Abstract
The hepatitis C virus (HCV) contains a plus-strand RNA genome. The 5′ noncoding region (NCR) of the viral genome functions as an internal ribosome entry site, and its unique 3′ NCR is required for the assembly of the replication complex during initiation of HCV RNA replication. Lohmann et al. (V. Lohmann, F. Korner, J.-O. Koch, U. Herian, L. Theilman, and R. Batenschlager, Science 285:110-113, 1999) developed a subgenomic HCV replicon system, which represents an important tool in studying HCV replication in cultured cells. In this study, we describe a cell-free replication system that utilizes cytoplasmic lysates prepared from Huh-7 cells harboring the HCV subgenomic replicons. These lysates, which contain ribonucleoprotein complexes associated with cellular membranes, were capable of incorporating [α32P]CTP into newly synthesized RNA from subgenomic replicons in vitro. Replicative forms (RFs) and replicative intermediates (RIs) were synthesized from the endogenous HCV RNA templates. Consistent with previous observations, RFs were found to be resistant to RNase A digestion, whereas RIs were sensitive to RNase treatment. The radiolabeled HCV RF-RI complexes contained both minus and plus strands and were specific to the lysates derived from replicon-expressing cells. The availability of a cell-free replication system offers opportunities to probe the mechanism(s) of HCV replication. It also provides a novel assay for potential therapeutic agents.
The hepatitis C virus (HCV), a hepacivirus of the family Flaviviridae, contains a single positive-strand RNA genome of 9600 nt (15). The genome consists of a 5′ noncoding region (NCR) (341 nucleotides [nt]), a region encoding a 3,010-amino-acid polyprotein, and a 3′ NCR. The 3′ NCR is composed of a short variable region, a U/(UC) motif, and a terminal 3′ X tail (98 nt) (17). The 5′ NCR and the 3′ X are the most conserved (>97%) elements of the HCV RNA genome. These cis elements have been shown to fold into stable secondary and tertiary structures that function as promoters of HCV gene expression (32). The 5′ NCR serves as an internal ribosome entry site (IRES) and directs translation initiation of the viral genome (32, 34, 35). The polyprotein is processed cotranslationally into three structural (C, E1, and E2) and six nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) polypeptides (for reviews, see references 2, 15, and 32 and references therein).
The NS5B protein has been shown to possess structural and functional similarities to other viral RNA-dependent RNA polymerases (29). The X-ray structure of NS5B revealed several unique structural features that are characterized by the presence of two loops from the finger and a thick thumb in a right-hand structure model (1, 7, 20). Bacterially expressed HCV NS5B has been used by a number of investigators to test various biochemical properties of this enzyme (21, 22, 25, 28, 38). These studies revealed that recombinant NS5B polymerase showed a lack of template specificity, and replication products of various lengths were produced. In cultured cells, however, NS5B not only forms ribonucleoprotein (RNP) complexes with viral NS proteins and 3′ cis elements (2) but is also capable of forming homo-oligomeric complexes (31). These RNP complexes play important roles during HCV replication, a process that leads to productive infection.
During the initiation of HCV replication, an RNP complex is formed at the HCV 3′ NCR of the viral genome. The HCV polymerase within the initiation complex serves as a catalytic subunit and synthesizes minus-strand RNA. The 3′-terminal region of this RNA (which is complementary to the 5′ NCR or IRES) promotes the assembly of the replication initiation complex to produce plus-strand viral RNA in an asymmetric fashion. At present, the identity of the viral and/or cellular factors that make up the replication initiation complex and the regulatory pathways that control the translation-replication molecular switch are not known. It is believed that HCV replication occurs on the endoplasmic reticulum (ER) membrane (2), consistent with the schemes utilized by other RNA viruses (19). ER-associated HCV gene expression induces ER stress and a cascade of signal transduction pathways. These pathways ultimately activate transcription factors that alter cellular metabolism (11, 33).
