Abstract
It was reported previously that four baby hamster kidney (BHK) proteins with molecular masses of 108, 60, 50, and 42 kDa bind specifically to the 3′-terminal stem-loop of the West Nile virus minus-stand RNA [WNV 3′(−) SL RNA] (P. Y. Shi, W. Li, and M. A. Brinton, J. Virol. 70:6278-6287, 1996). In this study, p42 was purified using an RNA affinity column and identified as TIAR by peptide sequencing. A 42-kDa UV-cross-linked viral RNA-cell protein complex formed in BHK cytoplasmic extracts incubated with the WNV 3′(−) SL RNA was immunoprecipitated by anti-TIAR antibody. Both TIAR and the closely related protein TIA-1 are members of the RNA recognition motif (RRM) family of RNA binding proteins. TIA-1 also binds to the WNV 3′(−) SL RNA. The specificity of these viral RNA-cell protein interactions was demonstrated using recombinant proteins in competition gel mobility shift assays. The binding site for the WNV 3′(−) SL RNA was mapped to RRM2 on both TIAR and TIA-1. However, the dissociation constant (Kd) for the interaction between TIAR RRM2 and the WNV 3′(−) SL RNA was 1.5 × 10−8, while that for TIA-1 RRM2 was 1.12 × 10−7. WNV growth was less efficient in murine TIAR knockout cell lines than in control cells. This effect was not observed for two other types of RNA viruses or two types of DNA viruses. Reconstitution of the TIAR knockout cells with TIAR increased the efficiency of WNV growth, but neither the level of TIAR nor WNV replication was as high as in control cells. These data suggest a functional role for TIAR and possibly also for TIA-1 during WNV replication.
Flaviviruses are transmitted between bird and mammalian hosts via mosquitoes or ticks. Flaviviruses, such as dengue virus, Japanese encephalitis virus, West Nile virus (WNV), St. Louis encephalitis virus, Murray Valley virus, and tick-borne encephalitis virus, can sometimes cause severe disease in infected humans (10, 25). The genomes of flaviviruses are single-stranded, positive-polarity RNAs of approximately 11 kb and encode a single large polyprotein that is posttranslationally processed by viral and cellular proteases into three structural proteins and seven nonstructural proteins (35). During the flavivirus replication cycle, which takes place in the cytoplasm of infected cells, the genomic RNA serves as the only viral mRNA and is also the template for transcription of the complementary minus-strand RNA. The minus-strand RNA in turn serves as a template for the synthesis of genomic RNA. Plus-strand synthesis is 10 to 100 times more efficient than minus-strand synthesis (35). The noncoding regions (NCRs) of the flavivirus genome contain terminal RNA structures that are conserved between divergent flaviviruses even though only short sequences in these regions are conserved (8, 9, 28, 37, 38). The terminal RNA structures located at the 3′ ends of the genome and complementary minus-strand RNAs differ from each other in shape and size. Deletion or mutation of either 3′-terminal structure in flavivirus infectious clones resulted in no progeny virus production, indicating that these regions are essential for virus replication (10a, 32, 46). However, specific cis-acting signal sequences within these structures have not yet been mapped or functionally analyzed. The WNV 3′-terminal RNA plus- and minus-strand structures have previously been reported to bind specifically to different sets of cell proteins (3, 38).
Understanding the mechanisms and components involved in the initiation and regulation of nascent viral-genome RNA synthesis from the minus-strand template is the ultimate goal of ongoing studies. The formation in solution of the 3′-terminal stem-loop structure of the WNV minus-strand RNA [WNV 3′(−) SL RNA] was previously confirmed by RNase structure probing (38). Three RNA-protein complexes (RPCs) were detected by gel shift mobility assays performed with baby hamster kidney (BHK) cytoplasmic extracts and the WNV 3′(−) SL RNA probe (38). The same pattern of RPCs was observed when WNV-infected or uninfected BHK S100 cytoplasmic cell extracts were used, suggesting that the proteins in these complexes were cellular proteins. UV-induced cross-linking studies indicated that the molecular masses of the RNA binding proteins in these complexes were 42, 50, 60, and 108 kDa. The specificities of these RNA-protein interactions were demonstrated by competition gel mobility shift and competition UV-induced cross-linking assays (38).
The identification of one of the WNV 3′(−) SL RNA binding proteins, p42, as TIAR is reported here. The closely related protein TIA-1 was also shown to bind specifically to the WNV 3′(−) SL RNA. Results from WNV growth studies in TIAR knockout and TIA-1 knockout cells suggest a functional role for these cell proteins in flavivirus replication.
MATERIALS AND METHODS
Cells.
BHK-21/WI2 cells (43) (referred to hereafter as BHK cells) were used to prepare S100 cytoplasmic extracts or ribosomal salt wash cell extracts. BHK, CV-1, and Vero cells were maintained at 37°C in a CO2 incubator in minimal essential medium supplemented with 10 μg of gentamicin/ml and 5 or 10% fetal calf serum.
TIAR knockout C57BL/6 mice and TIA-1 knockout BALB/C mice were prepared as described previously (2, 34). Embryo fibroblast cell lines were established from wild-type (W4 and TIA+/+43), TIAR knockout (NaR4 and TIAR−/−43), and TIA-1 knockout (a−/−43 and TIA−/−44) mouse embryos using the standard NIH 3T3 protocol.
To prepare control-reconstituted (Cont-REC) and TIAR-reconstituted (TIAR-REC) cell lines, TIAR knockout (TIAR−/−43) cells were transfected with a pSR-α-hygromycin vector containing full-length human TIAR cDNA (a gift from M. Streuli, Dana Farber Cancer Institute, Boston, Mass.) by the calcium phosphate method (36). Stable cell lines were established from clones that grew in the presence of hygromycin. Reconstituted cells were reselected by growth in hygromycin for 1 week prior to use in experiments. These cell lines were maintained in minimal essential medium supplemented with 10% fetal calf serum, 10 mM HEPES, and 10 μg of gentamicin/ml in a CO2 atmosphere at 37°C. The average total number of cells in a confluent monolayer in a T25 flask for control (W4) cells was 2.3 × 106, that for TIAR knockout (NaR4) cells was 2.2 × 106, that for TIA-1 knockout (a−/−43) cells was 3.1 × 106, that for Cont-REC cells was 2.5 × 106, and that for TIAR-REC cells was 2.45 × 106.
Viruses.
Stocks of WNV strain EG101 (titer = 2 × 108 PFU/ml) and Sindbis virus strain SAAR 339 (titer = 7 × 109 PFU/ml) were prepared as 10% (wt/vol) newborn mouse brain homogenates. A stock of vaccinia virus strain Wyeth was prepared as a CV-1 cell lysate (titer = 1.2 × 108 PFU/ml). A stock of herpes simplex virus type 1 [HSV-1; strain H129 (H1)] was prepared as a medium pool in Vero cells (titer = 1.6 × 108 PFU/ml), and vesicular stomatitis virus (VSV) strain Indiana was prepared as a medium pool in L cells (titer = 4.8 × 107 PFU/ml).
For virus growth experiments, confluent monolayers of wild-type or knockout cells in T25 flasks were infected with WNV at a multiplicity of infection (MOI) of 1, and culture fluid samples (0.5 ml) were harvested at different times postinfection (p.i.). An MOI of 1 was chosen so that no virus interference effects would be observed. At each time point, 0.5 ml of fresh medium was added to maintain a constant volume in the flask. WNV samples were titrated in duplicate on BHK cells by plaque assay, and the values were averaged. In rare cases, the titers from duplicate wells were not similar and the sample was retitrated. Monolayers of cells in T25 flasks were also infected at an MOI of 1 with Sindbis virus, vaccinia virus, HSV-1, or VSV. Virus yields at different times p.i. were determined by plaque assay. Sindbis virus was plaqued on BHK cells, vaccinia virus was plaqued on CV-1 cells, HSV-1 was plaqued on Vero cells, and VSV was plaqued on BHK cells.
In vitro transcription of 32P-labeled RNA probes and unlabeled RNA transcripts.
