Abstract
To investigate the association of the murine leukemia virus (MuLV) Env protein with lipid rafts, we compared wild-type and palmitoylation-deficient mutant Env proteins by using extraction with the mild detergent Triton X-100 (TX-100) followed by a sucrose gradient flotation assay. We found that the wild-type MuLV Env protein was resistant to ice-cold TX-100 treatment and floated to the top of the gradients. In contrast, we observed that the palmitoylation-deficient mutant Env protein was mostly soluble when extracted by ice-cold TX-100 and stayed at the bottom of the gradients. Both the wild-type and mutant Env proteins were found to be soluble when treated with methyl-β-cyclodextrin before extraction with ice-cold TX-100 or when treated with ice-cold octyl-β-glucoside instead of TX-100. These results indicate that the MuLV Env protein is associated with lipid rafts and that palmitoylation of the Env protein is critical for lipid raft association. Although the palmitoylation-deficient Env mutant was synthesized at a level similar to that of the wild-type Env, it was found to be expressed at reduced levels on the cell surface. We observed syncytium formation activity with both the wild-type and mutant Env proteins, indicating that palmitoylation or raft association is not required for MuLV viral fusion activity.
Lipid rafts, also known as detergent-insoluble glycosphingolipid-enriched domains, are specific domains on plasma membranes which are enriched in cholesterol and sphingolipids (16, 30, 31). The high concentration of sphingolipids, which contain long, saturated acyl side chains, confers a more ordered state to the lipid rafts, which are resistant to extraction with mild nonionic detergents such as Triton X-100 (TX-100) at low temperature (4°C) (5). Lipid rafts are known to play an important role in several biological processes, such as signal transduction, T-cell activation, protein sorting, and virus assembly and budding (4, 31). Classes of proteins which have been shown to be associated with lipid rafts include glycosylphosphatidylinositol (GPI)-linked proteins, Src family kinases, and palmitoylated proteins (4, 14, 15, 24).
The assembly and budding of several enveloped viruses occur selectively from lipid rafts on the surface of infected cells (1, 4, 13, 17, 26, 35, 36, 41). It has been reported that particles of human immunodeficiency virus type 1 (HIV-1) contain a high concentration of cholesterol, sphingomyelin, and proteins that are known to localize to lipid rafts (2, 3). Envelope (Env) proteins of several viruses have also been reported to be associated with lipid rafts (1, 13, 21, 26, 41). Acylation of the Env proteins, especially palmitoylation, is thought to be important for the targeting of the proteins into lipid raft microdomains on the cell surface (14, 24, 29). It has been shown that removal of the cytoplasmic tail or mutation of the three palmitoylated cysteine residues in the transmembrane (TM) domain and the cytoplasmic tail decreased the association of influenza virus hemagglutinin (HA) with lipid rafts and decreased the incorporation of HA into virions (41). In HIV-1, the palmitoylation of the cytoplasmic tail of Env is also critical for lipid raft association and viral infectivity. Replacement of both cysteine residues in the cytoplasmic tail dramatically decreased the association of HIV-1 gp160 with lipid rafts and resulted in a lower level of viral infectivity (26). However, acylated vesicular stomatitis virus (VSV) G protein and Rous sarcoma virus (RSV) Env protein were shown not to be associated with lipid raft domains (6, 19).
The murine leukemia virus (MuLV) Env protein is synthesized as a precursor protein that is processed by a cellular protease into two subunits: the surface subunit (SU), containing the receptor binding domain, and the TM subunit, which forms a complex with the SU protein and is involved in subsequent membrane fusion (22, 23, 37). The TM subunit contains three structural domains: an extracellular domain containing the highly hydrophobic N-terminal fusion peptide, a membrane-spanning region for anchorage to the cell membrane, and a cytoplasmic tail (7, 34). The C-terminal 16-amino-acid fragment, designated the R peptide, is cleaved during viral maturation, which activates viral fusion activity and infectivity (10, 22, 23, 39).
The Moloney MuLV (M-MuLV) Env protein is modified by palmitoylation of Cys 597 in the TM domain (40). To investigate whether the MuLV Env protein is associated with lipid rafts and to examine the role of palmitoylation in lipid raft association, we used a mutant MuLV in which the cysteine residue in the TM domain of the Env protein was replaced by a serine residue. We examined the lipid raft association, surface expression, and syncytium formation activities of the wild-type and mutant MuLV Env proteins.