Studies on the mechanism of replication have been hampered due to the lack of an efficient animal model or tissue culture system. However, the development of subgenomic HCV replicons and their derivatives (23) has offered opportunities to study HCV gene expression and its effects on intracellular events. The HCV replicon is a bicistronic RNA molecule, which contains an HCV IRES in front of the neomycin phosphotransferase gene. The second cistron in the RNA molecule contains an encephalomyocarditis virus IRES, followed by the HCV nonstructural proteins (NS3 through NS5B) and terminating with the HCV 3′ NCR (Fig. 1) (6, 23). Replicon RNA molecules are maintained during numerous passages after hepatocyte-derived Huh-7 cells are transfected with the in vitro-synthesized replicon RNAs. However, during this period, a number of adaptive mutations accumulate in the subgenomic replicon. The most frequent among these mutations were found to occur in NS3- and NS5A-coding sequences (6, 18, 24). A number of such tissue culture-adapted replicons acquire the ability to replicate at a significantly higher efficiency than the parent strain. In this manner, they disguise their original sequence identity that once represented HCV isolates extracted from infected patients. Despite their apparent usefulness, there are limitations in using the present system for addressing mechanistic questions relating to HCV RNA replication. Here, we describe a cell-free HCV replication system that utilizes cytoplasmic lysates prepared from Huh-7 cells harboring subgenomic replicons. Replicative intermediates (RIs) or replicative forms (RFs) that are specific to the HCV subgenomic replicon sequences are produced. The cell-free system described here opens up new avenues to investigate biochemical and molecular features of HCV replication. The system will permit further investigation of the functional roles of viral and/or cellular factors in mediating this process.
FIG. 1.
Genetic organization of the HCV subgenomic replicon. The bicistronic RNA molecule contains the HCV IRES and a few core-coding sequences, which are followed by neomycin phosphotransferase gene (Neor). These sequences are followed by the encephalomyocarditis virus IRES placed in front of HCV sequences encoding nonstructural protein NS3 to NS5B, terminating at the HCV 3′ NCR as described by Lohmann et al. (23).
MATERIALS AND METHODS
Plasmids and in vitro RNA transcription.
The plasmid SP1/DS BM4-5 contains HCV subgenomic replicon sequences derived from HCV-1b genotype and an upstream T7 promoter for in vitro RNA synthesis (Fig. 1). This replicon (BM4-5) contains an adaptive mutation in the NS5A region (13). The plasmid was linearized with ScaI and purified by elution from an agarose gel. The linearized plasmid was transcribed by using an AmpliScribe T7 transcription kit (Epicentre Technologies) according to the manufacturer's instructions. The plasmid pCNS5A-M3 encodes the N terminus of the HCV NS5A (amino acids 1973 to 2135) cloned between T7 and SP6 promoters (11). The plasmid was linearized with HindIII or XbaI and gel purified. The XbaI-linearized plasmid was transcribed as described above by T7 polymerase to produce plus-strand HCV RNA, whereas SP6 RNA polymerase was used to synthesize minus-strand HCV RNA from the HindIII-cut template DNA. The pSPX plasmid was linearized with FspI and transcribed by SP6 polymerase to produce a 450-nt-long hepatitis B virus (HBV) X RNA, which was used here as a negative control. All of the RNAs were checked for purity and integrity by standard procedures.
Preparation of cellular lysates and HCV replication assay.
The FCA4 cell line is derived from Huh-7 cells selected in the presence of G418 after RNA transfection with a subgenomic HCV replicon. These cells stably express the HCV replicon at a high efficiency and contain adaptive mutations (13). The cytoplasmic fraction of these cells was prepared by a modified protocol of Chandrika et al. (8). Briefly, FCA4 cells were grown in the standard Dulbecco’s modified Eagle medium (Invitrogen) containing 10% fetal bovine serum, 100 U of penicillin per ml, 100 μg of streptomycin sulfate per ml, and 500 μg of G418 per ml in 100-mm-diameter petri dishes. The cells were washed with cold washing buffer (150 mM sucrose, 30 mM HEPES [pH 7.4], 33 mM ammonium chloride, 7 mM KCl, 4.5 mM magnesium acetate), followed by treatment with lysolecithin buffer (250 μg/ml of washing buffer) for 1 to 2 min. Three milliliters of washing buffer was added to each culture plate. The buffer was removed by aspiration. The cells were collected by scraping in 120 μl of incomplete replication buffer (100 mM HEPES [pH 7.4]; 50 mM ammonium chloride; 7 mM KCl; 1 mM spermidine; 1 mM [each] ATP, GTP, and UTP; 10 μM CTP), transferred to a new tube, and lysed gently by pipetting at least 15 times. The cell suspension was centrifuged at 1,600 rpm in a microcentrifuge (Eppendorf) for 5 min at 4 C. The cytoplasmic fraction (supernatant) was aliquoted and stored at −70°C until used. The cytoplasmic extract (60 to 70 μl for each reaction) was incubated with [α-32P]CTP (30 μCi; 800 Ci/mmol) for 1 to 1.5 h at 30°C or as indicated in the figure legends. The replication reaction was terminated by adding sodium dodecyl sulfate (SDS)-containing STE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl, 0.5% SDS), followed by phenol-chloroform-isoamyl alcohol (25:24:1) extraction twice and water-saturated ether extraction two additional times. The RNAs were precipitated in ethanol. The centrifuged pellet was washed with 70% ethanol and resuspended in RNase-free water. The replication products were analyzed by native, denaturing formaldehyde, or denaturing methylmercury agarose gel electrophoresis followed by autoradiography as indicated. To visualize the RIs and RFs, native agarose gel electrophoresis was carried out, whereas the molecular sizes and the integrity of replication products were determined by formaldehyde or methylmercury agarose gel electrophoresis (5).