Plasmid p75nt(−)3′ was previously constructed by P.-Y. Shi et al. (38). A PCR product, PCRT73′(−)SL, which consisted of the 75 3′-terminal nucleotides of the WNV minus-strand RNA with 3 extra nucleotides at the 5′ end copied from the T7 promoter, was amplified from plasmid p75nt(−)3′ DNA using an M13 reverse primer (5′-CAGGAAACAGCTATGACCATG-3′) and a forward primer (5′-AGTAGTTCGCCTGTGTGAGC-3′). The 3′(−) SL RNA was transcribed from the amplified PCR DNA. The T7 polymerase used for in vitro RNA transcription was expressed from BL21 cells containing pAR1219 (kindly provided by F. W. Studier, Brookhaven National Laboratory) and purified as described by Davanloo et al. (11).
The methods used for in vitro transcription and gel purification of the 32P-labeled RNA probes and unlabeled competitor RNAs were described previously (38). Large-scale batches of unlabeled RNAs, needed for the RNA affinity columns, were prepared by scaling up the in vitro transcription reaction to 1 ml and extending the reaction time to 4 h.
RNA affinity column.
In vitro-transcribed WNV 3′(−) SL RNA was oxidized with periodate in the presence of NaOAc (pH 5) and then attached to an agarose adipic acid matrix as described by Blyn et al. (4, 5). The RNA matrix (1 ml) was poured into a 10-ml column and then equilibrated with column binding buffer (5 mM HEPES [pH 7.5], 25 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, and 2 mM dithiothreitol).
BHK S100 cell extracts prepared as described previously (38) were subjected to ammonium sulfate precipitation prior to passage through an RNA affinity column. Ammonium sulfate was first added to a final concentration of 16%. Ammonium sulfate was then added to the supernatant obtained from the first precipitation to a final concentration of 45%, and the resulting pellet was resuspended in storage buffer. The pellet fraction was preincubated with the nonspecific competitors poly(I · C) (1 mg/ml) and heparin (500 μg/ml) at 4°C for 10 min and then passed over the RNA affinity column three to five times. The column was then washed several times with column binding buffer and once with the same buffer containing 0.2 M NaCl. The bound proteins were eluted with column binding buffer containing 1 or 2 M NaCl. The eluted fractions were subjected to buffer exchange in a Centricon-30 concentration cell (Amicon). Aliquots of each fraction were analyzed for RNA binding activity by gel mobility shift assay. Proteins were initially detected by GoldBlot staining (Integrated Separation Systems). The proteins in the eluted fractions were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie blue staining. Protein bands were excised and peptides were generated by trypsin digestion. The peptides were separated by high-performance liquid chromatography, and the sequences of selected peptides were determined by automated liquid chromatography-tandem mass spectrometry (MS) using a Finnigan MAT LCQ ion trap mass spectrometer as described previously (12, 13, 39). The OWL nonredundant composite protein sequence database version 26.0 was used with the automated SEQUEST database-searching program (Finnigan MAT) to interpret the MS/MS spectra. This database contains all protein sequences from all public databases and is kept up to date on a continuous basis. The automated search algorithum generates and scores peptide matches with the MS/MS spectra. For those proteins with the highest matching scores, the spectra were manually evaluated to confirm the match and to identify the protein from which the peptides were generated (16).
RNA-protein interaction assays.
Gel mobility shift and UV-induced cross-linking assays were performed as described previously (38). Prior to use in these assays, the RNA probes were denatured by incubation at 90°C for 10 min followed by renaturation by slow cooling to 60°C and incubation at 60°C for ∼2 min. The probes were then kept on ice until they were used.
Immunoprecipitation of UV-cross-linked proteins.
Proteins in S100 cytoplasmic extracts were first cross-linked to 32P-labeled WNV 3′(−) SL RNA by exposure to UV light, and then unbound RNA was digested with RNase (38). The cross-linked proteins were then incubated for 2 h at 4°C with 1 μg of anti-TIAR antibody (6E3; murine monoclonal antibody immunoglobulin G2a [IgG2a] [1]) or anti-TIA-1 antibody (ML29; murine monoclonal antibody IgG1 [23, 40])/ml that had been preincubated with Sepharose A CL-4B beads (Pharmacia). The precipitated complexes were pelleted by centrifugation at 300 × g, washed twice with dilution buffer (0.1% Triton X-100 and 0.5% nonfat dry milk in TSA buffer [0.01 M Tris-HCl, pH 8.0, 0.14 M NaCl, 0.025% NaN3]), once with TSA buffer, and once with 0.05 M Tris-HCl (pH 6.8). The immunoprecipitated complexes were then separated by SDS-10% PAGE and visualized by autoradiography. The control antibodies used for immunoprecipitation of cross-linked proteins were a goat anti-EF-1α antibody kindly provided by W. W. Merrick, Case Western Reserve University, Cleveland, Ohio, and mouse anti-La antibody kindly provided by J. D. Keene, Duke University Medical Center, Durham, N.C.
Immunoblotting.
BHK cells from a confluent monolayer in a T75 flask were trypsinized, pelleted by centrifugation for 3 min at 150 × g, and washed three times with 1× phosphate-buffered saline (PBS). The cell pellet was resuspended in ice-cold lysis buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing freshly added protease inhibitors (1× Complete; Roche) and passed through a 21-gauge needle four times. Nuclei were removed from the cytoplasmic extracts by centrifugation at 10,000 × g for 10 min at 4°C. The total protein concentration in the extracts was determined using a Dc protein assay kit (Bio-Rad). The proteins in 20 μg of extract were separated by SDS-10% PAGE and then electrophoretically transferred to a nitrocellulose membrane (0.45-μm pore size; Bio-Rad). The membrane was blocked with BLOTTO A (10 mM Tris-HCl, pH 8.0, 150 mM NaCl [TBS]; 5% nonfat dry milk; 0.05% Tween 20) for 1 h at room temperature or overnight at 4°C and probed first with an anti-protein primary antibody and then with a horseradish peroxidase-conjugated secondary antibody diluted in BLOTTO A. The membrane was washed three times with 1× TBS containing 0.05% Tween 20 and then once with 1× TBS prior to incubation with chemiluminescence reagent (Santa Cruz Biotechnology) and detection of the proteins by autoradiography.
Mouse anti-TIAR monoclonal antibody 6E3 was used at 0.8 μg/ml, and goat anti-TIA-1 polyclonal antibody (Santa Cruz Biotechnology) was used at 0.5 μg/ml. Horseradish peroxidase-conjugated goat anti-mouse IgG and donkey anti-goat IgG (Santa Cruz Biotechnology) were used at 0.2 μg/ml.
Indirect immunofluorescence.
Cells were grown to about 50% confluence in the wells of a two-chamber Lab-Tek II slide (Nalge Nunc International) and infected with WNV at an MOI of 5. At various times after infection, the cells were fixed with 2% paraformaldehyde for 10 min at room temperature, permeabilized with ice-cold methanol for 10 min, stained with a 1:100 dilution of a hyperimmune mouse anti-WNV antibody (Walter Reed Army Institute of Research) for 1 h, and then washed three times with PBS. The cell nuclei were then stained with Hoechst dye (33258) and fluorescein isothiocyanate-goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) for 1 h and washed three times with PBS. The coverslips were mounted in vinol mounting medium (18) and viewed with a Nikon Eclipse 800 microscope equipped with epifluorescence optics and appropriate filters for detection of fluorescein isothiocyanate, Texas Red, or Hoechst dye.
Preparation of figures.
Autoradiographs of gels and membranes were scanned with an Arcus II Agfa scanner. The digitized images were adjusted using Adobe Photoshop (version 5.5) software on a Power PC Macintosh G3.
RESULTS
Purification of the WNV 3′(−) SL RNA binding proteins.
It was previously reported that three RPCs, RPC1, -2, and -3, were detected in gel mobility shift assays done with a 32P-labeled WNV 3′(−) SL RNA probe and BHK S100 extracts (38). UV-induced cross-linking assays indicated that these complexes contained four cell proteins (p42, p50, p60, and p108) that bind specifically to the WNV 3′(−) SL RNA. An RNA affinity column was used to purify these viral RNA binding proteins.
In a preliminary experiment, a BHK S100 cytoplasmic extract that was prepared from 10 T150 flasks of cells was subjected to precipitation with different concentrations of ammonium sulfate. The supernatant and pellet fractions were analyzed for viral RNA binding activity by gel mobility shift and UV-induced cross-linking assays. Although a pellet was obtained after precipitation with 16% ammonium sulfate, none of the four cell proteins that bound to the viral 3′ RNA were present in this pellet in detectable amounts (data not shown). After precipitation with 45% ammonium sulfate, there was good recovery of RPC2 and RPC3, but only a small amount of RPC1 was detected in the pellet fraction by gel mobility shift assay (data not shown). p60, p50, and p42, but only a small amount of p108, were detected in the pellet fraction with a UV-induced cross-linking assay (data not shown).
