann, P. Keikavoussi, H. Jonuleit, A. Bender, C. Maczek, D. Schreiner, P. von den Driesch, E. B. Brocker, R. M. Steinman, A. Enk, E. Kampgen, and G. Schuler. 1999. Vaccination with mage-3A1 peptide-pulsed mature, monocyte-derived dendritic cells expands specific cytotoxic T cells and induces regression of some metastases in advanced stage IV melanoma. J. Exp. Med. 190:1669-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timmerman, J. M., and R. Levy. 1999. Dendritic cell vaccines for cancer immunotherapy. Annu. Rev. Med. 50:507-529. [DOI] [PubMed] [Google Scholar]
- 35.Tokunaga, K., Y. Ishikawa, A. Ogawa, H. Wang, S. Mitsunaga, S. Moriyama, L. Lin, M. Bannai, Y. Watanabe, K. Kashiwase, H. Tanaka, T. Akaza, K. Tadokoro, and T. Juji. 1997. Sequence-based association analysis of HLA class I and II alleles in Japanese supports conservation of common haplotypes. Immunogenetics 46:199-205. [DOI] [PubMed] [Google Scholar]
- 36.Toshitani, K., V. Braud, M. J. Browning, N. Murray, A. J. McMichael, and W. F. Bodmer. 1996. Expression of a single-chain HLA class I molecule in a human cell line: presentation of exogenous peptide and processed antigen to cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 93:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uger, R. A., and B. H. Barber. 1998. Creating CTL targets with epitope-linked β2-microglobulin constructs. J. Immunol. 160:1598-1605. [PubMed] [Google Scholar]
- 38.Uger, R. A., S. M. Chan, and B. H. Barber. 1999. Covalent linkage to β2-microglobulin enhances the MHC stability and antigenicity of suboptimal CTL epitopes. J. Immunol. 162:6024-6028. [PubMed] [Google Scholar]
- 39.White, J., F. Crawford, D. Fremont, P. Marrack, and J. Kappler. 1999. Soluble class I MHC with β2-microglobulin covalently linked peptides: specific binding to a T cell hybridoma. J. Immunol. 162:2671-2676. [PubMed] [Google Scholar]
- 40.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 41.Wiertz, E. J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 42.Williams, D. B., B. H. Barber, R. A. Flavell, and H. Allen. 1989. Role of β2-microglobulin in the intracellular transport and surface expression of murine class I histocompatibility molecules. J. Immunol. 142:2796-2806. [PubMed] [Google Scholar]
- 43.Yang, O. O., P. T. Nguyen, S. A. Kalams, T. Dorfman, H. G. Gottlinger, S. Stewart, I. S. Chen, S. Threlkeld, and B. D. Walker. 2002. Nef-mediated resistance of human immunodeficiency virus type 1 to antiviral cytotoxic T lymphocytes. J. Virol. 76:1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, D., T. Shioda, A. Kato, M. K. Hasan, Y. Sakai, and Y. Nagai. 1997. Sendai virus-based expression of HIV-1 gp120: reinforcement by the V(−) version. Genes Cells 2:457-466. [DOI] [PubMed] [Google Scholar]