For transient-transfection experiments, the BM4-5 HCV replicon RNA (10 μg) was electroporated into Huh-7 cells as described by Lohmann et al. (24). The medium was changed at 24 h after electroporation to standard Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, 100 μg of streptomycin sulfate per ml, and 500 μg of G418 (Invitrogen) per ml. The culture was supplemented with fresh medium every other day for 3 to 4 weeks, and total G418-resistant colonies were harvested to prepare replication lysates as described above.
RNase sensitivity of the HCV replication products.
The replication assays were carried out as described above. The RNA products from the reactions were dissolved in 20 μl of RNase digestion buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 150 mM NaCl) and digested with 140 ng or 1 μg of RNase A (DNase and protease free) per ml. The digestion was carried out for 0, 1, 2.5, and 8 min at room temperature. Similar RNase digestion reactions were also carried out in buffer containing 300 mM NaCl. The control samples (untreated) were treated similarly but without RNase A. The reaction mixtures were extracted twice and precipitated as described above. The RNA preparations were dissolved in RNase-free water, fractionated by native agarose gel electrophoresis, dried at 50°C and autoradiographed.
To determine the stability of the replicon in the cell-free replication system, in vitro-transcribed BM4-5 RNA (3 μg) was added to the FCA4 replication lysates and incubated at 30°C for 0 to 15 min. The reactions were terminated and the products were extracted as described above. The RNA samples were analyzed by native agarose gel electrophoresis and visualized by ethidium bromide staining.
Actinomycin D and micrococcal nuclease treatment.
The effect of actinomycin D, an inhibitor of DNA-dependent RNA polymerases in HCV RNA replication, was determined by the addition of 225 μg of actinomycin D per ml directly to the FCA4 lysates during the replication assay. Similarly, HCV RNA replication was also carried out in the presence of micrococcal nuclease. Sixty-microliter lysates were treated with 1 U of micrococcal nuclease (Sigma) at 20°C for 20 min in the presence of 1 mM CaCl2. The reaction was stopped by adding 2 mM EGTA, and the products were subjected to replication assay as described above.
LiCl fractionation of HCV replication products.
Replication assays were carried out as described above. The RNA samples were adjusted to 2 M LiCl and incubated overnight at 4°C. Double-stranded RNA (RFs) is soluble in 2 M LiCl and was separated from insoluble single-stranded genomic RNA and partially single-stranded RI RNA molecules by centrifugation. The RF RNA was recovered by precipitation in ethanol. The samples were analyzed by native agarose gel electrophoresis.
Northern blot hybridization.
The replication products were synthesized in 400 μl of FCA4 lysates and fractionated by native agarose gel electrophoresis. The RF-RI radiolabeled bands were eluted from the agarose gel by using a QIAquick gel extraction kit (Qiagen Inc.) and used as hybridization probes. Ten micrograms of each of the RNAs was fractionated by formaldehyde-1% agarose gel electrophoresis. The RNAs were visualized by ethidium bromide staining and transferred to a nylon membrane overnight in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer. The membrane was treated with 40 ml of blocking buffer (50 mM sodium phosphate [pH 6.5], 50% deionized formamide, 5× SSC, 2.5× Denhardt's solution, 1% SDS, and 100 μg of salmon sperm DNA) for 4 h at 42°C. The eluted probe was hybridized with the blot overnight at 42 C in 10 ml of blocking buffer. The blot was washed in 2× SSC-0.2% SDS buffer and subjected to autoradiography.