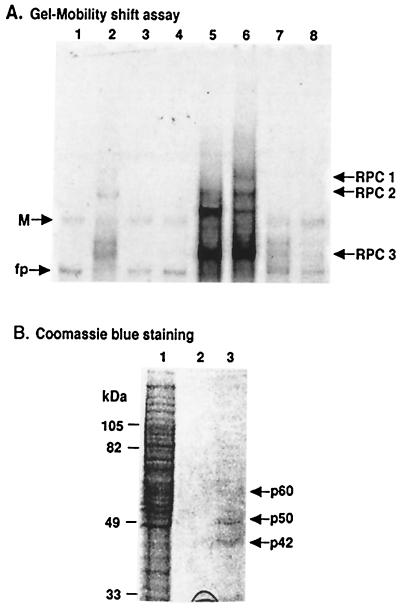
The proteins in the 45% ammonium sulfate pellet were resuspended in column binding buffer, incubated with nonspecific RNA competitors, and then passed through a WNV 3′(−) SL RNA affinity column several times (4, 5). The column was washed, and the bound proteins were eluted. Each eluted fraction was concentrated with a Centricon-30 and assayed for viral RNA binding activity using gel mobility shift and UV-induced cross-linking assays. Little or no specific binding activity was detected in the flowthrough fraction (Fig. 1A, lane 2) or the wash fractions (Fig. 1A, lanes 3 and 4) by gel mobility shift assay. The majority of the RNA binding activity was eluted with 2 M NaCl (Fig. 1A, lane 6). An aliquot of the eluted protein sample was separated by SDS-10% PAGE, electrophoretically transferred to a nitrocellulose membrane, and stained with GoldBlot (data not shown). Individual bands with molecular masses similar to those of three of the expected proteins (p60, p50, and p42), as well as some additional background bands, were observed. The p42 band was clearly the strongest of these. The remainder of the eluted protein sample was then electrophoresed on one lane of an SDS-10% PAGE gel and stained with Coomassie blue. p50 and p42 bands were clearly visible (Fig. 1B, lane 3). A very faint p60 band was also observed. The p42 and p50 bands were excised from the gel, and peptides were generated by trypsin digestion. The peptides were separated by high-performance liquid chromatography, and the sequences of selected peptides were determined by automated liquid chromatography-tandem mass spectrometry (Beckman Research Institute of the City of Hope). Insufficient unique sequence for p50 was obtained to allow the identification of this protein. The sequences of four peptides obtained from p42 were identical to sequences found in both TIA-1 and TIAR, while the sequences of two additional p42 peptides were unique to TIAR (Fig. 2). TIAR and TIA-1 are closely related RNA binding proteins that bind U-rich sequences interspersed with A's (15). Both proteins contain three N-terminal RNA recognition motif (RRM) domains, each approximately 100 amino acids in length, and a C-terminal auxiliary domain of approximately 90 amino acids (Fig. 2; (1). TIAR and TIA-1 show 80% overall amino acid identity, with the highest degree of similarity in RRM domain 3 (91% identity) and the lowest degree of similarity (about 50% identity) in the C-terminal auxiliary domain. The data suggest that p42 is TIAR. However, because of the high degree of sequence homology between TIAR and TIA-1 (Fig. 2), the possibility that TIA-1 also binds specifically to the WNV 3′(−) SL RNA could not be ruled out.
FIG. 1.
Analysis of WNV 3′(−) SL RNA binding proteins in fractions eluted from an agarose-adipic acid hydrazide RNA affinity column. (A) Gel shift assays. Lane 1, free probe; lane 2, final flowthrough fraction from the RNA affinity column; lane 3, first binding buffer wash fraction; lane 4, 0.2 M NaCl wash fraction; lanes 5 and 6, fractions eluted with 1 or 2 M NaCl, respectively; lanes 7 and 8, fractions eluted from a “beads-only” control column with 1 or 2 M NaCl, respectively. For each of the fractions, 1 μl of a total of 100 μl was analyzed on the gel. The positions of the three RPCs are indicated by arrows. M, multimer of the probe; fp, free probe. (B) Coomassie blue staining of the eluted fractions from an agarose-adipic acid hydrazide RNA affinity column. Lane 1, aliquot of sample loaded on the affinity column (10 μl out of 3 ml); lane 2, fraction eluted with 2 M NaCl from a beads-only control column (30 out of 100 μl); lane 3, fraction eluted with 2 M NaCl from the RNA affinity column (30 out of 100 μl). The positions of the eluted proteins are indicated by arrows. The protein markers are shown on the left of the gel.
FIG. 2.
Alignment of regions of the TIAR and TIA-1 amino acid sequences with peptides obtained from p42. The tall boxes enclose TIA-1 and TIAR sequences that are identical to peptide sequences obtained from RNA affinity column-purified p42. The short boxes enclose sequences found only in TIAR that are identical to p42 peptides. Deletions are indicated by dashes.
Studies to identify the other three cell proteins that bind to the WNV 3′ (−) RNA are in progress. Previous preliminary studies showed that neither anti-EF-1α nor anti-La antibody produced a supershift when added to S100 cytoplasmic extracts incubated with the WNV 3′(−) SL RNA probe (W. Li and M. A. Brinton, unpublished data).
Confirmation that TIAR and TIA-1 bind to the WNV 3′(−) RNA.
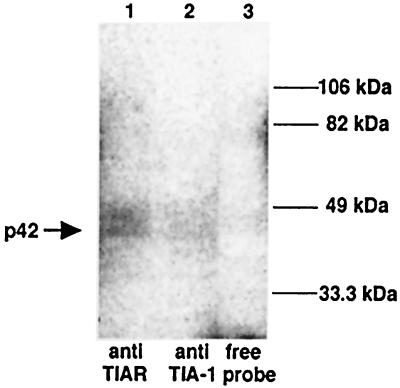
Anti-TIAR (6E3) and anti-TIA-1 (ML29) antibodies were used to immunoprecipitate UV-induced cross-linked WNV 3′(−) SL RPCs from BHK S100 cytoplasmic extracts after treatment with RNase. Even though four cell proteins (p108, p60, p50, and p42) that are able to cross-link to this viral RNA probe were present in these extracts, the anti-TIAR antibody precipitated only the p42-WNV 3′(−) SL RNA complex (Fig. 3, lane 1). The anti-TIA-1 antibody also precipitated the p42-WNV 3′(−) SL RNA complex (Fig. 3, lane 2). However, the band obtained after immunoprecipitation with anti-TIA-1 antibody was much weaker than that seen after immunoprecipitation with the anti-TIAR antibody. No immunoprecipitation bands with either of these antibodies were detected if the UV-induced cross-linking step was omitted (data not shown). Likewise, no immunoprecipitated bands were detected when the cross-linked complexes were mixed with antibodies to other cell proteins, such as anti-La and anti-EF-1α (data not shown).
FIG. 3.
Immunoprecipitation of UV-induced cross-linking RPCs. BHK S100 cytoplasmic extracts from BHK cells were incubated with a 32P-labeled WNV 3′(−) SL RNA probe. The complexes were cross-linked by exposure to UV light and were then precipitated with anti-TIAR or anti-TIA-1 antibody. The precipitates were analyzed by SDS-10% PAGE. Lane 1, anti-TIAR antibody added; lane 2, anti-TIA-1 antibody added; lane 3, free probe. The expected UV-induced cross-linked p42 product is indicated by an arrow. The protein markers are shown on the right of the gel.
In subsequent high-sensitivity Western blots, the ML29 anti-TIA antibody showed a small amount of cross-reactivity with TIAR. However, the 6E3 anti-TIAR antibody showed no cross-reactivity with TIA-1 (see Fig. 7). The weak RPC band immunoprecipitated by the ML29 antibody could therefore also have contained TIAR. However, these data did not rule out the possibility that both TIAR and TIA-1 can bind to the WNV 3′(−) SL RNA. Recombinant proteins were obtained to directly assess the ability of each of these proteins to bind to the WNV 3′(−) SL RNA.
FIG. 7.
