Abstract
The DNA-breaking and -joining steps initiating retroviral integration are well understood, but the later steps, thought to be carried out by cellular DNA repair enzymes, have not been fully characterized. Poly(ADP-ribose) polymerase 1 (PARP-1) has been proposed to play a role late during retroviral integration, because infection by human immunodeficiency virus (HIV)-based vectors was reported to be strongly inhibited in PARP-1-deficient fibroblasts. PARP-1, a nuclear enzyme, binds tightly to nicked DNA and synthesizes poly(ADP-ribose) as an early response to DNA damage. To investigate the role of PARP-1 in retroviral integration, we infected wild-type and PARP-1-deficient mouse embryonic fibroblasts (MEFs) separately with two HIV type 1-derived, vesicular stomatitis virus G-pseudotyped lentivirus vectors. Surprisingly, infection of both wild-type and PARP-1-deficient cells was observed with both vectors. Marker gene transduction and provirus formation by one vector was reduced by 45 to 75% compared to the wild type, but the other vector was unaffected by the PARP-1 mutant. In addition, PARP-1-deficient MEFs infected with Moloney murine leukemia virus showed no decrease in virus output after infection compared to the wild type. We conclude that PARP-1 cannot be strictly required for retroviral infection because replication steps, including integration, can proceed efficiently in its absence.
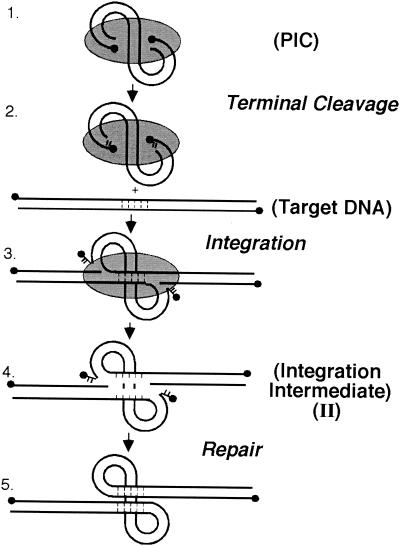
Integration of the human immunodeficiency virus type 1 (HIV-1) cDNA into the genome of the host cell is essential for virus replication. Initially, the virally encoded integrase enzyme binds the ends of the viral cDNA and removes two nucleotides from each 3′ end (Fig. 1, steps 1 and 2) (3, 9, 17, 23). Integrase then joins the recessed 3′ ends to the host DNA (Fig. 1, step 3) (4, 5, 9, 16). Completion of the integration reaction requires polymerization across the gap, removal of the frayed viral 5′ end, and sealing of the new DNA strand by ligation (Fig. 1, steps 4 and 5). In vitro, the final DNA repair steps are not carried out by purified HIV-1 integrase and naked target DNA. Thus, the result is a gapped intermediate in which the 5′ ends of the viral cDNA are not joined to the host DNA. Recent data support the idea that host DNA repair enzymes may be important for the final DNA repair activity (28).
FIG. 1.
DNA-breaking and -joining reactions involved in integration. The gray oval represents the protein factors of the preintegration complex (PIC). The solid circles represent the 5′ DNA ends. See text for explanation.
Poly(ADP-ribose) polymerase 1 (PARP-1) is a predominantly nuclear zinc-finger protein of 113 kDa that participates in DNA repair (for reviews, see references 10 and 11). PARP-1 activity is stimulated by a variety of DNA-damaging agents, including ionizing radiation, oxygen radicals, and alkylating agents. PARP-1 binds tightly to breaks in DNA and then uses NAD+ as a substrate to catalyze attachment of poly(ADP-ribose) polymers to nuclear proteins involved in chromatin architecture, DNA metabolism, or DNA repair, including PARP-1 itself. After automodification, PARP-1 dissociates from the DNA, providing access to other DNA repair factors. Although the molecular details are not well characterized, studies of PARP-1-deficient mice along with in vitro PARP-1 inhibition data implicate PARP-1 in events leading to DNA repair (20, 22, 27). As DNA breaks are known to stimulate PARP-1 activity, it has been proposed that PARP-1 may be involved in the resolution of the gapped intermediate of retroviral integration. Previous studies reported that PARP inhibitors blocked integration of transfected DNA into the mammalian genome and that efficient retroviral infection of mammalian cells can be blocked by inhibition of PARP activity by competitive inhibitors, antisense oligonucleotides, or overexpression of transdominant mutants (12, 14). However, other studies contend that HIV integration is not blocked by a PARP inhibitor in several cell types (2).
