Abstract
Hibiscus chlorotic ringspot virus (HCRSV) possesses a novel open reading frame (ORF) which encodes a putative 23-kDa protein (p23). We report here the in vivo detection of p23 and demonstrate its essential role in viral replication. The expression of p23 could be detected in protein extracts from transfected kenaf (Hibiscus cannabinus L.) protoplasts and in HCRSV-infected leaves. Further, direct immunoblotting of infected kenaf leaves also showed the presence of p23, and transient expression in onion and kenaf cells demonstrated that the protein is distributed throughout the cell. Site-directed mutagenesis showed that mutations introduced into the ORF of p23 abolished viral replication in kenaf protoplasts and plants but not in Chenopodium quinoa L. The loss of function of the p23 mutant M23/S33-1 could be complemented in trans upon the induced expression of p23 from an infiltrated construct bearing the ORF (pCam23). Altogether, these results demonstrate that p23 is a bona fide HCRSV protein that is expressed in vivo and suggest that p23 is indispensable for the host-specific replication of HCRSV. In addition, we show that p23 does not bind nucleic acids in vitro and does not act as a suppressor of posttranscriptional gene silencing in transgenic tobacco carrying a green fluorescent protein.
Hibiscus chlorotic ringspot virus (HCRSV) belongs to the Tombusviridae family of plant viruses. It is a member of the genus Carmovirus, which also includes Carnation mottle virus (16), Turnip crinkle virus (TCV) (6), Melon necrotic spot virus (35), Cardamine chlorotic fleck virus (40), Cowpea mottle virus (46), Saguaro cactus virus (48), Galinsoga mosaic virus (10), Japanese iris necrotic ring virus (44), and Pelargonium flower break virus (3). HCRSV is found in cultivated hibiscus hybrids worldwide. It induces chlorotic ringspots on naturally infected hibiscus leaves and causes local lesions on infected Chenopodium quinoa L. HCRSV has recently been reported to be a pathogen of aibika or bele (Abelmoschus manihot), the major crop cultivated in the South Pacific islands (5). A biologically active cDNA clone of HCRSV, p223, has previously been obtained and the genome organization of the virus described (19). HCRSV is composed of a single-stranded positive sense genomic RNA (gRNA) of ca. 4.0 kb; and two 3′-coterminal subgenomic RNAs (sgRNAs) of ca. 1.7 and 1.5 kb, respectively (19). The gRNA could potentially encode seven viral proteins. Two 5′-proximal open reading frames (ORFs) encode proteins of 28 kDa (p28) and a translational readthrough of 81 kDa (p81) which, by analogy with the ORF1 and -2 gene products of TCV (17, 49), are believed to form the putative viral RNA-dependent RNA polymerase (RdRp). The central region of the genome contains two overlapping small ORFs encoding the p8 and p9 proteins proposed to be involved in cell-to-cell movement. Based on immunoprecipitation with HCRSV antiserum (19), the 3′-proximal ORF encodes the 38-kDa viral coat protein (CP). In addition, comparison with other carmoviruses has revealed the existence of two novel ORFs in HCRSV which potentially encode a 23-kDa protein (p23) and a 25-kDa protein (p25) (Fig. 1A).
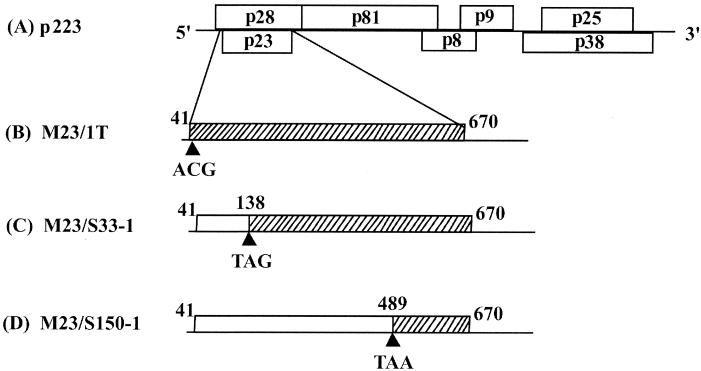
FIG. 1.
Schematic representation of HCRSV p23 mutants. Mutations were introduced into the full-length cDNA clone p223 (A). The point mutants of p23 were: M23/1T, nucleotide 41 (ATG→ACG), start codon→Thr(B); M23/S33-1, nucleotide 138 (TCG→TAG), amino acid 33 Ser→ stop codon (C); M23/S150-1, nucleotide 489 (TCA→TAA), amino acid 150 Ser→ stop codon (D).
The functions of the p28, p81, p8, p9, and p38 analogs have been studied extensively in TCV (17, 23, 25, 49). Although the replication of the TCV genome has been shown to involve the two viral RdRp proteins (17, 49), no reports have been published to identify any other novel proteins from this genus that are related to viral replication. Further, no proteins with significant homology to the deduced amino acid sequence of p23 could be identified from the EMBL GenBank sequence databases. Thus, the role of p23 in the pathogenesis of HCRSV was unknown. We report here the in vivo detection of p23 and demonstrate its absolute requirement for the host-specific replication of HCRSV.
MATERIALS AND METHODS
Construction of plasmids. (i) pCAL-c23CBP for the expression in Escherichia coli of a fusion protein between p23 and the calmodulin binding peptide.
Primers 1 (5′-CGGGATCCATGCTTTCTCAATTGCTTTCG-3′) and 2 (5′-CGGGATCCCGGGCGAGTACCCCTG-3′) were used to amplify the p23 coding region and terminal BamHI sites (underlined) were introduced to facilitate cloning. A full-length cDNA clone of HCRSV (p223) obtained previously (19) was used as a template for PCR by using Taq DNA polymerase (Promega). The amplified PCR fragment was digested with BamHI and cloned into BamHI-cut pCAL-c vector (Stratagene), with a calmodulin-binding peptide (CBP) tag at its C terminus, to yield pCAL-c23CBP.