MATERIALS AND METHODS
Cells and viruses.
NIH 3T3 cells and XC cells were obtained from the American Type Culture Collection (Rockville, Md.) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (GIBCO BRL). The cDNA of the wild-type M-MuLV and the palmitoylation-deficient mutant MuLV genome, in which the cysteine in the TM domain of the TM protein was replaced by a serine, were cloned into plasmid pGEM4 and were designated pMOV and pMOVC597S, respectively. The construction of these two plasmids has been described previously (40).
Protein expression, radioactive labeling, and immunoprecipitation.
NIH 3T3 cells expressing wild-type or palmitoylation-deficient mutant Env proteins were starved in Eagle's medium deficient in methionine and cysteine for 45 min, pulse-labeled with 100 μCi of [35S]Met-Cys (Du Pont NEN) in 600 μl of Eagle's deficient medium for 15 min, and then chased in DMEM medium for different times as indicated. The total supernatants were collected and incubated with lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 1% TX-100, 1% sodium deoxycholate [pH 7.5]) plus protease inhibitor (Roche), and the cells were biotinylated and lysed in lysis buffer and immunoprecipitated overnight with goat anti-MuLV Env antibody and protein A-agarose beads (Pierce) at 4°C. Samples were washed with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 1% TX-100, 1% sodium deoxycholate [pH 7.5]) three times and prepared in reducing gel-loading buffer (125 mM Tris-HCl [pH 7.5], 4% sodium dodecyl sulfate [SDS], and 20% glycerol, plus 10% β-mercaptoethanol). Samples were heated at 95°C for 5 min before they were loaded onto an SDS-10% polyacrylamide gel electrophoresis (PAGE) gel for subsequent autoradiography.
Solubilization assay.
NIH 3T3 cells transfected with pMOV or pMOVC597S were grown in 35-mm-diameter plates. After 72 h, cells were starved in Eagle's medium deficient in methionine and cysteine for 45 min, labeled with 100 μCi of [35S]Met-Cys (Du Pont NEN) in 600 μl of Eagle's deficient medium for another 45 min, and then chased in DMEM medium with 10% fetal calf serum for 4 h. After three washes with cold phosphate-buffered saline (PBS), cells were incubated with lysis buffer {0.5% TX-100, 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 0.1 M KCl, 3 mM MgCl2, 10 mM EGTA, 0.3 M sucrose, pH 7.5} plus protease inhibitor (Roche) in ice or at 37°C for 3 min. The soluble and insoluble fractions were collected into 1-ml volumes of lysis buffer. The soluble fraction was cleared by centrifugation at 400 × g for 2 min, and the insoluble fraction was homogenized by passage through a 25-gauge needle 20 times. Both fractions were immunoprecipitated with goat anti-MuLV Env antibody and protein A-agarose beads (Pierce) at 4°C overnight. Samples were washed with RIPA buffer three times and prepared in reducing gel-loading buffer as described above. Samples were heated at 95°C for 5 min before they were loaded onto an SDS-10% PAGE gel for subsequent autoradiography.
Sucrose gradient flotation assay.
Radioactively labeled NIH 3T3 cells expressing wild-type or mutant MuLV Env proteins or transfected by the DNA encoding the placental alkaline phosphatase (PLAP) protein, a raft marker protein, were washed twice with cold PBS and incubated with TNE buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 1 mM EDTA) containing 1% TX-100 in ice for 30 min as described previously (4, 9, 29, 41). The cell lysates were then passed through a 25-gauge needle 20 times and adjusted to 40% (wt/vol) of sucrose in TNE, layered on the bottom of an SW41 centrifuge tube, and overlaid with 30% sucrose-TNE and 5% sucrose-TNE (wt/vol). The gradients were centrifuged at 260,000 × g at 4°C for 18 h. One-milliliter fractions were collected from the top and subjected to immunoprecipitation with goat anti-MuLV Env antibody. Samples were analyzed with an SDS-12% PAGE gel and subsequent autoradiography.