Recently, Ha and coworkers used the vector HIV-EGFPΔE, pseudotyped with vesicular stomatitis virus G (VSV-G) envelope, to infect mouse embryo fibroblast (MEF) cells derived from wild-type and PARP-1-deficient mice (15). According to this report, tests at a multiplicity of infection (MOI) of 1 (as determined on Jurkat cells) yielded approximately 93% infection of wild-type fibroblasts after 48 h, compared to infection of only 4% of PARP-1-deficient fibroblasts. It was determined that infection was severely reduced and not merely delayed in the absence of PARP-1, because no further change was seen at 72 h. The reduction in the PARP-1 deletion cells was attributed to a lack of HIV-1 genome integration.
We have been carrying out a long-term study of the proteins involved in repairing integration intermediates (28), and so we sought to determine whether PARP-1 is strictly required for integration, as implied by Ha and coworkers (15). We carried out infections of the same wild-type MEF and PARP-1-deficient MEF cells with two different HIV-based vectors. We found a two- to fourfold reduction in titer in the PARP-1 deletion cells with one HIV-based vector, but with the other we saw no significant difference.
In order to study a more biologically relevant system, Moloney murine leukemia virus (MLV) was also used to infect wild-type and PARP-1-deficient MEFs. We found that both wild-type and PARP-1-deficient MEFs infected with MLV showed similar virus output after infection. We therefore conclude that PARP-1 cannot be strictly required for retroviral integration into the host DNA.
MATERIALS AND METHODS
Cells.
MEF cells derived from PARP-1 wild-type and PARP-1-deficient mice (originally described in reference 26) were obtained from S. H. Snyder and cultured as described previously (15).
Immunoprecipitation of PARP-1 from MEF cells.
Crude cytoplasmic extracts from both wild-type and PARP-1-deficient MEFs were subjected to immunoprecipitation with either a rabbit anti-PARP-1 antibody (Serotec Inc., Raleigh, N.C.) or normal rabbit serum. Immunoprecipitated material was separated by sodium dodecyl sulfate (SDS)-8 to 16% gradient polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane. The blot was probed with a monoclonal antibody against PARP-1 (C2-10; Alexis Corporation, San Diego, Calif.), incubated with donkey anti-mouse secondary horseradish peroxidase conjugate, and visualized using ECL Plus reagent (Amersham-Pharmacia Biotech, Piscataway, N.J.).
Determination of poly(ADP-ribose) synthesis in cell extracts.
In order to compare poly(ADP-ribose)-synthesizing activities of wild-type and PARP-1-deficient cells, 250 μg of protein from each extract was incubated for 10 min at 25°C in 100 μl of assay buffer (50 mM Tris-Cl [pH 8.0], 4 mM MgCl2, 0.2 mM dithiothreitol), with [α-32P]NAD+ (final concentration, 100 nCi/μl) and 2 μg of histones (as added PARP substrates) per ml, with or without the addition of 10 μg of sonicated salmon sperm DNA per ml. The reaction was stopped by the addition of 5% (wt/vol) trichloroacetic acid containing 1% (wt/vol) inorganic phosphate. The acid-insoluble material was washed three times in the same solution and one time in 95% ethanol, and the radioactivity was measured by scintillation counting.
Virus production and infection.
HIV-based vectors were made in two ways. In the first, p156RRLsinPPTCMVGFPPRE (referred to in this paper as HIV-sinPPT and described in references 13 and 29) was cotransfected with pDeltaR9 (21) and pVSV-G (Ling Li, personal communication) into subconfluent human embryonic kidney 293T cells by using calcium phosphate. The medium was replaced 14 to 18 h later with Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml), with the addition of 10 mM sodium butyrate, then replaced 6 h later with fresh medium without sodium butyrate. HIV-EGFPΔE plasmid DNA was obtained from R. F. Siliciano and cotransfected with pVSV-G as described previously (15). Stocks were harvested at 48 h posttransfection and then filtered through a 0.45-μm-pore-size filter, aliquoted, and stored at −80°C. Both 293T and wild-type MEFs were infected with serially diluted virus for 48 h, and the virus titer was determined by counting green centers. MEFs were seeded into 10-cm-diameter dishes on the day before infection to 20% confluence at the time of infection. Infections were performed with 10 μg of DEAE dextran per ml and HIV vector stock at MOIs of 1 and 10 as determined on wild-type MEFs. Mock infections were carried out using DEAE dextran only. Cells were harvested after 48 h and counted, and equal numbers of cells were replated for analysis at the 72-h time point.