(ii) pCam23 for replication complementation studies.
The p23 ORF with terminal SacΙ sites (underlined) was amplified by PCR by using Vent polymerase (New England Biolabs) with primers 3 (5′-CGAGCTCATGCTTTCTCAATTGCTTTCG-3′) and 4 (5′-CGAGCTCTCACGGGCGAGTACCCCTG-3′). The amplified fragment was inserted into SacI-cut pCass3 (31) to generate pCass23. Plasmid pCam23 was constructed by introducing the PvuII fragment from pCass23 into the binary vector pCAMBIA1300 (p1300) (kindly provided by R. A. Jefferson, CAMBIA [Australia]) digested with EcoRI/SmaI. The EcoRI site was blunt ended by treatment with Klenow fragment.
(iii) pCass23GFP for the p23 localization studies.
By using the infectious clone p223 as a template and primers 5 (5′-GCTCTAGAATGCTTTCTCAATTGCTTTCG-3′) and 6 (5′-CATGCCATGGCGGGCGAGTACCCCTG-3′), XbaI and NcoI sites (underlined) were introduced by PCR into the 5′ and 3′ ends of the p23 coding region, respectively. The amplified p23 fragment was then inserted into the XbaI- and NcoI-digested green fluorescent protein (GFP) construct pCassGFP (31), resulting in the fusion protein expression construct, pCass23GFP. Nucleotide sequences of all cloned inserts were verified by DNA sequencing after construction.
p23-CBP fusion protein expression, purification and antibody production.
The p23 ORF was expressed as a CBP-tagged fusion protein by using the Affinity Protein Expression System (Stratagene) according to the manufacturer's instructions. Cultures of E. coli BL21(DE3)plys (Stratagene) carrying pCAL-c23CBP were grown at 37°C in Luria-Bertani medium containing 100 μg of ampicillin and 34 μg of chloramphenicol ml−1. When the optical density at 600 nm had reached 0.6, the culture was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and reincubated for 4 h at 37°C. Bacteria expressing p23-CBP were harvested by centrifugation and resuspended in CaCl2 binding buffer (50 mM Tris-HCl, pH 8; 150 mM NaCl; 10 mM β-mercaptoethanol; 1 mM magnesium acetate; 1 mM imidazole; 2 mM CaCl2). After three cycles of freezing and thawing with liquid nitrogen, 9 volumes of solubilization buffer (50 mM KH2PO4, pH 10.7; 1 mM EDTA; 50 mM NaCl) were added and the suspension was incubated for 30 min at room temperature. The mixture was subsequently adjusted to pH 8 and incubated for a further 40 min at room temperature. After centrifugation at 14,000 × g for 15 min, the supernatant was loaded onto an equilibrated calmodulin affinity resin column. The proteins were released from the column matrix by adding elution buffer (50 mM Tris-HCl, pH 8; 10 mM β-mercaptoethanol; 2 mM EGTA; 150 mM NaCl) and analyzed by 18% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fractions containing purified p23-CBP protein were pooled (to yield 0.7 mg of protein) and emulsified with an equal volume of complete Freund’s adjuvant (Sigma) for direct intramuscular injection into a rabbit. Four booster injections of p23-CBP protein (0.7 mg each) with incomplete Freund’s adjuvant were subsequently given at 2-week internals, and antisera were collected after each injection.
Detection of p23 in kenaf protoplasts and plant tissues.
Kenaf (Hibiscus cannabinus L.) protoplasts (4 × 105) were inoculated with in vitro transcripts of p223 (tp223), using a polyethylene glycol-mediated method described previously (26). Mock (i.e., without viral transcripts) and HCRSV-transfected protoplasts were collected by centrifugation at 100 × g for 10 min at 0, 12, 24, and 36 h postinoculation (h p.i.). The pellets were resuspended in lysis buffer (0.5 mM dithiothreitol, 4 mM phenylmethylsulfonyl fluoride, 8 M urea, 1% Triton-100, 5% SDS, 20 mM HEPES-KOH [pH 7.6], 150 mM NaCl) and centrifuged at 700 × g for 5 min. The supernatant was then collected, and an equal volume of 2× Laemmli sample buffer (250 mM Tris-HCl [pH 6.8], 8% SDS, 40% glycerol, 0.01% bromphenol blue dye, 200 mM β-mercaptoethanol) was added. Mock-inoculated or HCRSV-infected kenaf leaves (1 g) were ground in 5 ml of CaCl2 binding buffer, the suspension was centrifuged at 4,000 × g for 10 min, and the pellet was washed with the same buffer twice before resuspension in 5 ml of lysis buffer and subjection to two freeze-thaw cycles. After centrifugation at 4,000 × g for 10 min, total proteins were precipitated with 5 ml of acetone and resuspended in 1× Laemmli sample buffer. These protoplast and leaf samples were subsequently analyzed by Western blots.
Western blots.
Protein extracts from kenaf protoplasts and leaves were subjected to SDS-18% PAGE. Proteins were transferred to a 0.2-μm (pore-size) positively charged polyvinylidene difluoride nylon membrane (Boehringer Mannheim) and blocked by immersion in 5% skimmed milk for 1 h at room temperature. The blot was then washed three times (10 min for each wash) at room temperature with Tris-buffered saline (TBS; 20 mM Tris-HCl [pH 7.5], 500 mM NaCl) containing 0.5% Tween 20. The blot was subsequently incubated with p23-CBP polyclonal antiserum (1:500) in TBS for 1 h at room temperature and washed three times (10 min each) in TBS containing 0.2% Tween 20 prior to incubation in horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma) diluted 1:10,000 in TBS (40 min at room temperature). The enhanced chemiluminescence detection system (Amersham Pharmacia Biotech) was used according to the manufacturer's instructions prior to exposing the blot to X-ray film for 1 min at room temperature.