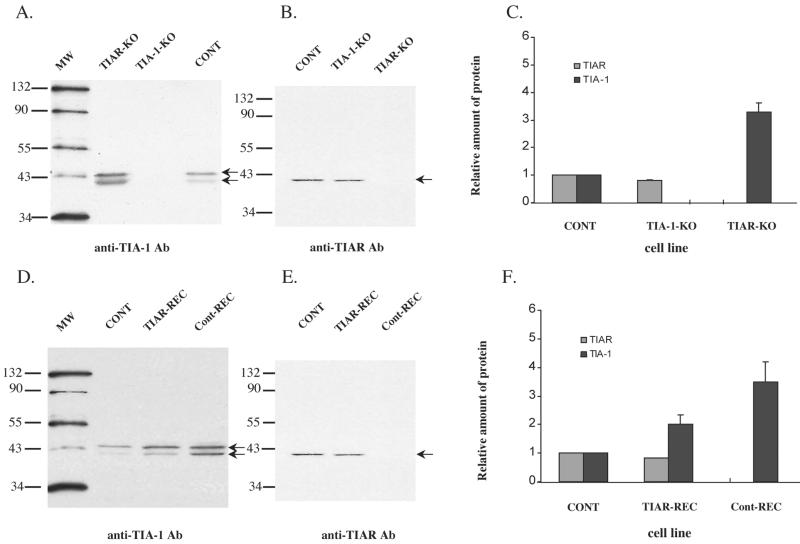
Western blot analyses of the amounts of TIAR and TIA-1 proteins in various cell lines. Twenty micrograms of total cell protein was run on each lane. (A, B, D, and E) Representative Western blots. The positions of the TIAR protein and two isoforms of the TIA-1 protein are indicated by arrows. Ab, antibody; KO, knockout. (C and F) Quantification of the relative amounts of protein in the various types of cells. The relative amount of each protein in the wild-type cells (CONT) was defined as 1. The relative amounts of the proteins in other cell lines were expressed as the ratio of the protein band intensity divided by the density of the band in wild-type cells. The values shown are means of the values obtained from three to five separate experiments. The error bars indicate the standard errors of the mean.
Analysis of the specificities of the viral RNA-cell protein interactions.
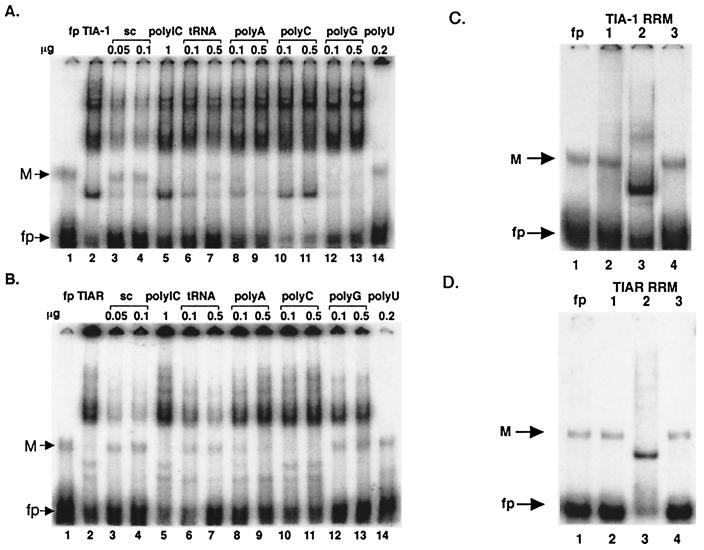
Purified glutathione S-transferase (GST)-TIAR and GST-TIA-1 fusion proteins (42) were tested for the ability to bind to the WNV 3′(−) SL RNA in a gel mobility shift assay. The results of preliminary experiments showed that at least a four-times-higher concentration of GST-TIA-1 than GST-TIAR was required to detect binding in gel mobility shift assays (data not shown). Therefore, different concentrations of GST-TIA-1 (200 nM) and GST-TIAR (50 nM) were used for the representative competition gel shift assays shown in Fig. 4A and B. Although the predominant gel shift band observed with both of the fusion proteins migrated to the middle of the gel, additional slower- and faster-migrating complexes were also observed. The slower-migrating bands most likely contained aggregated complexes, since the densities of these bands increased with increasing protein concentrations (data not shown). The faster-migrating bands most likely contained breakdown fragments that retained the RRM2 region containing the viral RNA binding site (Fig. 4C and D).
FIG. 4.
Analysis of the specificities of the interactions between the WNV 3′(−) SL RNA and recombinant TIAR or TIA-1 proteins. (A) Competition gel shift assays with a purified GST-TIA-1 fusion protein. Lane 1, free probe; lane 2, probe plus 200 nM purified GST-TIA-1 fusion protein; lanes 3 to 14, probe plus 200 nM purified GST-TIA-1 fusion protein and the indicated competitor RNA. sc, specific-competitor-unlabeled 75-nucleotide WNV 3′(−) SL RNA; M, multimer of the probe; fp, free probe. (B) Competition gel shift assays with a purified GST-TIAR fusion protein. Lane 1, free probe; lane 2, probe plus 50 nM purified GST-TIAR fusion protein; lanes 3 to 14, probe plus 50 nM purified GST-TIAR fusion protein and the indicated competitor RNA. (C) Purified GST fusion proteins (500 nM), each containing a single RRM domain of TIA-1, were analyzed by gel mobility shift assay. Lane 1, free probe; lane 2, probe plus GST-TIA-1 RRM1; lane 3, probe plus GST-TIA-1 RRM2; lane 4, probe plus GST-TIA-1 RRM3. M, multimer of the probe. (D) Purified GST fusion proteins (200 nM), each containing a single RRM domain of TIAR, were analyzed by gel mobility shift assay. Lane 1, free probe; lane 2, probe plus GST-TIAR RRM1; lane 3, probe plus GST-TIAR RRM2; lane 4, probe plus GST-TIAR RRM3.
Unlabeled WNV 3′(−) SL RNA (50 or 100 ng) was used as the specific competitor and competed efficiently with the labeled probe (Fig. 4A, lanes 3 and 4, and B, lanes 3 and 4). The nonspecific competitors, poly(I · C), poly(A), poly(G), and poly(C), showed little or no competition even at concentrations of 500 ng or 1 μg (Fig. 4A and B). tRNA (100 or 500 ng) partially competed, but with a lower efficiency than the specific competitor (Fig. 4A, lanes 6 and 7, and B, lanes 6 and 7). As expected from previous studies showing that TIAR and TIA-1 bound to U-rich sequences (15), poly(U) competed efficiently (Fig. 4A, lane 14, and B, lane 14). These data indicate that both the GST-TIAR and GST-TIA-1 proteins bind specifically to the WNV 3′(−) SL RNA.
Mapping the WNV 3′(−) SL RNA binding domain in the TIAR and TIA-1 proteins.
TIA-1 and TIAR each contain three RRM domains, each of about 100 amino acids (Fig. 2). To determine whether one of these RRM domains contains the major binding site for the WNV 3′(−) SL RNA, purified truncated GST fusion proteins, GST-TIA-1 RRM1, GST-TIA-1 RRM2, and GST-TIA-1 RRM3 (Fig. 4C, lanes 2 to 4) and GST-TIAR RRM1, GST-TIAR RRM2, and GST-TIAR RRM3 (Fig. 4D, lanes 2 to 4), were tested for the ability to bind to the WNV 3′(−) SL RNA in gel mobility shift assays. Only GST-TIA-1 RRM2 (Fig. 4C, lane 3) and GST-TIAR RRM2 (Fig. 4D, lane 3) were able to bind the WNV 3′(−) SL RNA. These data suggest that the major WNV 3′(−) SL RNA binding site in both TIAR and TIA-1 is RRM 2.
Determination of the relative dissociation constants (Kds) for the viral RNA-cell protein interactions.