For studies with MLV, MLV stocks were harvested from MLV-K producer cells and filtered before use (19). Wild-type and PARP-1-deficient MEFs were seeded at 105 cells into 12-well dishes on the day before infection. For infection, 50, 100, or 500 μl of MLV stock was added to wells and infection was allowed to proceed for 5 h, after which the cells were washed with 1× phosphate-buffered saline (PBS) and medium was replaced. At 48 h postinfection, 100 μl of viral supernatant was collected, filtered, and centrifuged at 14,000 rpm in an Eppendorf table top centrifuge for 1 h at 4°C. The supernatant was removed, leaving 10 μl, and the pellet was separated by SDS-PAGE and analyzed by Western blotting with anti-MLV capsid (Quality Biotech, Camden, N.J.) as described previously (18).
Flow cytometry.
MEFs were detached from plates by using trypsin-EDTA, washed twice with 1× PBS, and resuspended in 4% paraformaldehyde-1× PBS for 10 min at room temperature. After being washed with 1× PBS, MEFs were analyzed by fluorescence-activated cell sorting (FACS), and expression of green fluorescent protein (GFP) was quantitated with CellQuest software (BD Immunocytometry Systems, San Jose, Calif.). Gates were established such that the background was <0.1% of 30,000 events of the mock-infected culture.
Assay for integrated HIV-1.
MEFs were mock infected or infected with HIV-sinPPT or HIV-EGFPΔE virus at an MOI of 10, grown for 30 days to ensure that all of the extrachromosomal forms of viral DNA were lost, and then harvested, and genomic DNA was extracted. For fluorescence-monitored quantitative PCR (TaqMan) analysis, 250 ng of genomic MEF DNA was assayed for HIV-1 cDNA integration, normalizing with a separate standard curve for HIV-sinPPT or HIV-EGFPΔE plasmid. A primer set was used to amplify the late reverse transcriptase product amplicon internal to the viral cDNA as described previously (6).
For Southern blotting, 10 μg of genomic DNA isolated from HIV-sinPPT-infected MEF cells was digested with BamHI and EcoRI to isolate a 1.3-kb fragment containing the GFP-coding region. The DNA fragments were separated on a 0.8% agarose gel and then transferred overnight to a 0.45-μm-pore-size nylon transfer membrane, which was hybridized with a randomly primed anti-gfp probe.
RESULTS
Infection of wild-type and PARP-1-deficient MEF cells with HIV-1-based vectors.
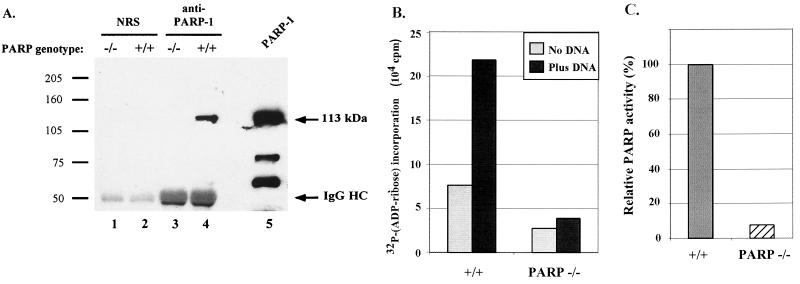
To begin our studies of the role of PARP-1 in integration, we obtained wild-type and PARP-1-deficient MEF cells (26) and confirmed through immunoprecipitation analysis that the PARP-1-deficient MEFs did not contain PARP-1 protein (Fig. 2A). Lysates were prepared from PARP-1 deletion and wild-type MEFs, and a polyclonal anti-PARP-1 antibody was used for immunoprecipitation. The captured proteins were then analyzed by Western blotting with a PARP-1 monoclonal antibody as a probe. Comparison of lanes 3 and 4 of Fig. 2A shows that the knockout cells lacked PARP-1 protein, as expected.
FIG. 2.