Whole-leaf immunoblots.
A protocol previously described for immunohistochemistry of tissue sections (30) was adapted to detect HCRSV p23 in situ in whole leaves. Briefly, either mock- or HCRSV-inoculated leaves were fixed in acid formaldehyde (FAA; 5% formaldehyde, 5% glacial acetic acid, 45% methanol) at 4°C overnight. The leaves were then dehydrated with a series of increasing concentrations of ethanol, cleared in Histolene (Sigma), and rehydrated through decreasing concentrations of ethanol. Fixed, cleared, and rehydrated leaves were then directly subjected to immunohistochemical detection by using antisera to p23-CBP. Leaves incubated with HCRSV CP antisera were used as controls.
Transient expression and localization of p23 in kenaf leaves.
The pCass23GFP construct was used for transient expression in kenaf leaves. Bombardments were performed by using the PDS-1000/He Biolistic Particle Delivery System (Bio-Rad Laboratories) based on previously described methods (31, 36). Briefly, expanded kenaf leaves were excised and placed on prewetted 3MM Whatman filter paper in a petri dish. Five milligrams of 1-μm-diameter gold particles, coated with 5 μg of the DNA construct, were used to biolistically bombard the leaves at 1,100 lb/in2. Bombarded leaves were subsequently incubated in a darkened high humidity chamber at room temperature. At 6, 12, 30, 48, 56, and 72 h p.i., the bombarded leaves were examined under blue-light excitation (489 nm) with an epifluorescence microscope (Nikon E800 M). A total of five replicates were carried out for each time point tested.
Construction of p23 mutants.
Three point mutants were constructed without changing the amino acid sequence of the p23-overlapping ORF for p28. For M23/1T, the initiation codon (ATG) of p23 was changed to ACG (Thr) by the PCR mutagenesis method used by Ding et al. (11). Two other mutants, M23/S33-1 and M23/S150-1, were similarly constructed such that nucleotide substitutions were made at positions 138 (TCG→TAG) or 489 (TCA→TAA) to introduce premature translational stops in the amino acid sequence at residues 33 or 150, respectively (Fig. 1B to D). Uncapped in vitro transcripts were synthesized from SmaI-linearized p223 or the mutant constructs by using T7 RNA polymerase (Promega). The transcripts were used to inoculate kenaf and C. quinoa protoplasts and leaves according to protocols described previously (18, 26).
Analysis of p23 mutants in protoplasts and leaves.
To extract total RNA from plants, mock- or p23 mutant-inoculated leaves (0.5 g) were ground in liquid nitrogen and transferred to 2-ml microtubes containing 1 ml of prewarmed 80°C phenol extraction buffer (0.1 M LiCl, 0.1 M Tris-HCl [pH 8], 10 mM EDTA, 1% SDS, 50% buffer-saturated phenol). The suspension was mixed with 500 μl of chloroform and, after thorough mixing, centrifuged for 5 min at room temperature. The supernatant was precipitated with an equal volume of 4 M LiCl at −20°C overnight. After centrifugation, the pellet was resuspended in 990 μl of suspension buffer (300 μl of Tris-EDTA [pH 7.5], 30 μl of 3 M sodium acetate [pH 5.2], 660 μl of 95% ethanol) and incubated for 20 min at −20°C. After centrifugation for 10 min at 4°C, RNA was resuspended in 50 μl of Tris-EDTA. C. quinoa protoplasts were isolated as described by Hans et al. (18). A digoxigenin (DIG)-labeled cRNA probe corresponding to the 3′ 0.8 kb of the HCRSV genome was generated from PstI-linearized p223 by using the T7 DIG RNA labeling kit (Boehringer Mannheim). For Northern blot analyses, equal amounts (5 μg) of total RNA were loaded for all samples.
Replication complementation between the p23 mutant M23/S33-1 and pCam23.
Kenaf plants with three pairs of leaves were infiltrated with Agrobacterium tumefaciens EHA105 carrying pCam23. Plants infiltrated with an empty vector (p1300) were used as negative controls. Subsequently, in vitro transcripts of M23/S33-1 (tM23/S33-1) were used to inoculate the infiltrated leaves. At 2, 7, and 14 days p.i., total RNA was extracted and subjected to reverse transcription-PCR (RT-PCR) and Northern blot analyses.
Testing for p23 nucleic acid-binding activity.
For radioactive RNA labeling, [32P]UTP was incorporated into the in vitro transcripts by using p223 as a template for T7 RNA polymerase. Cross-linking of p23 to radiolabeled RNA probes was carried out according to the method previously described (9), except that protein-RNA complexes were analyzed by SDS-8% PAGE and visualized by autoradiography. To synthesize double-stranded DNA (dsDNA) probes, an EcoRI/SpeI fragment from p223 was used as a template. [32P]dCTP was incorporated by incubation with Klenow fragment after restriction digestion. The single-stranded DNA (ssDNA) probes were obtained by heat denaturation of the synthesized dsDNA probes. The complexes of p23-ssDNA and p23-dsDNA were analyzed by 4% PAGE and visualized by autoradiography.
Testing for the ability of p23 to suppress PTGS.
A. tumefaciens EHA 105, carrying pCam23 expressing p23 or pBin-35S-mGFP5 expressing GFP (4) (upon induction), was used for posttranscriptional gene silencing (PTGS) suppression analyses in tobacco. Induction and infiltration of Agrobacterium into GFP transgenic plants (16C) was performed based on a protocol previously described (4). GFP fluorescence in whole plants was monitored by using a high-intensity UV lamp (model SB-100P/F, 365 nm; Spectroline). Plants coinfiltrated with pBin-35S-mGFP5 and A. tumefaciens expressing the 2b protein of Tomato aspermy virus (T2b) were used as a positive control (24). Total RNAs were extracted from the leaves at 3 and 6 days postinfiltration and subjected to Northern blot analysis. The blot was probed with digoxigenin-labeled full-length antisense GFP RNA. probes.