The distribution of the cell surface proteins was detected by a surface biotinylation assay. Radiolabeled NIH 3T3 cells were washed three times with ice-cold PBS-CM (PBS containing 0.1 mM CaCl2 and 1 mM MgCl2) and incubated with 0.5 mg of sulfsuccinimidyl 2-(biotinamido)ethyl-1,3′-dithiopropionate (NHS-SS-biotin; Pierce) in 1 ml of PBS-CM at 4°C for 30 min. Unreacted biotin was quenched by the addition of fresh DMEM. Cells were incubated with TNE buffer for 30 min in ice and then immunoprecipitated as described above. Samples were washed three times in RIPA buffer and then divided into two equal aliquots. One aliquot was used for immunoprecipitation, and the other was treated with 10 μl of 10% SDS and heated at 95°C for 5 min to release the Env proteins. The dissociated proteins were then dissolved in 1 ml of lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 1% TX-100, 1% sodium deoxycholate [pH 7.5]) plus protease inhibitor (Roche) and incubated with 15 μl of streptavidin-agarose (Pierce) for 5 h at 4°C. Biotinylated samples were washed three times with RIPA buffer and heated at 95°C for 5 min before analysis by SDS-PAGE.
Fusion assay of wild-type and mutant MuLV.
The fusion activities of MuLV Env proteins were determined by the following procedure. NIH 3T3 cells transfected with pMOV or pMOVC597S were overlaid with XC cells, a transformed rat cell line which has the receptors for MuLV Env proteins. Syncytium formation was observed 1 h later with a phase-contrast microscope. Syncytia were defined as giant cells with more than four nuclei within one single membrane. Ten fields from each sample were randomly selected, and the fusion activity was calculated based on the average value of the ratio of the number of nuclei in syncytia to the total number of nuclei in the same field.
RESULTS
Wild-type MuLV Env protein is resistant to TX-100 extraction.
As shown in Fig. 1, we investigated an MuLV mutant in which the palmitoylation site at Cys 597 in the TM domain of the M-MuLV Env protein was replaced by a Ser residue (C597S). Previous studies showed that the expression and fusion activity of the mutant Env protein were not significantly affected compared with those of the wild-type MuLV Env protein when expressed in a vaccinia T7 transient expression system (40).
FIG. 1.
Schematic diagram of the wild-type and palmitoylation-deficient mutant MuLV Env proteins. The designation of each Env protein is given on the left. Shaded boxes represent the extracellular domain, TM domain, cytoplasmic tail, and R peptide-coding region of the MuLV Env protein; the cleavage site before the R peptide is indicated with an arrow. The palmitoylation site (Cys 597) and the point mutation C597S are shown under each protein.
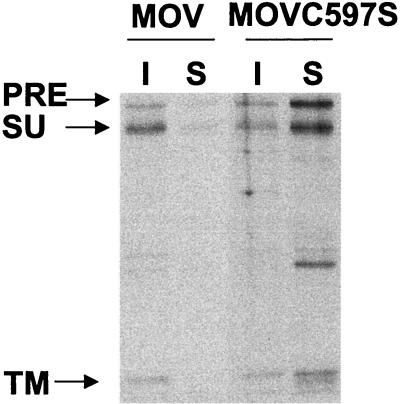
To investigate the association of the MuLV Env protein with lipid rafts, we first employed a solubilization assay to compare the solubilities of the wild-type and palmitoylation-deficient mutant MuLV Env proteins. As shown in Fig. 2, we found that the wild-type MuLV Env protein was predominantly in the insoluble fraction after treatment with ice-cold TX-100; about 80% of the total Env protein was TX-100 resistant. The Env protein became much more soluble when extracted at 37°C, and only about 5% of it was retained in the lipid raft fraction (data not shown). In contrast, the Triton solubility of the Env protein was greatly increased when the palmitoylated Cys residue was replaced by a Ser residue in the TM domain of the MuLV Env protein. Only about 20% of the mutant Env protein was associated with the insoluble fraction extracted by TX-100 in ice. These results indicate that the wild-type MuLV Env protein is resistant to TX-100 extraction at low temperature, while palmitoylation-deficient mutant Env is more sensitive to TX-100 extraction and became solubilized even at low temperature, suggesting the importance of palmitoylation in determining the lipid raft association of the Env protein.
FIG. 2.