PARP-1 protein is absent in PARP-1-deficient MEF cells. (A) Cytoplasmic extracts from wild-type (+/+) or PARP-1-deficient (−/−) MEF cells were immunoprecipitated with normal rabbit serum (NRS) (lanes 1 and 2) or anti-PARP polyclonal antibody (lanes 3 and 4), separated by SDS-8 to 16% PAGE along with 20 ng of purified bovine PARP protein as a standard (lane 5), and then transferred to a polyvinylidene difluoride membrane and probed with an anti-PARP monoclonal antibody. IgG HC, immunoglobulin G heavy chain. (B) Comparison of stimulation of PARP activity by DNA in wild-type and PARP-1-deficient MEF cell extracts. Cell extracts were incubated at 25°C for 10 min with [α-32P]NAD+ and 2 μg of histones per ml, with or without the addition of 10 μg of sonicated salmon sperm DNA per ml. [32P]ADP-ribose incorporation into the acid-insoluble material was measured by scintillation counting. (C) Relative PARP activity, expressed as the ratio between the radioactivity of the acid-insoluble material produced by the wild-type (+/+) and PARP-1-deficient (−/−) cell extracts (plotted to facilitate comparison with previous work [1]).
To test the PARP-dependent repair activity in our stocks of PARP-1-deficient and wild-type cells, the stimulation of PARP activity by broken DNA was assayed according to the method of Ame and coworkers (1). The PARP-1-deficient cells were found to be diminished in PARP activity to the degree expected from published characterizations of the PARP-1 knockout cells (Fig. 2B and C) (1, 24).
Next, we compared infections with two HIV-based vectors, the HIV-EGFPΔE vector used by Ha and coworkers (15) and the HIV-sinPPT vector described previously (13, 29). The HIV-EGFPΔE vector has a deletion in the envelope gene and a substitution of the gfp gene in this region (15). The HIV-sinPPT vector contains (i) deletions in the long terminal repeats (LTRs) blocking gene expression after integration; (ii) the central polypurine tract normally present in the integrase-coding region, which has been proposed to bolster infection (7), and (iii) a gfp gene under control of the cytomegalovirus promoter (for further details see references 13 and 29).
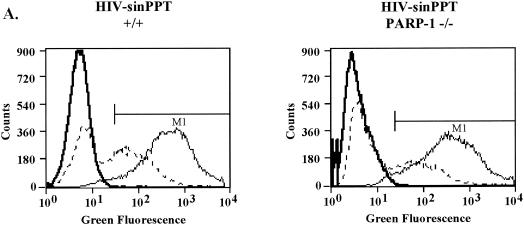
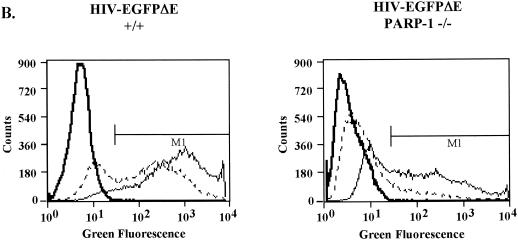
We exposed the MEF cells to the HIV vector particles, each pseudotyped with the VSV-G envelope protein, at MOIs of 1 and 10 (as determined on wild-type MEFs). At 48 h after infection, aliquots of cells were analyzed by FACS to quantify expression of the GFP marker transduced by each vector (Fig. 3A to C). Sample FACS assays are shown in Fig. 3A and B, and data are summarized in Fig. 3C.
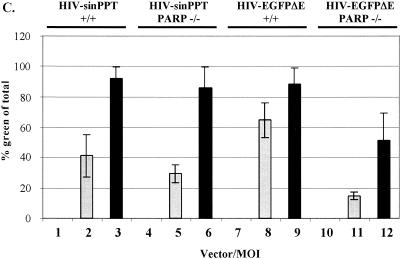
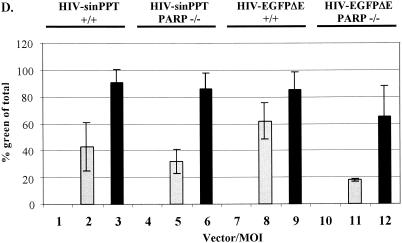
FIG. 3.