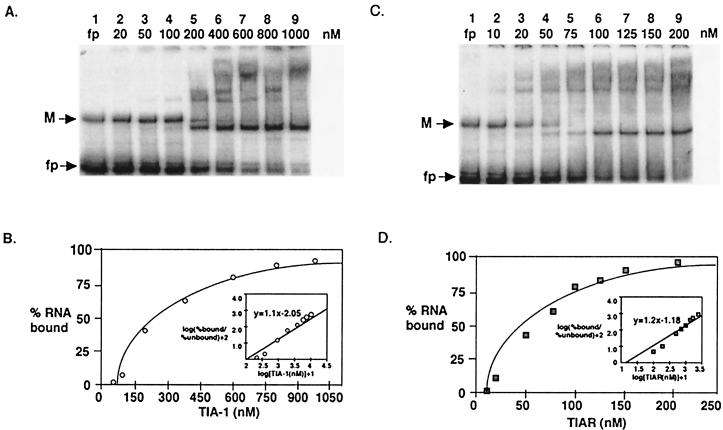
Gel mobility shift assays were performed using different amounts of GST-TIA-1 RRM2 or GST-TIAR RRM2 protein and a constant amount of the 32P-WNV 3′(−) SL RNA. Although gel shift bands were observed with 10 nM GST-TIAR RRM2, bands for GST-TIA-1 RRM2 were first observed when 50 nM protein was used (Fig. 5A and C). A theoretical saturation binding curve was generated by plotting the percentage of bound WNV 3′(−) SL RNA versus the concentration of either GST-TIA-1 RRM2 or GST-TIAR RRM2. The data from the saturation binding curve were transformed as described previously (29, 44). The relative Kd for the interaction between GST-TIA-1 RRM2 and the WNV 3′(−) SL RNA was estimated to be about 1.12 × 10−7 M (Fig. 5A), while the relative Kd for the interaction between GST-TIAR RRM2 and the WNV 3′(−) SL RNA was estimated to be about 1.5 × 10−8 M (Fig. 5C). The slope (n) of the line represents the ratio of GST-TIA-1 RRM2 or GST-TIAR RRM2 molecules to WNV 3′(−) SL RNA molecules in each RPC (Fig. 5B and D, insets). For both proteins, the slope was calculated to be about 1 (1.1 for TIA-1 and 1.2 for TIAR), suggesting that approximately one TIAR or TIA-1 molecule binds to each WNV 3′(−) SL RNA molecule. Similar Kd and n values were obtained from four independent experiments with standard deviations of ±15 nM and ±0.15 for GST-TIA-1 RRM2 and ±5 nM and ±0.1 for GST-TIAR RRM2, respectively. These data indicate that the relative binding activity of TIAR RRM2 for the WNV 3′(−) SL RNA is >10 times higher than that of the TIA-1 RRM2 for the same RNA. Although the RRM2 domain was shown to contain the main binding site for the viral RNA (Fig. 4), both proteins also contain two additional RRM domains that are likely to participate in stabilizing the RNA-protein interaction. The relative binding activities of the complete proteins for the viral 3′ RNA, therefore, would be expected to be somewhat higher. In previous filter binding studies with a selected synthetic U-rich RNA, the Kd of the TIAR RRM2 fragment was calculated to be 5 × 10−8 M while the Kd of full-length TIAR was calculated to be 8 × 10−9 M (15). Kds for the interaction between TIA-1 or TIA-1 RRM2 and U-rich RNAs were not reported previously.
FIG. 5.
Quantification of RNA-protein interactions. (A) Representative gel mobility shift assay done with increasing amounts of the GST-TIA-1 RRM2 protein and a constant amount of WNV 3′(−) SL RNA. Lane 1, free probe; lanes 2 to 9, probe plus GST-TIA-1 RRM2 in the amounts indicated. M, multimer of the probe; fp, free probe. (B) The percent 32P-WNV 3′(−) SL RNA bound was plotted against the concentration of TIA-1 to generate a theoretical saturation binding curve. (Inset) Data from the saturation binding curve were transformed as described previously (29, 44). The stoichiometry of the interaction of TIA-1 with the WNV 3′(−) SL RNA, as determined by the slope of the line in the inset graph, was about 1:1. The dissociation constant was calculated using the equation log (percent bound/percent unbound) + 2 = n{log[TIA-1 (nM)] + 1} − log Kd. The Kd was estimated to be 112 nM for TIA-1. (C) Representative gel mobility shift assay done with increasing amounts of GST-TIAR RRM2 protein and a constant amount of WNV 3′(−) SL RNA. Lane 1, free probe; lanes 2 to 9, probe plus GST-TIAR RRM2 in the amounts indicated. (D) The percent 32P-WNV 3′(−) SL RNA bound was plotted against the concentration of TIAR to generate a theoretical saturation binding curve. (Inset) The data from the saturation binding curve were transformed. The Kd was estimated to be 15 nM for TIAR. The stoichiometry of the interaction of TIAR with the WNV 3′(−) SL RNA, as determined by the slope of the lines of the inset graph, was about 1:1.
Effect of TIAR and TIA-1 on the replication of WNV.
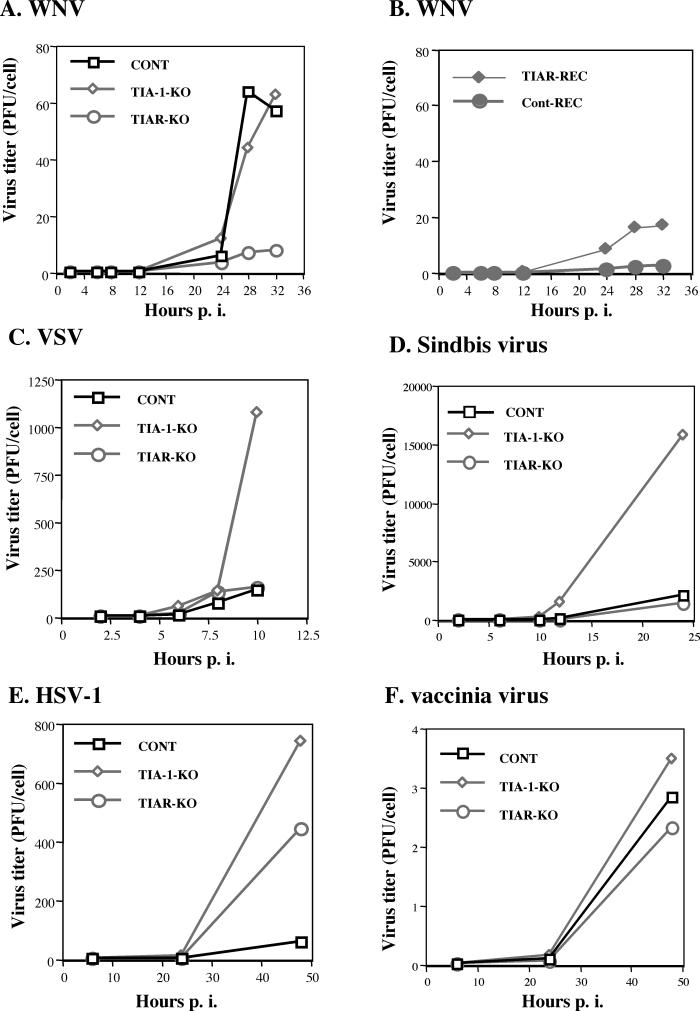
As one means of assessing the effect of the TIAR and TIA-1 proteins on WNV replication, virus growth was compared in TIAR knockout, TIAR-REC, TIA-1 knockout, and control murine embryo fibroblast cell lines. Confluent cell monolayers were infected with WNV at an MOI of 1. Culture fluid samples were taken at 2, 8, 12, 24, 28, and 32 h p.i. Representative growth curves of WNV in control (W4; TIAR+/+ TIA-1+/+), TIAR knockout (NaR4; TIAR−/− TIA-1+/+), and TIA-1 knockout (a−/−43; TIAR+/+ TIA-1−/−) cells are shown in Fig. 6A. The virus titers were expressed as PFU per cell because the various cell lines grew to different but characteristic densities when confluent. The actual virus titers in PFU per milliliter were >10,000 times higher than the PFU-per-cell titers.
FIG. 6.
Growth of virus in TIAR and TIA-1 knockout cell lines. Confluent monolayers of wild-type (CONT) (W4; TIAR+/+ TIA-1+/+), TIA-1 knockout (TIA-1-KO) (a−/−43; TIAR+/+ TIA-1−/−), and TIAR knockout (TIAR-KO) (NaR4; TIAR−/− TIA-1+/+) cells were infected with WNV (A), VSV (C), Sindbis virus (D), HSV-1 (E), or vaccinia virus (F) at an MOI of 1, and confluent monolayers of TIAR-REC or Cont-REC cells were infected with WNV (B) at an MOI of 1. Culture fluid samples were taken at the indicated hours p.i. and titered in duplicate by plaque assay in the cell lines indicated in Materials and Methods. Each growth curve was repeated two or three times, and representative growth curves are shown. All virus titers were expressed as PFU per cell because the various cell lines grew to different but characteristic densities when confluent. All peak virus titers were in the range of 106 to 107 PFU/ml.