RESULTS
Expression of p23 in E. coli and its detection in infected kenaf protoplasts and plant tissues.
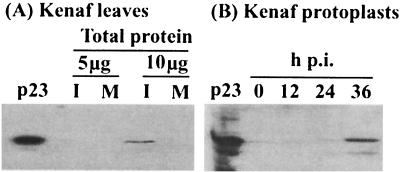
The p23 ORF, fused with a CBP tag, was expressed in E. coli and purified for use as an antigen in rabbits to raise antisera against p23 for immunoblot detection. The p23-CBP fusion protein was purified as an antigen to raise antisera in rabbits. Using the generated antisera, a protein corresponding to the molecular weight of the expressed p23 could be detected in 10 μg of protein extracted from infected kenaf leaves at 18 days p.i. but was absent from equivalent loadings of protein extracted from mock-inoculated plants (Fig. 2A), indicating that p23 is expressed in vivo, albeit at a low level. A protein of similar molecular weight was also detected in total protein extracts from transfected protoplasts at 36 h p.i. but could not be detected at earlier time points (Fig. 2B). Immunoblots of whole leaves also showed the presence of p23 in vivo where signal was detected only in the infected kenaf leaf but not in the mock-inoculated leaf at 18 days p.i. (Fig. 3). These results provide evidence that p23 is expressed in vivo and support a previous report that p23 can be translated in vitro by using wheat germ extracts or a rabbit reticulocyte system (19).
FIG. 2.
Immunoblot detection of p23 in infected kenaf protoplasts and leaves. (A) Total proteins (5 or 10 μg) from infected or mock-inoculated leaves. I represents HCRSV-infected leaves. M represents mock-inoculated leaves. The purified p23 protein expressed from E. coli was used as a positive control. (B) Equal loading of total proteins (5 μg) isolated from protoplasts inoculated with tp223 at 0, 12, 24, and 36 h p.i.
FIG. 3.
Whole-leaf immunoblot detection of p23. Fixed, cleared leaves were subjected to immunohistochemical detection. Alternate substrates were used for the detection of p23 (blue) and coat protein (red). Mock-inoculated control leaves (A) show only background coloration, whereas HCRSV-infected leaves (B) show intense punctate blue staining or intense red staining to indicate the presence of p23 or coat protein, respectively.
p23 is distributed throughout HCRSV-transfected cells.
The in vivo localization of p23 was investigated by using GFP as a visual reporter (7, 14, 32). Unlike transiently expressed free GFP (from pCassGFP) which, as early as 6 h posttransfection (h p.t.), gave rise to a cytoplasmic distribution of fluorescent signal, moderately enhanced in both the nucleus and along cytoplasmic filaments; a delayed expression (24 h p.t.) was seen for p23-GFP. The fusion protein was found to accumulate throughout both the nuclear and the cytoplasmic compartments of kenaf leaf epidermal cells (data not shown). Further, the number of cells transfected with pCassGFP which showed fluorescent signal reached an equilibrium by 24 h p.t., whereas the number of fluorescing cells transfected with pCass23GFP continued to rise up to 48 h p.t. Similar results were obtained when p23, transiently expressed as a C-terminal fusion to GFP (pCassGFP23), was used for bombardments (data not shown).
p23 is essential for HCRSV replication in kenaf.
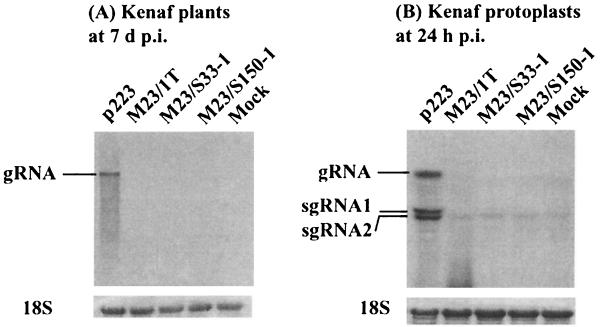
In order to explore the possible functions of p23, three point mutations were introduced into the p23 coding sequence without altering the amino acid sequence of the overlapping ORF p28. Every p23 mutant examined failed to replicate in both kenaf protoplasts and whole plants. No symptoms were observed, and progeny RNA remained undetectable in the inoculated and upper leaves of kenaf at 2 days p.i. (data not shown) and at 7 days p.i. when wild-type control infections were detectable (Fig. 4A). In addition, neither of the two sgRNAs could be detected in infected leaves, suggesting a low level of accumulation. Further, at 24 h p.i., gRNA and the two sgRNAs of HCRSV were detectable in protoplasts inoculated with in vitro transcripts of a biologically active cDNA clone of HCRSV (p223) but not in protoplasts inoculated with any of the described mutants (Fig. 4B).
FIG. 4.
Northern blot analyses of kenaf protoplasts and plants inoculated with in vitro transcripts of the p23 mutants. (A) Kenaf leaves at 7 days p.i. with tp223, tM23/1T, tM23/S33-1, and tM23/S150-1, respectively. (B) Transfected protoplasts at 24 h p.i. The 18S RNA bands (at the bottom) indicate equal RNA loadings.