Triton solubility of the wild-type and palmitoylation-deficient mutant MuLV Env proteins. NIH 3T3 cells were transfected with pMOV or pMOVC597S. After 72 h, cells were labeled with 100 μCi of [35S]Met-Cys and then chased for 4 h. After three washes with cold PBS, cells were incubated with lysis buffer (0.5% TX-100, 10 mM PIPES, 0.1 M KCl, 3 mM MgCl2, 10 mM EGTA, 0.3 M sucrose, pH 7.5) in ice for 3 min. The soluble (S) and insoluble (I) fractions were collected into 1-ml volumes of lysis buffer. The soluble fraction was cleared by centrifugation at 400 × g for 2 min, and the insoluble fraction was homogenized by passage through a 25-gauge needle 20 times. Both fractions were immunoprecipitated and analyzed by SDS-PAGE. The percentage distribution of Env protein was quantitated using phosphorimager analysis. PRE, Env precursor protein.
Palmitoylation is critical for lipid raft association of MuLV Env protein.
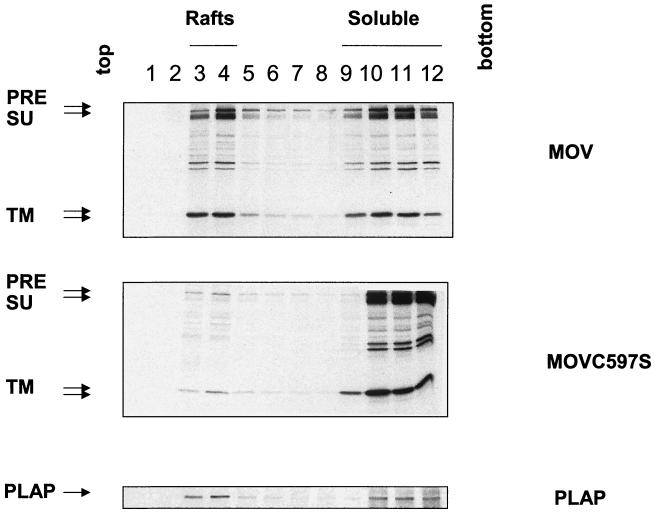
To further determine the association of the MuLV Env protein with lipid rafts, we isolated lipid raft microdomains by using a sucrose flotation gradient assay and compared the distributions of the wild-type and palmitoylation-deficient mutant Env proteins. As shown in Fig. 3, we found the wild-type MuLV Env protein in the low-density sucrose fractions after extraction with ice-cold TX-100; about 30% of the total Env protein was TX-100 resistant, which would be expected if the protein was sorted into lipid raft microdomains. In contrast, the Triton solubility of the Env protein was greatly increased when the palmitoylated Cys residue was replaced by a Ser, and less than 5% of the mutant Env protein was associated with the low-density fractions when extracted with ice-cold TX-100. PLAP, a GPI-anchored protein which is known to be associated with lipid rafts, was used as a marker and was found to be distributed in the low-density fractions after treatment with ice-cold TX-100. These results indicate that the wild-type MuLV Env protein is associated with lipid rafts and that palmitoylation of the Env protein is a determinant of efficient lipid raft association. The percentages of the insoluble proteins observed by using the different procedures are similar to the results reported previously with other viral glycoproteins (26, 41).
FIG. 3.
Lipid raft association of the MuLV Env protein. NIH 3T3 cells expressing wild-type or palmitoylation-deficient MuLV Env or transfected with the PLAP gene were labeled with 100 μCi of [35S]Met-Cys and then chased for 4 h. After washing with cold PBS, cells were scraped and incubated with TNE buffer containing 1% TX-100 in ice for 30 min. The cell lysates were homogenized by passage through a 25-gauge needle 20 times before being mixed with sucrose in TNE buffer and adjusted to 40% (wt/vol) sucrose in TNE; 30% and 5% sucrose in TNE were then overlaid on the top, and ultracentrifugation was performed at 260,000 × g for 18 h at 4°C. Twelve 1-ml fractions were immunoprecipitated and analyzed by SDS-PAGE. The percentage of Env protein distribution was quantitated using phosphorimager analysis. PRE, Env precursor protein.