RESULTS AND DISCUSSION
Cell-free HCV replication.
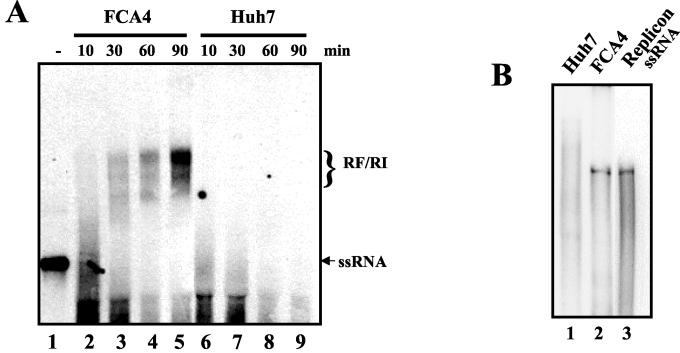
The cell-free replication system provides an excellent tool to study viral gene expression. A HeLa cell lysate-based coupled translation-replication (CTR) system that supports poliovirus gene expression (translation and replication) and allows de novo maturation of infectious viral particles has been previously described (4, 5, 26). We investigated whether similar CTR lysates would also support replication of in vitro-synthesized full-length or truncated HCV RNAs representing different genotypes. Interestingly, these lysates and those supplemented with Huh-7 cytoplasmic fractions were able to translate the HCV RNA but failed to support HCV RNA replication (data not shown). These results suggested that the CTR lysates possibly lacked factors and conditions required for HCV replication. Therefore, we focused on a liver-derived Huh-7 cell line (designated FCA4 cells) that harbored HCV replicons (13). We reasoned that FCA4 cytoplasmic lysates containing endogenous HCV replication complexes might permit detection of ongoing replication in vitro. To initiate these studies, we prepared FCA4 and Huh-7 (control) lysates with a modified protocol that has been previously described (8). The lysates were incubated in reaction mixtures containing [α32P]CTP, and the synthesis of RNA was monitored by the incorporation of radioactivity into newly synthesized RNA (Fig. 2A). HCV RNA synthesis was detected by the appearance of slowly migrating RNA species that resembled RFs. The intensity of radiolabeled RNA bands increased considerably by 90 min (lanes 2 to 5). These RNA bands were not observed in Huh-7 lysates that were prepared in a similar manner (lanes 6 to 9). In all cases, the prominent RNA bands migrated much slower than the positive-sense single-stranded replicon RNA marker (lane 1). Further analysis of the HCV replication products was carried out by denaturing methylmercury agarose gel electrophoresis. These results show that the major labeled replication products migrate similarly to that of HCV replicon single-stranded RNA (ssRNA) that was synthesized in vitro by T7 polymerase (Fig. 2B). Since the denatured products have the same molecular mass as the ssRNA marker, it can be safely assumed that predominant labeled RNA bands shown in Fig. 2A represents HCV RFs and are synthesized by de novo synthesis in the cell-free FCA4 extracts. In fact, other investigators have also found that HCV NS5B alone is capable of initiating RNA synthesis by a de novo mechanism in vitro (25, 38). Similar RNA products have been characterized as RIs or RFs for poliovirus and flaviviruses (3, 4, 9, 10, 12). Based on these assessments, we conclude that the RNA species observed here are synthesized from endogenous replicon RNA template and represent both RFs and RIs.
FIG. 2.
(A) Cell-free HCV replication assay. FCA4 (lanes 2 to 5) or Huh-7 (lanes 6 to 9) lysates were incubated with [32P]CTP for 10, 30, 60, or 90 min as indicated. The RNAs from these lysates were purified and fractionated by agarose gel electrophoresis in Tris-acetate-EDTA buffer. The gel was stained with ethidium bromide to visualize the RNA bands. The band intensities of rRNAs in each lane were found to be similar. Lane 1, in vitro-transcribed replicon that was labeled with the [α32P]CTP and used as an RNA marker. The labeled products are indicated as replicative forms (RI/RF). ssRNA, position of replicon. The gel was dried and exposed to X-ray film for 14 h. (B) Determination of the molecular weight of the HCV replication products synthesized in the FCA4 lysates. The replication assay was carried out with FCA4 lysates as described above, and the migration of replication products was compared with that of the in vitro-labeled BM4-5 RNA marker. The samples were denatured and electrophoresed on a methylmercury agarose gel, and the gel was dried and autoradiographed.