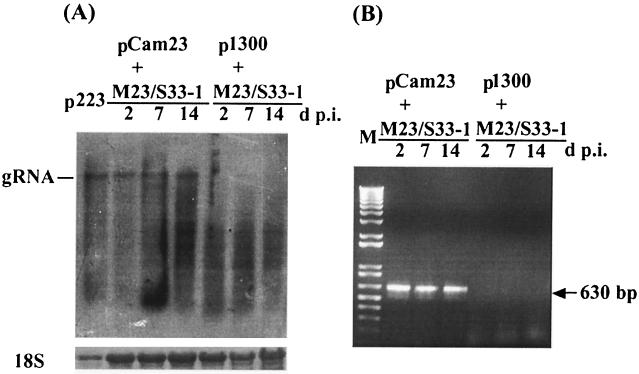
Complementation experiments confirmed the requirement of p23 for the replication of HCRSV in kenaf. Northern blot and RT-PCR analyses of leaf extracts collected at 2, 7, and 14 days p.i. showed the detection of progeny gRNA in leaves inoculated with tM23/S33-1 after infiltration with Agrobacterium carrying pCam23 but not with Agrobacterium carrying the p1300 vector alone (Fig. 5). These results indicate that the replicative function of p23 mutant M23/S33-1 could be restored by the in trans expression of p23. However, no gRNA could be detected in the upper new leaves (data not shown), suggesting that the mutant failed to move systemically even upon complementation by pCam23 in the infiltrated leaves. Direct sequencing of the PCR fragments showed that the progeny RNA from M23/S33-1 retained the point mutation, suggesting that the replicational rescue of the mutant was due to complementation and not reversion. Taken together, these data demonstrate that p23 is essential for the replication of HCRSV in kenaf.
FIG. 5.
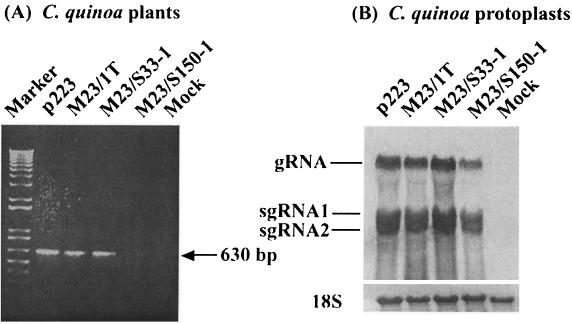
Complementation between the p23 mutant M23/S33-1 and a rescue construct expressing p23, pCam23. tM23/S33-1 was used to inoculate kenaf plants after infiltration with pCam23. Total RNA was extracted at 2, 7, and 14 days p.i. and subjected to Northern blot and RT-PCR analyses. (A) Northern blot with a DIG-labeled cRNA probe specific to the 3′ 0.8 kb of the HCRSV sequence. Genomic RNA was detected in leaves infiltrated with pCam23 but not with p1300. (B) RT-PCR analysis with p23 specific primers detected a 630-bp band in leaves infiltrated with pCam23 but not with p1300. M, DNA marker.
p23 is dispensable for HCRSV replication in C. quinoa.
C. quinoa plants inoculated with tp223 showed an expected local lesion response at 10 d p.i. and a similar response was observed on leaves inoculated with in vitro transcripts of M23/1T (tM23/1T). In contrast, chlorotic local lesions appeared in plants inoculated with in vitro transcripts of M23/S33-1 as early as 5 days p.i. but did not appear in leaves inoculated with in vitro transcripts of M23/S150-1 (tM23/S150-1) even after 40 days (data not shown). Consistently, RT-PCR revealed that a 630-bp fragment corresponding in size to the p23 ORF was obtained from progeny RNA isolated from the local lesions induced by tp223, tM23/1T, and tM23/S33-1, respectively, but not from RNA isolated from symptomless leaves inoculated with tM23/S150-1 (Fig. 6A). Direct sequencing of the amplified PCR products showed that the progeny RNA from the p23 mutants did not revert to wild-type (data not shown). However, all of the p23 mutants could replicate in C. quinoa protoplasts at a similar level to tp223 (Fig. 6B). These data indicate that p23 is not necessary for replication in C. quinoa, although a role in movement cannot be ruled out.
FIG. 6.
Infectivity of HCRSV p23 mutants in C. quinoa. (A) One percent agarose gel electrophoresis of RT-PCR products amplified from total RNA (isolated from local lesions appearing in C. quinoa) with p23 specific primers. (B) Northern blot of p23 mutants in protoplasts. Total RNA was extracted at 24 h p.i. The 18S RNA bands indicate similar RNA loading.
p23 is not a suppressor of PTGS in tobacco and does not bind to nucleic acids in vitro.
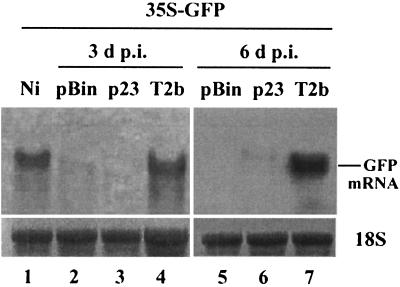
The possibility that p23 acts as a suppressor of PTGS was investigated in GFP transgenic tobacco plants (line 16C) (4). Fluorescence was monitored in GFP transgenic tobacco plants coagroinoculated with pCam23 and pBin-35S-mGFP5 at 3, 6, 9, and 14 d p.i. Observation of leaves and whole plants under UV light clearly showed that, when compared to plants coagroinoculated with p1300 and pBin-35S-mGFP5 (as negative controls) or T2b and pBin-35S-mGFP5 (as positive controls), p23 was unable to suppress the silencing of the GFP transgene (data not shown). Furthermore, the expression of p23 had no effect on GFP mRNA accumulation (Fig. 7, lanes 3 and 6) compared to the positive control T2b, which suppressed PTGS and resulted in higher accumulation levels of GFP mRNA in the infiltrated plants at 3 and 6 days p.i. (Fig. 7, lanes 4 and 7). These results strongly indicate that p23 does not act as a suppressor of PTGS. Predicted protein sequence analysis indicated that p23 contains an N-proximal leucine-rich domain reminiscent of both a nucleic acid-binding signal and a nuclear-cytoplasmic shuttling signal (2, 13). However, in our experiments, a transiently expressed p23-GFP fusion protein did not appear to specifically accumulate in the nuclei of transfected cells and E. coli expressed p23, when UV cross-linked to radiolabeled RNAs, did not show an electrophoretic mobility shift compared to controls either without p23 or with bovine serum albumin (data not shown). This suggests that p23 does not bind to RNA in vitro. In addition, no binding of p23 to either ssDNA or dsDNA could be detected (data not shown). These data indicate that E. coli-expressed p23 does not bind nucleic acids under our experimental conditions.