The peak titer of WNV produced by TIAR knockout cells was significantly (six- to eightfold) lower than that produced by control cells (Fig. 6A). WNV grew to comparable peak titers in TIA-1 knockout cells and control cells, but the peak virus level was not attained until 6 h later in TIA-1 knockout cells. Each growth curve was repeated two or three times, and although the absolute values of the titers obtained varied somewhat from experiment to experiment, the relative differences between the virus yields produced by the different cell types were consistent from experiment to experiment. Similar results were also obtained with an additional set of separately derived control and knockout cell lines (data not shown), suggesting that the decrease in WNV replication observed in the knockout cells was not due to a peculiarity of a single knockout cell line. The drop in the WNV titer in the control cells at 28 h was consistently observed and likely represented a period during which the production of virus from the cells infected by the inoculum was waning and virus production from secondarily infected cells was not yet at the peak level.
The efficiency of infection of these cells with WNV was investigated by indirect fluorescence. Control, TIAR knockout, and TIA-1 knockout cells were infected with WNV for 24, 28, or 32 h, fixed, and then stained with Hoechst dye and anti-WNV antibody. At 24 h p.i., bright virus-specific perinuclear staining was observed in about 40% of the control and TIA-1 knockout cells. However, the stained perinuclear rings and areas in the control cells were generally wider than those in the TIA-1 knockout cells. Although a similar percentage of TIAR knockout cells showed virus-specific perinuclear staining at 24 h, the fluorescence in these cells was faint and the areas of staining were focal (data not shown). The intensity of the perinuclear staining in the infected TIAR knockout cells increased somewhat by 28 h p.i., and thin perinuclear rings were observed in some cells. At 32 h, although the intensity and distribution of the fluorescence had increased in the WNV-infected TIAR knockout cells, only about 10 to 20% of the cells contained broad, brightly stained perinuclear rings (data not shown). These results suggest that WNV infects similar numbers of cells in the three types of cultures but that virus replication is most efficient in the control cells, slightly less efficient in the TIA-1 knockout cells, and least efficient in the TIAR knockout cells.
Growth of other types of viruses in TIAR knockout and TIA-1 knockout cells.
To determine whether other types of viruses also showed reduced growth in TIAR knockout cells, control, TIAR knockout, and TIA-1 knockout cells were infected with Sindbis virus, vaccinia virus, VSV, or HSV-1 at an MOI of 1. Sindbis virus is another plus-strand RNA virus but from the alpha togavirus family. VSV, a rhabdovirus, is a minus-strand RNA virus, while vaccinia virus, a poxvirus, is a DNA virus. Similar to WNV, these three viruses replicate in the cytoplasm of infected cells. HSV-1, a herpesvirus, is a DNA virus that replicates in the nucleus. Culture fluid samples were harvested at various times after infection and titered by plaque assay. Representative growth curves obtained for each of the viruses are shown in Fig. 6C through F. All virus titers were expressed as PFU per cell because the various cell lines grew to different but characteristic densities when confluent. VSV (Fig. 6C) and Sindbis virus (Fig. 6D) grew to similar titers in TIAR knockout and control cells, whereas in TIA-1 knockout cells both of these viruses grew to significantly higher titers, suggesting that the presence of TIA-1 had a negative effect on the growth of these viruses. HSV-1 also grew to significantly higher levels in TIA-1 knockout cells than in control cells (Fig. 7E). However, the growth of HSV-1 in TIAR knockout cells was also more efficient than in control cells but not as efficient as in TIA-1 knockout cells. The efficiencies of growth of vaccinia virus (Fig. 6F) in all three types of cells were similar. Because the majority of the vaccinia progeny virus are cell associated, the extracellular virus titers detected were significantly lower than those for the other viruses. Each growth curve was repeated two or three times, and although the absolute values of the titers obtained varied somewhat from experiment to experiment, the relative differences between the virus yields produced in the different cell types by the different viruses were consistent. These results indicate that the growth of WNV, but not that of the other viruses tested, was less efficient in the TIAR knockout cells.
WNV growth in TIAR-REC cells.
To further investigate the effect of TIAR on viral growth, the growth of WNV in a TIAR-reconstituted stable cell line, TIAR-REC, was tested. Another stable cell line, Cont-REC, which had been transfected with the same vector but did not express TIAR at detectable levels (Fig. 7E), was used as a control for possible nonspecific effects of the vector. Figure 6B shows representative WNV growth curves obtained with TIAR-REC and Cont-REC cells. Although the peak titer of WNV produced by TIAR-REC cells was consistently higher than that produced by Cont-REC cells, it did not reach as high a level as that normally produced by control cells (Fig. 6A and B).
Comparison of relative amounts of TIAR and TIA-1 proteins in various cell lines.
The relative amounts of TIAR and TIA-1 in cytoplasmic extracts from each of the cell lines were estimated by immunoblotting using protein-specific antibody. Previous studies showed that two isoforms generated by alternative splicing exist for both TIA-1 and TIAR (1). The two TIA-1 isoforms, 42-kDa TIA-1a and 40-kDa TIA-1b, differ from each other by an 11-amino-acid deletion. These isoforms are usually found in cells in a 1:1 ratio. The two TIAR isoforms, 42-kDa TIARa and 40-kDa TIARb, differ from each other by a 17-amino-acid deletion. Because TIARb is six times more abundant in cells than TIARa, it is the only isoform that is detected by Western blotting.
As expected, no TIA-1 protein was detected in cytoplasmic extracts from TIA-1 knockout cells (Fig. 7A) and no TIAR protein was detected in cytoplasmic extracts from TIAR knockout cells (Fig. 7B). The level of the TIAR protein in cytoplasmic extracts from TIA-1 knockout cells (a−/−43; TIAR+/+ TIA-1−/−) was slightly decreased compared to the level of this protein in control cells (Fig. 7B and C). In contrast, the amount of TIA-1 protein in the cytoplasm of TIAR knockout cells (NaR4; TIAR−/− TIA-1+/+) was significantly increased (3.3-fold) compared to that present in the control (W4; TIAR+/+ TIA-1+/+) cells (Fig. 7A and C). These data indicate that the level of TIA-1 is down-regulated by TIAR. No significant differences in the cytoplasmic levels of either of these proteins were observed at either 5 or 8 h p.i. with WNV in the various cell lines tested (data not shown).
The amount of TIAR protein detected in the reconstituted TIAR-REC cells was about 80% of that detected in control (W4) cells (Fig. 7E and F), while no TIAR protein was detected in the nonexpressing Cont-REC cells (Fig. 7E). The amount of TIA-1 protein in the TIAR-REC cells was 2-fold higher, while the amount of TIA-1 protein in Cont-REC cells was 3.5-fold higher, than that in control (W4) cells (Fig. 7D and F). The Cont-REC cells were considered the best controls for the TIAR-REC cells, since both of these cell lines had been transfected with the expression vector and selected for drug resistance. Comparison of the protein levels showed that Cont-REC cells contained higher levels of TIA-1 than TIAR-REC cells. The extent of the increase in TIA-1 levels corresponded to the extent of the decrease in TIAR levels. These data further demonstrate the regulation of TIA-1 levels by TIAR.
Comparison of TIAR and TIA-1 cDNA sequences from cells obtained from flavivirus-resistant and -susceptible mice.
A single dominant gene that maps to chromosome 5 confers a flavivirus resistance phenotype in mice. Data from previous studies showed that resistant mice, as well as cells obtained from a number of different tissues in resistant mice, produced significantly lower titers of flaviviruses than did congenic susceptible mice or cells and that genomic RNA levels, but not minus-strand viral RNA levels, were lower in resistant cells (7; Y. Li and M. A. Brinton, unpublished). Since both TIAR and TIA-1 bind to the WNV 3′(−) SL RNA and this SL is located at the site of initiation of genomic RNA synthesis, it was of interest to determine whether the sequences of TIAR and TIA-1 cDNAs differed in cells from resistant C3H/He and congenic susceptible C3H.RV mice. Cell RNAs were extracted from resistant and susceptible embryo fibroblasts with TRIZOL-LS (Gibco-Life) according to the manufacturer's instructions. Using primers designed from mouse (strain 129 SVJ) TIA-1 and TIAR cDNA sequences previously reported by Beck et al. (1), cDNAs were amplified by reverse transcription-PCR from cell mRNA and TA cloned into pCR 2.1-TOPO (Invitrogen). At least three cDNA clones for each isoform were sequenced. The sequences obtained for the two TIAR isoform cDNAs and for the two TIA-1 isoform cDNAs from resistant C3H.RV cells were identical to those of the comparable isoforms obtained from susceptible C3H/He cells. These sequences were also identical to the sequences for these proteins from 129 SVJ mice (accession numbers U55861 and U55862) previously reported by Beck et al. (1). As assessed by Western blotting, the expression levels of the TIAR and TIA-1 proteins in resistant and susceptible cells were similar (data not shown). These data indicate that neither TIAR nor TIA-1 is the product of the Flv gene.