Characterization of the replication products.
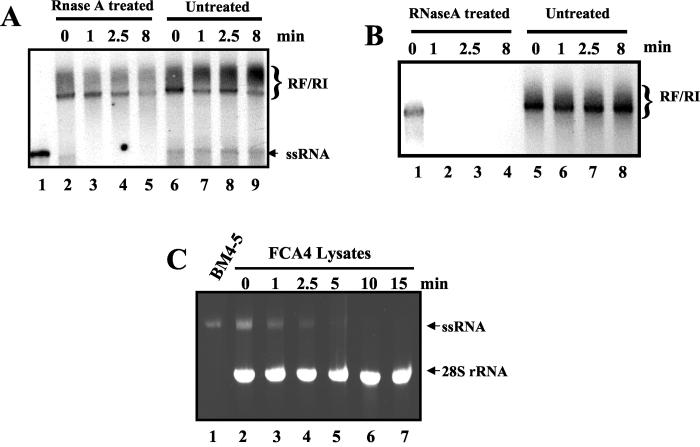
We examined the RNase A sensitivity of the putative HCV RFs and compared it to the sensitivity of 28S rRNA present in the same reaction mixture. The RF complexes were found to be sensitive to the RNase A treatment at a higher concentration (1 μg/ml) of the enzyme (Fig. 3B). At lower concentrations of RNase A (0.14 μg/ml), however, the HCV RF exhibited moderate resistance to the RNase treatment (Fig. 3A). A radiolabeled RNA (marked ssRNA) that migrated similarly to the replicon RNA marker (lane 1) was found to be highly sensitive (lanes 2 to 5). This sensitivity was similar to that of rRNA (not shown). In most experiments, this form is visible only as a faint band after a longer exposure to X-ray film.
FIG. 3.
RNase A sensitivity of the replication products. (A) RNA replication reactions, performed using FCA4 lysates. RNA products from the reactions were purified and resuspended in 20 μl of buffer containing 150 mM NaCl. The samples were treated with RNase A (DNase and protease free; 140 ng/ml) for 0, 1, 2.5, or 8 min at room temperature. The digestion was terminated by phenol extraction as described in Materials and Methods. The final products were resuspended in water and fractionated by 1% agarose gel electrophoresis to visualize RFs and RIs. The gel was dried and subjected to autoradiography. (B) Determination of the RNase A sensitivity of the replication products was carried out as described for panel A except that the RNase A concentration was raised to 1 μg/ml in this assay. (C) Determination of the stability of the HCV replicon in the cell-free replication system. In vitro-transcribed BM4-5 RNA (3 μg) was incubated in FCA4 replication lysates (lanes 2 to 7) for 0, 1, 2.5, 5, 10, and 15 min. The RNA was extracted from each of the samples and analyzed on a native (1%) agarose gel.
To characterize the stability of HCV ssRNA in replication lysates, the in vitro-transcribed single-stranded replicon RNA was incubated in FCA4 lysates (Fig. 3C). Interestingly, within 5 to 10 min, the replicon RNA was completely degraded and the RNA bands could not be detected by ethidium staining, whereas 28S rRNA remained fully intact under these conditions (Fig. 3C). These results further confirmed the instability of replicon in the ssRNA form.
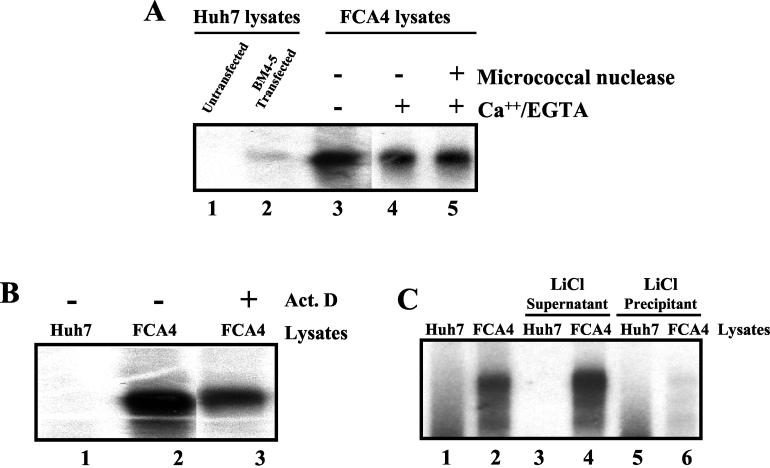
Next, we conducted a series of experiments to investigate the specificity of HCV RNA synthesis in the cell-free replication system. Huh-7 cells were transiently transfected with the in vitro-transcribed HCV subgenomic replicon RNA (BM4-5) in the presence of Geneticin (G418). The cytoplasmic lysates derived from a pool of G418-resistant transfected Huh-7 cells, untransfected Huh-7 cells, and FCA4 cells were subjected to the replication assay (Fig. 4A). As expected, the labeled RNA products were detected only in the transiently transfected Huh-7 (lane 2) and FCA4 (lane 3) lysates and not in the untransfected lysates (lane 1). These observations further support the view that the replicon RNA is being used as the template for RNA synthesis. The rationale of using transient transfection is to rule out any concern that the HCV replication system we have developed is specific only to FCA4 lysates, which are derived from one clone.