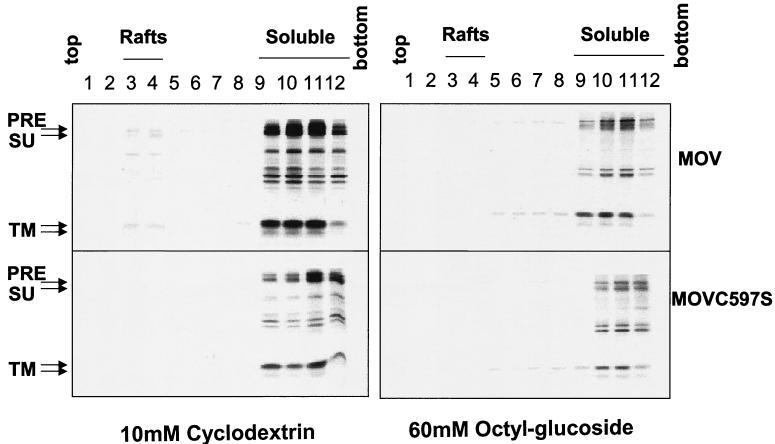
Both the wild-type and the mutant Env proteins became much more TX-100 soluble when cells were treated with 10 mM methyl-β-cyclodextrin, which is known to deplete cholesterol from the plasma membrane, or extracted with TNE buffer containing 60 mM octyl-β-glucoside, which destroys lipid rafts. Less than 5% of the Env protein was retained in the lipid raft fractions, as shown in Fig. 4, suggesting that the resistance of the MuLV Env protein to the nonionic detergent is due to its association with lipid rafts and that the Env proteins become soluble once the raft complexes are disrupted.
FIG. 4.
Solubilization of the MuLV Env protein in the presence of methyl-β-cytodextrin or octyl-β-glucoside. Radiolabeled NIH 3T3 cells expressing wild-type or mutant MuLV Env were treated with 10 mM methyl-β-cyclodextrin at 37°C for 1 h and then incubated with TNE-1% TX-100 in ice for 30 min, or cells were incubated with TNE containing 60 mM octyl-β-glucoside in ice for 30 min. The cell lysates were homogenized, and ultracentrifugation was performed at 260,000 × g for 18 h at 4°C. Twelve 1-ml fractions were immunoprecipitated and analyzed by SDS-PAGE. PRE, Env precursor protein.
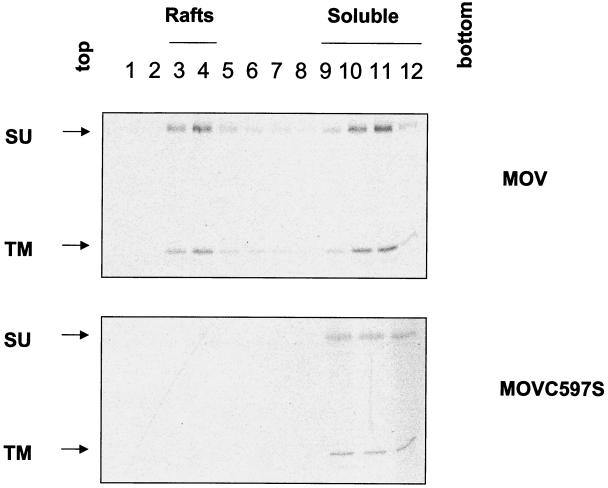
The results of a cell surface biotinylation assay followed by the sucrose flotation gradient also supported the conclusion that the wild-type MuLV Env protein is associated with lipid rafts, as shown in Fig. 5. There is evidence that lipid rafts are the sites for the assembly and budding of enveloped viruses on the cell surface, and they may play a role in the concentration of viral structural components during virus maturation (1, 11).
FIG. 5.
Surface distribution of the MuLV Env proteins. Radiolabeled NIH 3T3 cells were incubated with 0.5 mg of NHS-SS-biotin (Pierce) in 1 ml of PBS-CM at 4°C for 30 min. Unreacted biotin was quenched by the addition of fresh DMEM. Cells were incubated with TNE buffer for 30 min in ice and then immunoprecipitated as described above.
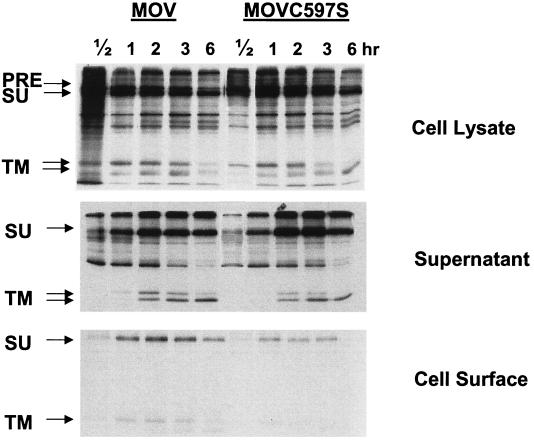
Surface expression of the wild-type and mutant MuLV Env proteins.