FIG. 7.
Northern blot showing GFP accumulation in the infiltrated leaves of the N. benthamiana GFP transgenic plants. Total RNAs were extracted from the leaves 3 and 6 days after infiltration with pBin35S-mGFP5 (pBin; lanes 2 and 5), pBin35S-mGFP5/pCam23 (p23; lanes 3 and 6), pBin-35S-mGFP5/T2b (T2b; lanes 4 and 7), or noninfiltrated leaves (Ni; lane 1), respectively, and monitored by Northern blot analysis. The blot was hybridized with digoxigenin-labeled GFP full-length antisense RNA probes. 18S RNA indicates equal RNA loading.
DISCUSSION
Herein we have described the in vivo expression of the HCRSV p23 protein. Although sequence predictions indicate a mass of 23 kDa and in vitro translation products support this prediction (19), the molecular mass of p23 as detected in infected protoplasts and plants is ca. 27 kDa and is similar to that of its fusion protein p23-CBP when expressed in E. coli. Such discrepancies are frequently the result of posttranslational modifications and, indeed, by using the PROSITE motif search program (1), p23 was shown to have seven potential protein kinase C phosphorylation sites as well as three casein kinase II phosphorylation sites and two N-myristoylation sites. p23 accumulated to a detectable level in infected kenaf protoplasts only at 36 h p.i., whereas we have previously shown that HCRSV gRNA and sgRNAs could be detected in infected kenaf protoplasts as early as 6 h p.i. (26). We therefore suggest that p23 does not accumulate to a detectable level in vivo during the early stages of viral replication and, as we could only detect p23 from 10 μg of total cellular protein even late in infections of kenaf, we further suggest that the accumulation of the p23 protein remains very low throughout the course of an HCRSV infection.
Mutational analyses demonstrated that p23 is essential for the replication of HCRSV in a host-specific manner. Since the ORF for p23 overlaps with ORF p28, the expression of p23 may be the result of a ribosomal frameshift. Viral proteins from Red clover necrotic mosaic virus and Cocksfoot mottle sobemovirus, which have been shown to employ frameshifts for expression, have also been shown to be involved in viral replication and downregulation of the expression of other essential genes (22, 29). In contrast, the translation of the p22-nested p19 gene of TBSV is influenced by the nucleotide sequence context surrounding the p22 start codon. In this case, context-dependent ribosomal leaky scanning leads to increased translation from the downstream p19 start codon. This cotranslational regulation of gene expression results in different levels of p19 and p22 protein accumulation, which influences host-dependent onset of symptoms (39). The possibility of a similar expressional regulation mechanism for the two nested genes of HCRSV (p23 and p28) remains to be investigated. Scanning through the sequenced genomes of other carmovirus members reveals no similarly sized ORFs in the corresponding location of any other viruses in the genus. Thus, although the precise mechanism by which p23 affects the host-specific replication of HCRSV remains to be elucidated, several possibilities can be postulated.
First, it is possible that p23 interacts with specific host factors to regulate the host replicational machinery involved in viral replication. Several plant species contain specific resistance genes corresponding to individual viral proteins, and it has recently been shown that the replication of tobamoviruses is inhibited by mutations in specific host genes and that this regulation is achieved through interactions between viral proteins and a certain host factor(s) (45). In TBSV, host-specific symptom determination via the 19-kDa protein (p19) enables the systemic spread of the virus in certain host plants (8, 37, 38, 39) through a mechanism associated with p19 suppression of gene silencing (33, 34). However, although gene silencing and its suppression are similarly employed in a variety of host-virus interactions to regulate gene expression (4, 21), p23 does not appear to be a suppressor of PTGS, as tested in GFP transgenic tobacco. Attempts to investigate p23 suppression of PTGS in kenaf have failed due to premature cell death of the kenaf leaves following Agrobacterium coinfiltration; therefore, the possibility that p23 suppresses PTGS in kenaf requires further examination.
A second possibility is that p23 might function as a cis- or trans-activator to either directly or indirectly regulate the expression of genes involved in viral replication. Such a host-specific effect has been reported for ORF C2 mutants of Tomato yellow leaf curl virus (TYLCV) (47), wherein C2 is essential for the infection of tomato by TYLCV, but not of tobacco. In addition, the C2 ORF also plays a role in the transactivation of viral gene expression and is involved in host and tissue-specific infection (12, 47). A similar regulatory process has been postulated for a cis-acting repressor element located within, or near to, the ORF C2 of Tomato golden mosaic virus and is believed to repress the expression of the CP gene and thereby mediate virus-host interactions (41, 42, 43). Further investigations should reveal if p23 exploits a similar mechanism. As evidenced by the p23 mutagenesis experiments, there is an absolute requirement for p23 during HCRSV infection in kenaf but not in C. quinoa, raising the possibility that p23 shares a similar regulatory function to that of TYLCV ORF C2. In addition, we have observed that HCRSV CP also plays a role in host-specific infection (27). Perhaps p23 confers its functional effect on the host-specific replication of HCRSV by interacting with the viral CP.
Finally, it is also possible that p23 may interact with the viral RdRp complex of p28/p81 to regulate viral replication. However, such speculations remain to be resolved through investigations of protein-protein interactions between p23 and any other viral or host components.