FIG. 4.
Effects of actinomycin D and micrococcal nuclease on HCV replication. (A) HCV replication assay of lysates from transiently transfected Huh-7 cells with HCV subgenomic replicon RNA (BM4-5) and FCA4 cell lysates treated with micrococcal nuclease. Agarose gel electrophoresis of the products of a replication assay carried out as described in Materials and Methods with lysates prepared from Huh-7 cells (lane 1), Huh-7 cell lysates transiently transfected with BM4-5 RNA subgenome (lane 2), FCA4 cell lysates (lane 3), FCA4 lysates treated with Ca2+ and subsequently with EGTA in a mock reaction (lane 4), and FCA4 cell lysates treated with micrococcal nuclease and Ca2+ and subsequently with EGTA (lane 5) is shown. (B) Replication of HCV RNA is resistant to actinomycin D. Agarose gel electrophoresis of the products of a replication assay with Huh-7 cell lysates (lane 1), FCA4 cell lysates (lane 2), and FCA4 cell lysates in the presence of actinomycin D (Act. D) (lane 3) is shown. (C) Analysis of the products synthesized in the replication assay. The RNAs labeled in Huh-7 (lane 1) or FCA4 (lane 2) cell lysates were fractionated with 2 M LiCl as described in Materials and Methods. The insoluble and soluble fractions were analyzed by native agarose gel electrophoresis. Lanes 3 and 4, LiCl-soluble forms of the replication assay products; lanes 5 and 6, nucleic acid products that are insoluble in LiCl.
The FCA4 lysates were treated with actinomycin D at a concentration that inhibits DNA-dependent RNA synthesis. Actinomycin D treatment had no inhibitory effect on the synthesis of RFs or RIs (Fig. 4B, lane 3), suggesting that the HCV polymerase is responsible for the synthesis of RFs. The minor band present in lane 2 (below the major labeled product) is occasionally observed in some of the samples because of the partial degradation of the major products during the extraction procedure. The FCA4 lysates were also treated with micrococcal nuclease in the presence of CaCl2. The micrococcal nuclease activity in the FCA4 lysates was then quenched by the addition of EGTA (Fig. 4A, lane 4). Although micrococcal nuclease degrades both DNA and RNA, these lysates were still competent for RF synthesis (lane 5), similar to the untreated sample (lane 4). This result suggests that the replicon RNA in FCA4 lysates is present in a nuclease-resistant replication complex. To test this possibility, we separated the FCA4 lysates into an RNP-containing pellet and a soluble supernatant. This approach has been successfully employed to pellet crude poliovirus replication complexes from the HeLa CTR system. Such complexes were shown to be efficient in RNA synthesis (5). With this strategy, the pellets prepared from FCA4 lysates were resuspended in the incomplete replication buffer and subjected to the replication assay. Under these conditions, the resuspended RNP complexes were able to synthesize RFs and RIs but with lower efficiency than for the standard FCA4 reaction. However, when a similar reaction mixture was supplemented with heterologous HeLa S10 lysates, RF and RI synthesis resumed efficiently (unpublished results). These results further indicate that soluble cellular factors and the replicon RNP complexes are important for HCV RNA synthesis.
Double-stranded RNA replication products (RFs) have been shown to be soluble in 2 M LiCl and can be separated from insoluble ssRNA molecules by centrifugation. We employed this technique to further characterize the nature of HCV replication products synthesized in the FCA4 lysates. Most of the labeled RNAs were found to be partitioned in the soluble fraction (supernatant) after treatment with 2 M LiCl (Fig. 4C, lane 4). However, the insoluble fraction (lane 6) contains very little of the labeled RNA, which was visible only after prolonged exposure. In the control Huh7 samples, similar labeled RNA products were not detected in either soluble or insoluble fractions (lanes 3 and 5). These results suggest that majority of the HCV products synthesized in the cell-free system were in double-stranded RNA forms.