Infection of PARP-1-deficient MEF cells with HIV-sinPPT or HIV-EGFPΔE. (A and B) Sample FACS profiles from infection of wild-type (+/+) (left panels) and PARP-1-deficient (−/−) (right panels) MEF cells with HIV-sinPPT (A) or HIV-EGFPΔE (B), analyzed after 48 h. M1 indicates a cutoff in the fluorescence signal so that 99.9% of uninfected cells were excluded. Thick lines mock infection; dashed lines, MOI of 1; thin lines, MOI of 10. (C to E) Summary of FACS measurements after 48 h (C) and 72 h (D) and cell numbers present 48 h after infection (E). Bars 1, 4, 7, and 10, mock infections (open bars); bars 2, 5, 8, and 11, infections at an MOI of 1 (shaded bars); bars 3, 6, 9, and 12, infections at an MOI of 10 (black bars). Error bars indicate standard deviations.
For the wild-type fibroblasts, infection with HIV-sinPPT at MOIs of 1 and 10 yielded an average of 40 and 95% infection, respectively (Fig. 3C, bars 2 and 3). To our surprise, infecting the PARP-1-deficient MEF cells at MOIs of 1 and 10 yielded an average of 30 and 85% infection (Fig. 3c, bars 5 and 6). Thus, results of infections with the HIV-sinPPT vector did not show a significant decrease in the PARP-1-deficient MEFs.
To determine whether this effect was due to our use of the HIV-sinPPT vector, we obtained the HIV-EGFPΔE plasmid and produced viral stocks for a side-by-side comparison. After exposure to HIV-EGFPΔE virus at an MOI of 1 or 10 for 48 h, an average of 65 and 90% of the wild-type fibroblasts were infected, respectively (Fig. 3C, bars 8 and 9). In the PARP-1-deficient MEF cells, 16 and 50% of cells were infected, respectively (Fig. 3C, bars 11 and 12). Thus, we detected a decrease in infectivity between the wild-type and PARP-1 deletion MEF cells infected with HIV-EGFPΔE (45 to 75% over several data sets [Fig. 3 and data not shown]), but our results did not reproduce the 96% decrease in infection of the PARP-1 deletion cells reported previously.
To check whether greater differences between PARP-1 deletion and wild-type cells were evident after longer times, we extended this analysis to include a 72-h time point, as described by Ha et al. (15). Figure 3D shows that results similar to those at the 48-h time point were obtained.
We also considered the possibility that the previously reported reduced infection of PARP-1-deficient cells might be due to increased toxicity of infection in these knockout cells. According to this idea, there would be apparently reduced infection in PARP-1-deficient MEFs because infected knockout cells were preferentially killed in the infection process. We counted cell numbers at 48 h (Fig. 3E) and 72 h (not shown) and did indeed observe some cytotoxicity in infections with the HIV-EGFPΔE vector, but cell death was similar in wild-type and PARP-1 deletion cells. Thus, differential killing of PARP-1 deletion and wild-type cells does not seem to explain the apparent difference in titer. We conclude that the deletion of the gene for PARP-1 either had no effect on infection (for the HIV-sinPPT vector) or reduced infection only 45 to 75% (for the HIV-EGFPΔE vector).
Integration of HIV-1 cDNA into PARP-1-deficient MEF cells infected with HIV-sinPPT and HIV-EGFPΔE.
Cells infected at an MOI of 10 were grown for 30 days, a time known to be sufficient to dilute out unintegrated viral cDNA forms (6). At this time cells were harvested, genomic DNA was extracted, and the HIV cDNA forms quantitated by fluorescence-monitored PCR (TaqMan). In this method, the DNA is amplified by a primer set internal to the viral DNA. A third probe oligonucleotide anneals between the amplification primers. The probe contains a reporter-quencher pair that is separated by the exonuclease activity of Taq polymerase during amplification; thus, amplification leads to an increase in fluorescence intensity. Viral cDNA copies can then be quantified by reading the fluorescence of the released reporter during each PCR cycle and comparison to a standard curve.