The lack of infectivity for the p23 mutant in kenaf could be complemented in trans by the infiltration of pCam23, confirming the requirement of p23 for HCRSV replication in kenaf. Within the genus Carmovirus, only the TCV p28 and its readthrough protein p81 have been shown to be required for viral replication (17). However, sequence analysis of p23 revealed no conserved domain except for an N-proximal leucine zipper-like motif. Leucine zipper domains have been shown to be involved in viral replication by mediation of protein-protein interactions (15, 20, 28). In addition, our results showed that p23 does not bind nucleic acids in vitro. Nevertheless, this failure might be due to the denatured state of the protein during the experiment and/or to possible conformational changes in the expressed p23 during its expression and purification from E. coli. Further, any interaction between p23 and other viral or host proteins remains to be investigated. Hence, we cannot rule out the possibility that the leucine zipper-like motif in p23 may have a functional role in vivo.
The involvement of p23 in host determination is unique among Carmoviruses, making this the first report of a Carmovirus-encoded protein with a host-specific replicative function. However, the mechanism by which p23 affects viral replication and whether the requirement of p23 for HCRSV replication is directly correlated to its evolutionary adaptation to kenaf remain to be elucidated.
Acknowledgments
This research was supported by National University of Singapore research grant R-154-111-112. X.-Z.L. was a recipient of a Ph.D. research scholarship from the National University of Singapore.
We thank David Baulcombe of the John Innes Centre, Norwich, United Kingdom, for the 16C GFP transgenic tobacco line and Tianwei Lin of The Scripps Research Institute, La Jolla, Calif., for help in preparing the graphics.
REFERENCES
- 1.Bairoch, A., P. Bucher, and K. Hofmann. 1997. PROSITE is a database of functional motifs: ScanProsite finds all functional motifs in your sequence that are annotated in the ProSite database. Nucleic Acids Res. 25:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begitt, A., T. Meyer, M. van Rossum, and U. Vinkemeier. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 97:10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthome, R., C. Kusiak, J. P. Renou, J. Albouy, M. A. Freire, and S. Dinant. 1998. Relationship of the pelargonium flower break carmovirus (PFBV) coat protein gene with that of other carmoviruses. Arch. Virol. 143:1823-1829. [DOI] [PubMed] [Google Scholar]
- 4.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6742. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Brunt, A. A., and N. J. Spence. 2000. The natural occurrence of Hibiscus chlorotic ringspot virus (Carmovirus; Tombusviridae) in aibika or bele (Abelmoschus manihot) in some South Pacific Island countries. Online New Dis. Rep. [Online.] 1:"http://www.bspp.org.uk/ndr/2000/2000.
- 6.Carrington, J. C., L. Heaton, D. Zuidema, B. I. Hillman, and T. J. Morris. 1989. The genome structure of turnip crinkle virus. Virology 170:219-226. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1995. Green fluorescence protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 8.Chu, M., B. Desvoyes, M. Turina, R. Noad, and H. B. Scholthof. 2000. Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology 266:79-87. [DOI] [PubMed] [Google Scholar]
- 9.Citovsky, V., D. Knorr, D. Schuster, and P. Zambryski. 1990. The protein P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60:637-647. [DOI] [PubMed] [Google Scholar]
- 10.Ciuffreda, P., L. Rubino, and M. Russo. 1998. Molecular cloning and complete nucleotide sequence of galinsoga mosaic virus genomic RNA. Arch. Virol. 143:173-180. [DOI] [PubMed] [Google Scholar]
- 11.Ding, S. W., W. X. Li, and R. H. Symons. 1995. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 14:5762-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dry, I., L. Krake, P. Mullineaux, and A. Rezaian. 2000. Regulation of tomato leaf curl viral gene expression in host tissues. Mol. Plant-Microbe Interact. 13:529-537. [DOI] [PubMed] [Google Scholar]
- 13.Forgues, M., A. J. Marrogi, E. A. Spillare, C. G. Wu, Q. Yang, M. Yoshida, and X. W. Wang. 2001. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J. Biol. Chem. 276:22797-22803. [DOI] [PubMed] [Google Scholar]
- 14.Fujii, G., R. Tsuchiya, E. Ezoe, and S. Hirohashi. 1999. Analysis of nuclear localization signals using green fluorescent protein-fusion protein library. Exp. Cell Res. 251:299-306. [DOI] [PubMed] [Google Scholar]
- 15.Gachon, F., G. Gaudray, S. Thebault, J. Basbous, J. A. Koffi, C. Devaux, and J. Mesnard. 2001. The cAMP response element binding protein-2 (CREB-2) can interact with the C/EBP-homologous protein (CHOP). FEBS Lett. 27:57-62. [DOI] [PubMed] [Google Scholar]
- 16.Guilley, H., J. C. Carrington, E. Balazs, G. Jonard, K. Rochard, and T. J. Morris. 1985. Nucleotide sequence and genome organization and carnation mottle virus RNA. Nucleic Acids Res. 13:6663-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker, D. L., T. D. Petty, N. Wei, and T. J. Morris. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 186:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Hans, F., M. Fuchs, and L. Pinck. 1992. Replication of grapevine fanleaf virus satellite RNA transcripts in Chenopodium quinoa protoplasts. J. Gen. Virol. 73:2517-2523. [DOI] [PubMed] [Google Scholar]
- 19.Huang, M., D. C. Y. Koh, L. J. Weng, M. L. Chang, Y. K. Yap, L. Zhang, and S. M. Wong. 2000. Complete nucleotide sequence and genome organization of hibiscus chloritic ringspot virus, a new member of the genus Carmovirus: evidence for the presence and expression of two novel open reading frames. J. Virol. 74:3149-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishizuka, T., T. Satoh, T. Monden, N. Shibusawa, T. Hashida, M. Yamada, and M. Mori. 2001. Human immunodeficiency virus type 1 tat binding protein-1 is a transcriptional coactivator specific for transcription. Mol. Endocrinol. 15:1329-1343. [DOI] [PubMed] [Google Scholar]
- 21.Kasschau, K. D., and J. C. Carrington. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71-81. [DOI] [PubMed] [Google Scholar]
- 22.Kim, K. H., and S. A. Lommel. 1994. Identification and analysis of the site of −1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology 200:574-582. [DOI] [PubMed] [Google Scholar]
- 23.Kong, Q. Z., J. W. Oh, C. D. Carpenter, and A. E. Simon. 1997. The coat protein of turnip crinkle virus is involved in subviral RNA-mediated symptom modulation and accumulation. Virology 238:478-485. [DOI] [PubMed] [Google Scholar]
- 24.Li, H. W., A. P. Lucy, H. S. Guo, W. X. Li, L. H. Ji, S. M. Wong, and S. W. Ding. 1999. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, W. Z., F. Qu, and T. J. Morris. 1998. Cell-to-cell movement of turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology 244:405-416. [DOI] [PubMed] [Google Scholar]
- 26.Liang, X. Z., S. W. Ding, and S. M. Wong. 2002. Development of a kenaf (Hibiscus cannabinus L.) protoplast system for replication study of Hibiscus chlorotic ringspot virus. Plant Cell Rep. 20:982-986. [Google Scholar]
- 27.Liang, X. Z., B. T. K. Lee, and S. M. Wong. Covariation in the capsid protein of hibiscus chlorotic ringspot virus induced by serial passaging in a host restricting movement leads to avirulence in its systemic host. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 28.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 29.Lucchesi, J., K. Makelainen, A. Merits, T. Tamm, and K. Makinen. 2000. Regulation of −1 ribosomal frameshifting directed by cocksfoot mottle sobemovirus genome. Eur. J. Biochem. 267:3523-3529. [DOI] [PubMed] [Google Scholar]
- 30.Lucy, A. P. 1996. Pathways to systemic invasion of plants by maize streak, and other viruses. Ph.D. thesis. University of East Anglia, East Anglia, United Kingdom.