RNA products contain both positive- and negative-strand HCV RNA.
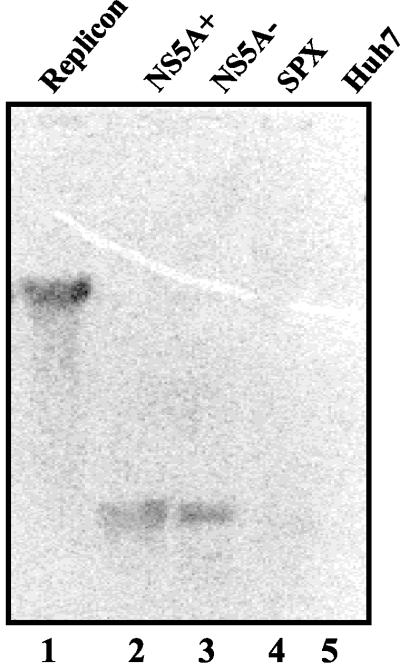
Northern blot hybridization analysis was carried out to determine the composition of RFs. The in vitro radiolabeled replication products were eluted from an agarose gel and used as a probe for hybridization in a Northern blot which included the following RNAs (Fig. 5): in vitro-transcribed plus-strand full-length replicon RNA (lane 1), an N-terminal portion of the NS5A-coding region representing either plus (NS5A+)- or minus (NS5A−)-polarity RNA (lanes 2 and 3, respectively), RNA derived from the HBV X gene of similar length (lane 4), and Huh-7 cytoplasmic RNAs (lane 5). The HCV replicon RNA (lane 1) but not the control RNAs (lanes 4 and 5) displayed hybridization with the radiolabeled replication products. Interestingly, both the NS5A+ and NS5A− RNAs also exhibited hybridization with the probe. This interaction appears to be highly specific to the HCV RNA, because cellular and HBV RNAs failed to hybridize with the radiolabeled replication products. These results demonstrate the presence of plus and minus strands of HCV RNA within the replication products. In the present assay system, it is not possible to measure the ratio of plus to minus strands present in the RF complex due to the limitations of the experimental system and the instability of ssRNAs in the lysates (see below). These results demonstrate that both strands are synthesized in replication lysates as evidenced by Northern blot hybridization.
FIG. 5.
Northern blot hybridization analysis of replication products. The in vitro-transcribed unlabeled RNAs representing the HCV subgenomic replicon (lane 1), the N-terminal half of the NS5A-coding region with plus (lane 2) or minus (lane 3) polarity, a nonspecific HBV RNA (lane 4) with a length similar to that of NS5A, and Huh-7 cytoplasmic RNA (lane 5) were fractionated by formaldehyde-agarose gel electrophoresis. The integrity of RNAs was determined by ethidium bromide staining before transfer to the nylon membrane. The 32P-labeled replication products synthesized in FCA4 lysates were eluted from a separate agarose gel and used to probe the blot. The blot was autoradiographed for 2 days.
Attempts to demonstrate trans-replication were unsuccessful when replication was assayed from the full-length HCV RNA or truncated subgenomic replicons containing 5′ IRES and 3′ NCR elements. We observed an immediate degradation of exogenously added HCV RNA in both the FCA4 and Huh-7 lysates. This was not surprising, because Han and Barton (14) have recently reported the degradation of HCV genomic RNA due to the RNase L activity present in the HeLa cell lysates. Attempts to overcome such problems are under way. The same problem was also encountered when 5′ IRES or 3′ NCR RNA molecules were used as competitors at various concentrations in trans-replication assays.
The presence of faint single-stranded subgenome-length RNA only in some replication assays (Fig. 3A) (3, 9) suggests that FCA4 lysates are competent for asymmetric replication. The fact that these ssRNAs are barely visible may reflect their highly unstable nature in the absence of structural proteins in the lysates. HCV subgenomic replicons are devoid of sequences encoding structural proteins. Chu and Westaway observed a similar decrease of ssRNA during in vitro replication of Kunjin virus, a flavivirus (9). The HCV core protein, for instance, which has been shown to bind the 5′ NCR (37) and initiate events in packaging and assembly of HCV particles, may offer protection against cellular RNase. Viral RNA replication, packaging of genomic RNA, and maturation and assembly of viral particles are viewed as continuous processes (19, 27). The replication system described here obviously lacks these crucial components and steps necessary for packaging and assembly. Therefore, it is possible that the lysates prepared from cells expressing a full-length replicon (16, 30) producing core and envelope proteins may alleviate this problem.