DISCUSSION
p42, one of the four cell proteins previously reported to bind specifically to the WNV 3′(−) SL RNA, has been identified as TIAR/TIA-1. This is the first report of the identification of a host protein that interacts specifically with the 3′ SL of a flavivirus minus-strand RNA, the site of initiation for nascent genome RNA synthesis. TIA-1 and TIAR are closely related multifunctional RNA binding proteins (21, 41) that have at least partial redundancy in their cellular functions (19, 34). The data obtained suggest that the binding of TIAR to the WNV 3′(−) SL RNA is functionally important for viral replication.
Evolutionary conservation of TIA-1/TIAR.
TIAR and TIA-1 are evolutionarily conserved proteins; homologs in different mammalian species show 96 (TIA-1) and 99% (TIAR) identity (1), while homologs in divergent species, such as Drosophila melanogaster (6, 24) and Caenorhabditis elegans (45), each show about 46% amino acid identity with human TIA-1 and TIAR. Because flaviviruses replicate efficiently in a large number of divergent host species and cycle between invertebrate and vertebrate hosts during their natural transmission cycles, it is expected that these viruses would need to interact with evolutionarily conserved cell proteins to replicate efficiently in different hosts. TIAR and TIA-1 proteins were initially discovered in T cells (hence their names) but have since been found to be expressed in good quantities in many tissues, including brain, spleen, and macrophages (1), which are sites of flavivirus replication in vivo.
Cellular localization of TIA-1/TIAR.
Both TIAR and TIA-1 shuttle between the nucleus and the cytoplasm in viable cells. Flaviviruses replicate in the cytoplasm. Interestingly, the level of TIAR in the cytoplasm of BHK cells was about 10 times higher than in several mouse embryo fibroblast cell lines (data not shown), and WNV grows to about 10-times-higher titers (peak titer, about 107.5 PFU/ml) in BHK cells than in the mouse cell lines (peak titer, about 106.5 PFU/ml).
RNA binding characteristics of TIA-1/TIAR.
In selection and amplification experiments with pools of randomized synthetic RNAs, both TIAR and TIA-1 bound with high affinity to RNAs that contained one or more short uridylate stretches (usually three or more) (15). Replacement of the U's in these stretches with C's eliminated the protein-RNA interaction (15). Although both proteins selected RNAs containing stretches of U's, the RNA sequences selected by TIA-1 were not identical to those selected by TIAR. Although the RRM2 domains in both proteins mediated specific binding to the uridylate-rich RNAs, the presence of the other two RRM domains increased the affinity of the interaction with the U-rich RNAs (15). In the 3′ NCR of tumor necrosis factor alpha (TNF-α) mRNA, a large fragment of AU-rich sequence containing clustered AUUUA pentamers was required for TIAR/TIA-1 binding, although the binding sites within this RNA were not finely mapped (19, 34).
The data presented here indicate that both TIAR and TIA-1 can bind specifically to the WNV 3′(−) SL RNA and that the RRM2 domain mediates this interaction. Since poly(U) competed efficiently with the WNV 3′(−) SL RNA in the competition gel mobility shift assays (Fig. 4), it is expected that the viral sequence(s) recognized by TIAR and TIA-1 contains U's. Although the WNV 3′(−) SL RNA is not AU rich, two of the single-stranded loops in this structure contain the sequences UAAU and UUAAU. These sequences are also conserved in the predicted single-stranded loops in the SLs of other mosquito-borne flaviviruses (data not shown). The observed Kd for the interaction between TIAR RRM2 and the WNV 3′(−) SL RNA was 1.5 × 10−8 M, which is similar to the Kd reported for the interaction between the TIAR RRM2 and U-rich synthetic RNA sequences and also to the Kds for other functionally relevant RNA-protein interactions, such as the interaction between the cellular U1A protein and the U1 RNA (20). The binding activity of the TIA-1 RRM2 (Kd, 10−7 M) for the WNV 3′(−) SL RNA was about 10 times lower than that of the TIAR RRM2. The putative viral binding sites do not contain stretches of at least three U's, as do the optimal selected synthetic sequences (15). Mapping studies are under way to identify the individual nucleotides in the WNV 3′(−) RNA required for binding by each of these proteins.
Comparison of the TIA-1 and TIAR RRM2 domain sequences (Fig. 2) indicated that they differ at 8 amino acid residues and that TIAR also contains an 11-amino-acid deletion at the beginning of RRM2 (Fig. 2) (1). The 10-fold-lower binding activity of TIA-1 for the WNV 3′(−) SL RNA would be expected to result in TIAR outcompeting TIA-1 for binding to the viral RNA affinity column and would significantly reduce the likelihood of detecting unique TIA-1 peptides in the protein eluted from the viral-RNA affinity column. However, the interaction of TIA-1 with the WNV 3′(−) SL RNA both in the presence and absence of TIAR may be functionally important for viral replication. Both TIAR and TIA-1 were found to be present in complex 1, which binds to the 3′ NCR of the TNF-α mRNA (19, 34).
Cellular functions of TIA-1/TIAR.
A number of cellular functions have been attributed to the RNA binding properties of TIA-1/TIAR. Both TIAR and TIA-1 regulate the generalized translational arrest that occurs following an environmental stress. Stress-induced phosphorylation of the translation initiation factor eIF-2α is followed by recruitment of poly(A)+ RNA into cytoplasmic stress granules by TIAR and TIA-1 (23). Stress granules and polysomes appear to be in equilibrium in cells (22). TIA-1/TIAR also function as specific translational silencers (19, 34). For example, TNF-α translation is blocked by the binding of TIAR and TIA-1 to specific U-rich sequences in the 3′ NCR of this mRNA. Upon stimulation with lipopolysaccharides, this translational repression is overcome by the binding of an additional protein, p55, to the 3′ NCR of the TNF-α mRNA (27). TIA-1 and TIAR have recently been shown to function as alternative splicing regulators by binding to specific U-rich intron (IAS1) sequences adjacent to cryptic 5′ splice sites and enhancing the use of these 5′ splice sites (14, 26). Such intron sequences exist in a subset of pre-mRNAs, including those of TIA-1 and TIAR, and it is thought that both proteins can regulate their own expression at the level of splicing, as well as the expression of some other proteins (17). The yeast protein Nam8p, a component of the U1 snRNP, is distantly related to TIA-1 and TIAR. It is interesting that even though the majority of the known cellular functions of TIAR and TIA-1 involve interactions with cellular mRNAs, it is the terminal region of the WNV minus strand, not the positive-strand genome, that interacts with these proteins.
Although both TIAR and TIA-1 were previously implicated as effectors of apoptotic cell death, the specific roles of these proteins in apoptosis have not as yet been delineated. Introduction of purified TIAR or TIA-1 into the cytoplasm of thymocytes permeabilized with digitonin resulted in fragmentation of genomic DNA into nucleosome-sized oligomers (21, 41). Increased amounts of TIAR were translocated from the nucleus to the cytoplasm in response to exogenous triggers of apoptosis (40). TIA-1 is phosphorylated by a serine/threonine kinase that is activated during Fas-mediated apoptosis (42). Although not rigorously tested, no evidence of apoptosis was observed when rodent cells infected with WNV were examined, at intervals up to 32 h after infection, after fixation and nuclear staining with Hoechst dye (data not shown). A previous study with WNV indicated that apoptosis occurred by 72 h in infected human mononuclear (K562) cells and mouse neuroblastoma (Neuro 2a) cells via the BAX pathway (33).
Both TIAR and TIA-1 appear to play important roles in embryo development (2). However, the specific functions of these proteins during development are not known. It was not possible to produce double-knockout mice because of lethality prior to embryonic day 8 (N. Kedersha and P. Anderson, unpublished data). Without a more complete understanding of the normal cell functions of TIAR/TIA-1, it is not possible to rule out the possibility that secondary effects of cell manipulation by gene knockout or cDNA reconstitution could be responsible for some of the observed effects on virus replication.