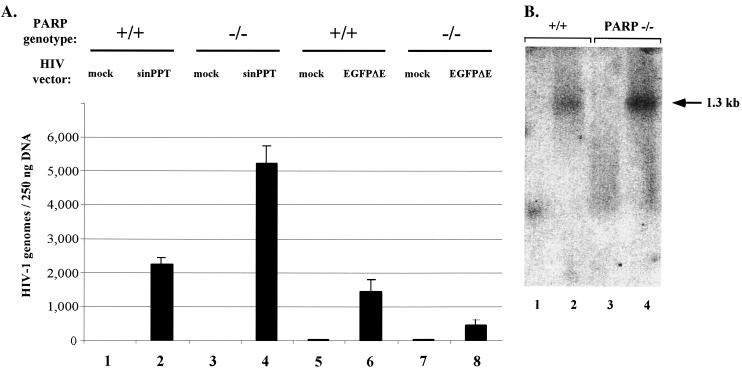
Viral genomes were detected in both wild-type and PARP-1-deficient MEF genomic DNAs infected with HIV-sinPPT (Fig. 4A, bars 2 and 4) and HIV-EGFPΔE (Fig. 4A, bars 6 and 8) virus but not in mock infections (Fig. 4A, bars 1, 3, 5, and 7). For the HIV-sinPPT vector, the knockout actually shows about twofold higher numbers of integrated proviruses (Fig. 4A, compare bars 2 and 4). For the HIV-EGFPΔE virus, there was an approximately 70% decrease in the number of proviruses in the PARP-1 deletion cells (Fig. 4A, compare bars 6 and 8). The results for the HIV-sinPPT case were confirmed by using Southern blot analysis with probing for the presence of gfp from the viral vector in the host genome. Genomic DNA was purified from the infected cells cultured for 30 days and cut with restriction enzymes to liberate a 1.3-kb GFP-containing fragment from the HIV-sinPPT vector. Analysis with an anti-gfp probe detected a 1.3-kb band in both the wild-type and PARP-1 deletion DNA samples (Fig. 4B, lanes 2 and 4) but not in those isolated from mock infections (Fig. 4B, lanes 1 and 3). The amount of viral cDNA in PARP-1-deficient cells was approximately twofold higher than that in wild-type cells as determined by PhosphorImager analysis and quantitation using ImageQuant software. Because the FACS data with the HIV-sinPPT vector show similar gfp expression in both cell types, the approximately twofold difference in viral cDNA is of questionable significance.
FIG. 4.
DNA integration assayed by fluorescence-monitored PCR and Southern blotting. (A) Measurement of integrated DNA copies in genomic DNA by fluorescence-monitored PCR (TaqMan) for wild-type (+/+) (bars 1, 2, 5, and 6) and PARP-1-deficient (−/−) (bars 3, 4, 7, and 8) MEF cells. TaqMan results for bars 1 though 4 were determined from a standard curve using copy standards from the HIV-sinPPT vector plus 250 ng of MEF wild-type genomic DNA. Similarly, results for bars 5 through 8 were determined from a standard curve using HIV-EGFPΔE copy standards plus 250 ng of MEF wild-type DNA. Infections were as follows: bars 1, 3, 5, and 7, mock; bars 2 and 4, HIV-sinPPT, bars 6 and 8, HIV-EGFPΔE. All samples were amplified using a primer set internal to the viral cDNA (see Materials and Methods). Error bars indicate standard deviations. (B) Southern blot analysis using an anti-gfp probe to detect the presence of a 1.3-kb fragment containing the GFP-coding region in genomic DNA isolated from HIV-sinPPT-infected MEFs. Lanes 1 and 3, mock infections. A 1.3-kb band is detected in HIV-sinPPT-infected wild-type (+/+) (lane 2) and PARP-1-deficient (−/−) (lane 4) MEF cells.
Infection of wild-type and PARP-1-deficient MEF cells with MLV.
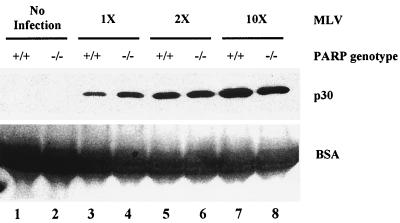
To examine the effect of infection with a more biologically relevant retrovirus, MEFs were incubated with three different input amounts of MLV and washed with PBS, and then fresh medium was added to the cells. At 48 h postinfection, the supernatant from the infections was harvested and analyzed for viral output by assaying for the presence of MLV capsid (p30). MLV capsid protein was not found in uninfected samples (Fig. 5, lanes 1 and 2) but was detected in supernatants from both wild-type and PARP-1-deficient infections (Fig. 5, lanes 3 to 8). Upon quantitation of capsid protein from several data sets, the supernatant from the PARP-1-deficient infection showed approximately equal viral output compared to the wild type, regardless of the MOI. We conclude that PARP-1-deficient MEFs are as susceptible to infection by MLV as the wild-type cells.