Replication of other flaviviruses (Kunjin virus and dengue virus type 2) has been extensively studied in cell-free systems by using either extract from virus-infected cells or a crude membrane-bound replicase complex (3, 9, 10, 36). Those studies have also provided evidence for the synthesis of three RNA species, similar to the data presented here, i.e., a minor genome-length RNA, a partially nuclease sensitive heterogeneous RI, and an RF that appears to migrate as a compact band just below the heterogeneous radiolabeled products (Fig. 3A). In our experiments the radiolabeled products in the FCA4 lysates were composed of several species. The gel mobility of these products was increased when replication was carried out for a longer time (Fig. 2A). Future work will focus on characterizing these multiple RNA forms resulting from RNA replication. These data suggest that the predominant RF species synthesized in the cell-free system may represent a mixture of HCV RNAs that contains both positive- and negative-strand RNAs.
In summary, our results demonstrate that lysates prepared from stably expressing subgenomic HCV replicons are capable of RNA synthesis in vitro. The prominent products of HCV replication are RFs consisting of both positive- and negative-strand RNAs. The in vitro replication system will be useful for characterizing the biochemical properties of the HCV replicase complex and for addressing mechanistic aspects of HCV replication. The system further offers opportunities to screen antiviral compounds with inhibitory actions during RNA replication.
Acknowledgments
We are highly indebted to C. Seeger of Fox Chase Cancer Research Institute, Philadelphia, Pa., for his kind and generous gifts of HCV replicon plasmid SP1/DS BM4-5 and the FCA4 cell line. We thank Ken Murray for methylmercury gel electrophoresis of our sample. We are also grateful to David Barton of our institution for his constant advice throughout this work.
N.A. was supported by an American Liver Foundation Ira M. Jacobson MD Liver Scholar Award, an American Cancer Society institutional research grant, the Cancer League of Colorado, and The Milheim Foundation for Cancer Research. K.D.T. was funded by a training grant (T3-DK07038) from NIH. This work was supported by an NIH grant (DK061566) to A.S.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, M. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomeusz, A. I., and P. J. Wright. 1993. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch. Virol. 128:111-121. [DOI] [PubMed] [Google Scholar]
- 4.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrika, R., S. M. Horikami, S. Smallwood, and S. A. Moyer. 1995. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213:352-363. [DOI] [PubMed] [Google Scholar]
- 9.Chu, P. W. G., and E. G. Westaway. 1985. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 140:68-79. [DOI] [PubMed] [Google Scholar]
- 10.Chu, P. W. G., and E. G. Westaway. 1987. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology 157:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, G., G. Waris, R. Tanveer, and A. Siddiqui. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc. Natl. Acad. Sci. USA 98:9599-9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grun, J. B., and M. A. Brinton. 1986. Characterization of West Nile virus RNA-dependent RNA polymerase and cellular terminal adenylyl and uridylyl transferases in cell-free extracts. J. Virol. 60:1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, J.-T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, J. Q., and D. J. Barton. 2002. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8:512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houghton, M. 1996. Hepatitis C virus, p. 1035-1058. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 16.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh-7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolykhalov, A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, M. M. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 20.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohmann, V., A. Roos, F. Korne, J.-O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilman, and R. Batenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, R. J. Lemm, and R. Colonno. 2001. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 27.Nugent, C. I. N., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh, J.-W., T. Ito, and M. M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 30.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in culture cells. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 32.Rijnbrand, R. C. A., and S. M. Lemon. 1999. Internal ribosome entry site-mediated translation in hepatitis C virus replication, p. 85-116. In C. H. Hegedorn and C. M. Rice (ed.), The hepatitis C viruses. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 33.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 76:7453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You, S., and R. Padmanabhan,. 1999. A novel in vitro replication system for Dengue virus: initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology -150293:141 73: 427-435. [DOI] [PubMed] [Google Scholar]
- 38.Zhong, W., A. S. U, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]