A pulse-chase assay was used to investigate whether the palmitoylation mutation affects the transport and stability of the MuLV Env protein. As shown in Fig. 6, the expression of the MuLV Env protein in the whole-cell lysate and the release of the SU protein in the medium were not significantly affected by the palmitoylation mutation. However, the surface expression level of the mutant Env was reduced dramatically, especially after a longer chase time, indicating that the palmitoylation-deficient mutant Env protein is not transported as efficiently to the cell surface or is not as stable as the wild-type Env on the cell surface. This observation is similar to that for the RSV envelope glycoprotein described in a recent study, in which it was found that two palmitoylated cysteine residues in the TM domain are required for protein stability and virus infectivity (20), and probably explains the reduced incorporation of the palmitate-deficient MuLV Env in virus particles as described previously (40).
FIG. 6.
Mutant MuLV Env protein exhibits reduced surface expression. NIH 3T3 cells transfected with pMOV or pMOVC597S were pulse labeled with 100 μCi of [35S]Met-Cys for 15 min and then chased for 0.5, 1, 2, 3, or 6 h. At the end of the chase, the supernatants were collected and incubated with lysis buffer, and cells were biotinylated and immunoprecipitated with antibodies against the MuLV Env protein plus protein A-agarose beads at 4°C overnight. The samples were prepared with reducing sample buffer and analyzed with an SDS-12% PAGE gel. PRE, Env precursor protein.
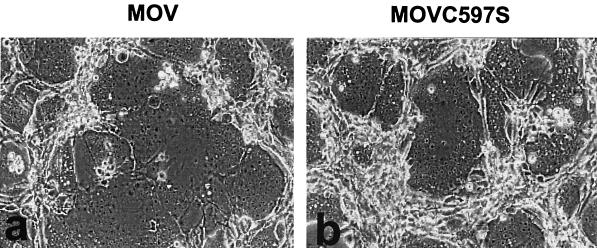
Syncytium formation by the Env palmitoylation mutant.
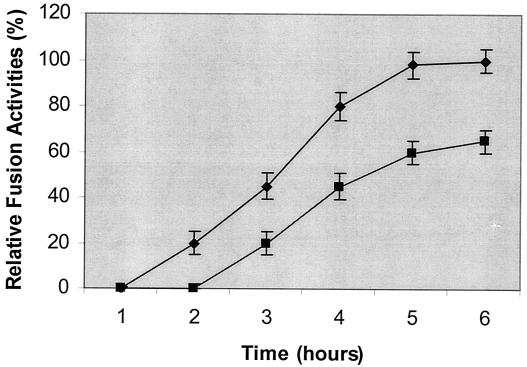
To evaluate the effect of the palmitoylation site mutation on fusion activity, we used a syncytium formation assay. NIH 3T3 cells expressing the wild-type or mutant MuLV Env proteins were overlaid with XC cells, which contain the cell receptors for the MuLV Env protein. As shown in Fig. 7, cells infected with either the wild-type or the mutant MuLV had significant syncytium formation activities; giant multinucleated cells were formed between the effector NIH 3T3 cells expressing the wild-type or mutant MuLV and the target XC cells. To investigate the kinetics of syncytium formation by the wild-type and mutant MuLVs, we overlaid the NIH 3T3 cells expressing wild-type or mutant MuLV with XC cells and monitored syncytium formation at intervals after cocultivation. As shown in Fig. 8, syncytium formation was first observed with wild-type MuLV at about 2 h after they were overlaid with XC cells and reached a maximal level after 5 h. The fusion kinetics of the MuLV mutant was somewhat slower than that of wild-type MuLV; syncytium formation was first observed at about 3 h after they were overlaid with the XC cells and reached a maximal level after 5 to 6 h. The maximal syncytium size induced by the mutant MuLV was smaller than that induced by the wild-type virus at the end point. These results indicate that the palmitate-deficient Env protein retains its membrane fusion activity. The smaller syncytia observed with the Env mutant may be due to the reduced level of Env found on the cell surface, as described above.
FIG. 7.