Effect of TIA-1/TIAR on virus replication.
Interestingly, of the five types of viruses tested in the TIAR and TIA-1 knockout cells, only the growth of the flavivirus, WNV, was decreased in cells lacking TIAR. In contrast, the growth of the four other types of viruses was more efficient in one or both types of knockout cells than in control cells. These data suggest that in the control cells, the presence of one or both of these proteins has a negative effect on the production of the viruses. However, the negative effect that the loss of TIAR and, to a lesser extent, TIA-1 have on WNV replication suggests that these proteins provide a necessary function for WNV during its replication cycle.
One possible reason why the growth of WNV was not reduced to a greater extent in TIAR knockout cells could be that the TIA-1 protein, which is present in increased amounts in the TIAR knockout cells (Fig. 7), can substitute for TIAR by providing the function needed by WNV. However, WNV replication in cells lacking TIAR was never as efficient as when TIAR was present. Also, although the efficiency of virus replication increased when TIAR knockout cells were reconstituted with vector-expressed TIAR (Fig. 6 and 7), neither the amount of TIAR nor the efficiency of WNV replication reached control cell levels in these reconstituted cells. WNV replication in a double-knockout cell line, if such a cell line were available, would be expected to be much less efficient than in the single-knockout cells.
The only known function of the flavivirus minus-strand RNA is as a template for the synthesis of nascent genomic RNA. Specific binding of TIA-1/TIAR to the 3′ terminus of the viral minus-strand RNA template appears to play a positive role in virus replication. Possible functions of this interaction include assisting in the formation or stabilization of the 3′-terminal (−) SL in replication complexes and/or in the recognition of the minus template by the polymerase.
Whether the ability of TIA-1/TIAR to associate with stress granules is utilized by flaviviruses and/or the other types of viruses tested is not known. Flavivirus infections do not shut off host cell translation (28), and flaviviral nonstructural proteins and double-stranded RNA (indicative of viral replication intermediates) have been colocalized to redistributed endoplasmic reticulum, trans-Golgi, and intermediate-compartment membranes (30, 31). Further studies are needed to investigate whether stress granules are present in flavivirus-infected cells and, if so, whether they are associated with viral replication complexes. If the binding of TIA-1/TIAR to viral minus-strand RNAs in replicative intermediates results in their colocalization with stress granules, this could provide an environment in which the translation of growing nascent plus strands would be inhibited. Alternatively, the binding of TIAR/TIA-1 to the WNV 3′(−) RNA may reduce the likelihood of the formation of stress granules in infected cells.
Acknowledgments
This work was supported by Public Health Service research grants AI18382 from the National Institute of Allergy and Infectious Diseases and GM54896 from the General Medicine Institute. W.L. was supported in part though a Research Program Enhancement Award from the GSU Office of Research and Sponsored Programs. The Beckman Research Institute facility was funded by National Institutes of Health Cancer Center Support Grant CA33572.
REFERENCES
- 1.Beck, A. R., Q. G. Medley, S. O'Brien, P. Anderson, and M. Streuli. 1996. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 24:3829-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, A. R., I. J. Miller, P. Anderson, and M. Streuli. 1998. RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl. Acad. Sci. USA 95:2331-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell, J. L., and M. A. Brinton. 1995. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 69:5650-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, S., and H. M. Bourbon. 1993. The developmentally-regulated Drosophila gene rox8 encodes an RRM-type RNA binding protein structurally related to human TIA-1-type nucleolysins. Nucleic Acids Res. 21:3699-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinton, M. A. 1997. Host susceptibility to viral disease, p. 303-328. In N. Nathanson et al (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa.
- 8.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162:290-299. [DOI] [PubMed] [Google Scholar]
- 9.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153:113-121. [DOI] [PubMed] [Google Scholar]
- 10.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 10a.Cahour, A., A. Pletnev, M. Yazeille-Falcoz, L. Rosen, and C. J. Lai. 1995. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology 207:68-76. [DOI] [PubMed] [Google Scholar]
- 11.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 81:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, M. T., and T. D. Lee. 1998. Rapid protein identification using a microscale electrospray LC/MS system on an ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 9:194-201. [DOI] [PubMed] [Google Scholar]
- 13.Davis, M. T., and T. D. Lee. 1997. Variable flow liquid chromatography-tandem mass spectrometry and the comprehensive analysis of complex protein digest mixtures. J. Am. Soc. Mass Spectrom. 8:1059-1069. [Google Scholar]
- 14.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dember, L. M., N. D. Kim, K. Q. Liu, and P. Anderson. 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 271:2783-2788. [DOI] [PubMed] [Google Scholar]
- 16.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 17.Forch, P., and J. Valcarcel. 2001. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis 6:463-468. [DOI] [PubMed] [Google Scholar]
- 18.Fukui, Y., S. Yumura, and T. K. Yumura. 1987. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and their changes during chemotaxis. Methods Cell Biol. 28:347-356. [DOI] [PubMed] [Google Scholar]
- 19.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 20.Jessen, T. H., C. Oubridge, C. H. Teo, C. Pritchard, and K. Nagai. 1991. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 10:3447-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawakami, A., Q. Tian, X. Duan, M. Streuli, S. F. Schlossman, and P. Anderson. 1992. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 89:8681-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedersha, N., S. Chen, N. Gilks, W. Li, I. J. Miller, J. Stahl, and P. Anderson. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13:195-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, Y. J., and B. S. Baker. 1993. Isolation of RRM-type RNA-binding protein genes and the analysis of their relatedness by using a numerical approach. Mol. Cell. Biol. 13:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 26.Le Guiner, C., F. Lejeune, D. Galiana, L. Kister, R. Breathnach, J. Stevenin, and F. Del Gatto-Konczak. 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276:40638-40646. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, T., C. Gueydan, G. Huez, J. J. Toulme, and V. Kruys. 1998. Mapping of a minimal AU-rich sequence required for lipopolysaccharide-induced binding of a 55-kDa protein on tumor necrosis factor-alpha mRNA. J. Biol. Chem. 273:13781-13786. [DOI] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippencott Williams and Wilkins, Philadelphia, Pa.
- 29.Lu, C. D., J. E. Houghton, and A. T. Abdelal. 1992. Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. J. Mol. Biol. 225:11-24. [DOI] [PubMed] [Google Scholar]
- 30.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 32.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parquet, M. C., A. Kumatori, F. Hasebe, K. Morita, and A. Igarashi. 2001. West Nile virus-induced bax-dependent apoptosis. FEBS Lett. 500:17-24. [DOI] [PubMed] [Google Scholar]
- 34.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, C. M. 1996. Flaviviridae: the viruses and their replication. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Shi, P. Y., M. A. Brinton, J. M. Veal, Y. Y. Zhong, and W. D. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 35:4222-4230. [DOI] [PubMed] [Google Scholar]
- 38.Shi, P. Y., W. Li, and M. A. Brinton. 1996. Cell proteins bind specifically to West Nile virus minus-strand 3′ stem-loop RNA. J. Virol. 70:6278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swiderek, K. M., M. T. Davis, and T. D. Lee. 1998. The identification of peptide modifications derived from gel-separated proteins using electrospray triple quadrupole and ion trap analyses. Electrophoresis 19:989-997. [DOI] [PubMed] [Google Scholar]
- 40.Taupin, J. L., Q. Tian, N. Kedersha, M. Robertson, and P. Anderson. 1995. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl. Acad. Sci. USA 92:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian, Q., M. Streuli, H. Saito, S. F. Schlossman, and P. Anderson. 1991. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67:629-639. [DOI] [PubMed] [Google Scholar]
- 42.Tian, Q., J. Taupin, S. Elledge, M. Robertson, and P. Anderson. 1995. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J. Exp. Med. 182:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaheri, A., W. D. Sedwick, S. A. Plotkin, and R. Maes. 1965. Cytopathic effect of rubella virus in BHK21 cells and growth to high titers in suspension culture. Virology 27:239-241. [DOI] [PubMed] [Google Scholar]
- 44.Weeks, K. M., and D. M. Crothers. 1992. RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry 31:10281-10287. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, R., R. Ainscough, K. Anderson, C. Baynes, M. Berks, J. Bonfield, J. Burton, M. Connell, T. Copsey, J. Cooper, et al. 1994. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature 368:32-38. [DOI] [PubMed] [Google Scholar]
- 46.Zeng, L., B. Falgout, and L. Markoff. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 72:7510-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]