FIG. 5.
MLV viral output in wild-type and PARP-1-deficient MEFs. MEFs were mock infected or infected with 50 μl (1×), 100 μl (2×), or 500 μl (10×) of MLV stock. At 48 h postinfection, 100 μl of viral supernatant was collected, pelleted, and separated by SDS-12% PAGE, which was analyzed with anti-MLV capsid antibody by Western blotting (upper panel). MLV p30 (capsid) is detected by Western blotting in supernatant from both wild-type (+/+) and PARP-1-deficient (−/−) cells infected with various amounts of MLV (lanes 3 through 8) but not in uninfected supernatant (lanes 1 and 2). The Ponceau S stain of the blot (lower panel) shows bovine serum albumin from the fetal bovine serum in the medium, which serves as a loading control.
DISCUSSION
A previous study has reported that PARP-1 is required for infection by HIV-based vectors (15); however, our results in a side-by-side comparison using the same infection conditions suggest that this is not universal for all HIV-based vectors. A two- to fourfold decrease in infection by the HIV-EGFPΔE vector was observed in the PARP-1-deficient MEFs compared to the wild type, but it was not significantly different in the case of the HIV-sinPPT vector. In addition, in our hands, the qualitative difference between wild-type and PARP-1 deletion cells was consistently less than previously reported for the HIV-EGFPΔE vector. We readily detected integrated proviral DNA in both wild-type and PARP-1-deficient cells.
The HIV-sinPPT and HIV-EGFPΔE vectors are both VSV-G-pseudotyped derivatives of the NL4-3 isolate, but they differ in their composition. The HIV-EGFPΔE virus was constructed from the NL4-3 proviral clone by removing the env sequence and replacing it with an in-frame enhanced GFP sequence (the VSV-G envelope is expressed from a separate plasmid). The HIV-sinPPT virus was produced from a three-component system in which HIV gag-pol (Δψ), a vector encoding GFP and containing a deletion in the 3′ LTR, and VSV-G envelope are expressed from separate plasmids. After a round of reverse transcription, the self-inactivating (sin) deletions appear in both LTRs of the HIV-sinPPT cDNA and block gene expression from the LTRs after integration. The HIV-sinPPT vector also contains the central polypurine tract present in the integrase-coding region of wild-type HIV-1, which has been proposed to bolster infection (7). Although some cytotoxicity was observed in infections with the HIV-EGFPΔE vector (Fig. 3E, bars 7 to 12), cell death was similar in both wild-type and PARP-1 deletion cells, so differential toxicity does not seem to explain the difference. How the difference in vectors explains the difference in the requirement for PARP-1 is unclear.
Due to the discrepancy in the results between the VSV-G-pseudotyped HIV vectors, we chose to infect the wild-type and PARP-1-deficient MEFs with MLV, a retrovirus that naturally infects murine cells. MLV infected both wild-type and PARP-1-deficient cells, as evidenced by quantification of viral antigen in culture supernatants 48 h later (Fig. 5). In several data sets, we observed approximately equal amounts of viral output in the PARP-1-deficient cells and the wild type. Thus, we conclude that PARP-1 is not required for infection of murine cells by MLV.
It is not possible to completely discount the involvement of PARP activity in the integration process, because the absence of PARP-1 may be complemented by other PARP proteins. PARP-1-deficient MEFs retain a low level of PARP activity that is stimulated by DNA damage (Fig. 2B and C), presumably contributed by the PARP-2 protein, which is known to act in response to DNA damage (1, 24). Cells also contain several other PARP proteins, which could theoretically contribute to retroviral replication (PARP-3, tankyrase-1, tankyrase-2, and VPARP) (8, 25). We also cannot rule out the possibility that the subline of the PARP-1 deletion cells tested here has developed a compensating mutation, bypassing the need for PARP-1, that does not affect PARP activity.
In summary, although we cannot rule out the participation of PARP activity in the integration process, we conclude that PARP-1 cannot strictly be required for integration of retroviral cDNA into the genomes of murine cells, because VSV-G-pseudotyped HIV-1 vectors and MLV can integrate in its absence.
Acknowledgments
We thank R. Siliciano and S. Snyder for materials and members of the Salk Institute Infectious Disease Laboratory for helpful discussions.