- 31.Lucy, A. P., H. S. Guo, W. X. Li, and S. W. Ding. 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padgett, H. S., B. L. Epel, T. W. Kahn, M. Heinlein, Y. Watanabe, and R. N. Beach. 1996. Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J. 10:1079-1088. [DOI] [PubMed] [Google Scholar]
- 33.Qiu, W., J. W. Park, and H. B. Scholthof. 2002. Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol. Plant-Microbe Interact. 15:269-280. [DOI] [PubMed] [Google Scholar]
- 34.Qu, F., and T. J. Morris. 2002. Efficient infection of Nicotiana benthamiana by tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant-Microbe Interact. 15:193-202. [DOI] [PubMed] [Google Scholar]
- 35.Riviere, C. J., and D. M. Rochon. 1990. Nucleotide sequence and genomic organization of melon necrosis spot virus. J. Gen. Virol. 71:1887-1896. [DOI] [PubMed] [Google Scholar]
- 36.Sanford, J. C., F. D. Smith, and J. A. Russell. 1993. Optimizing the biolistic process for different biological applications. Methods Enzymol. 217:483-509. [DOI] [PubMed] [Google Scholar]
- 37.Scholthof, H. B., K. B. G. Scholthof, and A. O. Jackson. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholthof, H. B., K. B. G. Scholthof, M. Kikket, and A. O. Jackson. 1995. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213:425-438. [DOI] [PubMed] [Google Scholar]
- 39.Scholthof, H. B., B. Desvoyes, J. Kuecker, and E. Whitehead. 1999. Biological activity of two tombusvirus proteins translated from nested gene is influenced by dosage control via context-dependent leaky scanning. Mol. Plant-Microbe Interact. 12:67-69. [Google Scholar]
- 40.Skotnicki, M. L., A. M. Mackenzie, M. Torronen, and A. J. Gibbs. 1993. The genomic sequence of cardamine chlorotic fleck carmovirus. J. Gen. Virol. 74:1933-1937. [DOI] [PubMed] [Google Scholar]
- 41.Sunter, G., and D. M. Bisaro. 1991. Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180:416-419. [DOI] [PubMed] [Google Scholar]
- 42.Sunter, G., and D. M. Bisaro. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunter, G., and D. M. Bisaro. 1997. Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP) evidence for activation and depression mechanisums. Virology 232:269-280. [DOI] [PubMed] [Google Scholar]
- 44.Takemoto, Y., T. Kanehira, M. Shinohara, S. Yamashita, and T. Hibi. 2000. The nucleotide sequence and genome organization of Japanese iris necrotic ring virus, a new species in the genus Carmovirus. Arch. Virol. 145:651-657. [DOI] [PubMed] [Google Scholar]
- 45.Yamanaka, T., T. Imai, R. Satoh, A. Kawashima, M. Takahashi, K. Tomita, K. Kubota, T. Meshi, S. Naito, and M. Ishikawa. 2002. Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J. Virol. 76:2491-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You, X. J., J. W. Kim, G. W. Sturat, and R. F. Bozarth. 1995. The nucleotide sequence of cowpea mottle virus and its assignment to the genus Carmovirus. J. Gen. Virol. 76:2841-2845. [DOI] [PubMed] [Google Scholar]
- 47.Wartig, L., A. Kheyr-Pour, E. Noris, F. De Kouchkovsky, F. Jouanneau, B. Gronenborn, and I. Jupid. 1997. Genetic analysis of the monopartite tomato yellow leaf curl geminivirus: roles of V1, V2, and C2 ORFs in viral pathogenesis. Virology 223:132-140. [DOI] [PubMed] [Google Scholar]
- 48.Weng, Z. M., and Z. G. Xiong. 1997. Genome organization and gene expression of saguaro cactus carmovirus. J. Gen. Virol. 78:525-534. [DOI] [PubMed] [Google Scholar]
- 49.White, K. A., J. M. Skuzeski, W. Z. Li, N. Wei, and T. J. Morris. 1995. Immunodetection, expression strategy and complementation of turnip crinkle virus p28 and p88 replication components. Virology 211:525-534. [DOI] [PubMed] [Google Scholar]