This work was supported by NIH grants GM56553 and AI34786 to F.B., the James B. Pendleton Charitable Trust, the Berger Foundation, and the family of Cornelia Mackey.
REFERENCES
- 1.Ame, J. C., V. Rolli, V. Schreiber, C. Niedergang, F. Apiou, P. Decker, S. Muller, T. Hoger, J. Menissier-de Murcia, and G. de Murcia. 1999. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274:17860-17868. [DOI] [PubMed] [Google Scholar]
- 2.Baekelandt, V., A. Claeys, P. Cherepanov, E. De Clercq, B. D. Strooper, B. Nuttin, and Z. Debyser. 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74:11278-11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman, F. D. 1999. Host proteins in retroviral cDNA integration. Adv. Virus Res. 52:301-317. [DOI] [PubMed] [Google Scholar]
- 4.Bushman, F. D., and R. Craigie. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. USA 88:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushman, F. D., T. Fujiwara, and R. Craigie. 1990. Retroviral DNA integration directed by HIV integration protein in vitro. Science 249:1555-1558. [DOI] [PubMed] [Google Scholar]
- 6.Butler, S., M. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV cDNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 7.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription: a termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 8.Cook, B. D., J. N. Dynek, W. Chang, G. Shostak, and S. Smith. 2002. Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22:332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 10.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249-268. [PMC free article] [PubMed] [Google Scholar]
- 11.de Murcia, G., and J. Menissier de Murcia. 1994. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 19:172-176. [DOI] [PubMed] [Google Scholar]
- 12.Farzaneh, F., G. N. Panayotou, L. D. Bowler, B. D. Hardas, T. Broom, C. Walther, and S. Shall. 1988. ADP-ribosylation is involved in the integration of foreign DNA into the mammalian cell genome. Nucleic Acids Res. 16:11319-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Follenzi, A., L. E. Ailes, S. Bakovic, M. Gueuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 14.Gaken, J. A., M. Tavassoli, S.-U. Gan, S. Villian, I. Giddings, D. C. Darling, J. Galea-Lauri, M. G. Thomas, H. Abedi, V. Schreiber, H. Menissier de Murcia, M. K. L. Collins, S. Shall, and F. Farzaneh. 1996. Efficient retroviral infection of mammalian cells is blocked by inhibition of poly (ADP-ribose) polymerase activity. J. Virol. 70:3992-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha, H. C., K. Juluri, Y. Zhou, S. Leung, M. Hermankova, and S. H. Snyder. 2001. Poly(ADP-ribose) polymerase 1 is required for efficient HIV-1 integration Proc. Natl. Acad. Sci. USA 98:3364-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz, R. A., G. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 17.Katzman, M., R. A. Katz, A. M. Skalka, and J. Leis. 1989. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 63:5319-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, L., J. M. Olvera, K. Yoder, R. S. Mitchell, S. L. Butler, M. R. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, A. D., and I. M. Verma. 1984. Two base changes restore infectivity to a noninfectious molecular clone of Moloney murine leukemia virus (pMLV-1). J. Virol. 49:214-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison, C., G. C. Smith, L. Stingl, S. P. Jackson, E. F. Wagner, and Z. Q. Wang. 1997. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat. Genet. 17:479-482. [DOI] [PubMed] [Google Scholar]
- 21.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 22.Shall, S., and G. de Murcia. 2000. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460:1-15. [DOI] [PubMed] [Google Scholar]
- 23.Sherman, P. A., and J. A. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 87:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shieh, W. M., J. C. Ame, M. V. Wilson, Z. Q. Wang, D. W. Koh, M. K. Jacobson, and E. L. Jacobson. 1998. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 273:30069-30072. [DOI] [PubMed] [Google Scholar]
- 25.Smith, S. 2001. The world according to PARP. Trends Biochem. Sci. 26:174-179. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Z. Q., B. Auer, L. Stingl, H. Berghammer, D. Haidacher, M. Schweiger, and E. F. Wagner. 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9:509-520. [DOI] [PubMed] [Google Scholar]
- 27.Wang, Z. Q., L. Stingl, C. Morrison, M. Jantsch, M. Los, K. Schulze-Osthoff, and E. F. Wagner. 1997. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11:2347-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoder, K., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zufferey, R., T. Dull, R. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72:9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]