Syncytium formation by the MuLV Env protein. NIH 3T3 cells transfected with pMOV (a) or pMOVC597S (b) were overlaid with XC cells. Syncytium formation was observed 1 h later with a phase-contrast microscope. Syncytia were defined as giant cells with more than four nuclei.
FIG. 8.
Kinetics of the fusion activities of the wild-type and mutant MuLVs. XC cells were overlaid on the NIH 3T3 cells expressing wild-type (⧫) or mutant (▪) MuLV. Syncytium formation was monitored with a phase-contrast microscope at intervals after cocultivation. Fusion activity is designated as follows: 100% is the maximal level found for the wild-type MuLV, with more than 80% of nuclei present in syncytia, and the levels for other samples were compared with that for the wild-type MuLV and shown as percentages of the maximal fusion activity. Error bars denote standard deviations.
DISCUSSION
We found that the wild-type MuLV Env protein was resistant to TX-100 extraction at low temperature, suggesting its association with lipid raft microdomains. This conclusion was further supported by results showing that treatments, using methyl-β-cyclodextrin or octyl-β-glucoside, known to disrupt lipid rafts caused increased solubility of MuLV Env in TX-100. The palmitoylated cysteine residue in the TM domain of the Env protein was found to play an important role in determining the lipid raft association of Env because the palmitoylation-deficient mutant Env protein showed greatly decreased raft association compared with that of the wild-type Env. We also found that the surface expression of the palmitoylation-deficient mutant Env protein was lower than that of the wild-type Env but that it retained membrane fusion ability, indicating that palmitoylation of the Env protein is not required for efficient membrane fusion.
Our results with MuLV are similar to those from studies of other enveloped viruses which appear to bud from specific lipid raft microdomains on the infected cell surface (13, 26, 28, 41). In contrast, the VSV glycoprotein was found to be TX-100 soluble and largely excluded from lipid rafts (8). Both VSV and MuLV bud from the basolateral side of polarized epithelial cells (25, 28, 33), indicating that lipid raft association does not play an essential role in determining the site of virus budding in polarized cells. Our previous studies of MuLV showed that the palmitoylation mutant virus incorporated a lower level of Env protein and had somewhat lower infectivity (40). The Gag protein of retroviruses is important in virus assembly and is associated with lipid rafts by itself (11). It is also thought that the Gag protein can interact with Env through the cytoplasmic tail of the Env protein (38). However, we found that even in the presence of the Gag protein, the Env palmitoylation mutant still showed decreased lipid raft association, which indicates that the interaction of the cytoplasmic tail of the MuLV Env protein with Gag is not adequate for efficient lipid raft association. The interaction between the palmitoylated site and the lipid bilayer seems to be critical for lipid raft association and detergent insolubility and may affect the distribution of Env in the plasma membrane.
In this study, we also found that the surface expression of the palmitoylation-deficient mutant Env was significantly reduced, especially after a 6-h chase. An apparent discrepancy between this result and that of a previous study (40) likely resulted from the different expression systems used in these studies. In previous studies, surface expression of Env was analyzed in a transient vaccinia T7 expression system in HeLa T4 cells, while in the present study, we analyzed Env expression in the context of MuLV-infected NIH 3T3 cells. Furthermore, we observed that the level of the TM protein of mutant MuLV in the medium was reduced while that of the SU protein was not significantly affected. Since the SU protein in the medium represents both shed and virus particle-bound proteins, the amount of the TM protein is more accurate for determining the incorporation of Env protein into virus particles. This result is consistent with the previous observation that mutant Env was incorporated into virions less efficiently (40). These results indicate that the increased amount of the SU protein detected in the medium may result from increased shedding. The reduced level of mutant Env protein incorporation and expression on the cell surface is likely to be a result of decreased transport to the cell surface or the decreased stability of Env on the cell surface. Combined with the results presented in Fig. 2, this shows that mutant Env is less efficiently processed than wild-type Env. These results indicate that palmitoylation has a role in the surface expression of the MuLV Env protein. In a separate assay of the surface distribution of Env, we also found that not only is the majority of the surface-expressed mutant Env protein soluble upon TX-100 treatment but the total amount of it is less than that of wild-type Env. Similar results were also observed with the RSV envelope protein, in which a palmitoylation-deficient protein showed decreased stability on the cell surface (20). Considering that the wild-type palmitoylated RSV envelope protein is not associated with lipid rafts, palmitoylation of the viral envelope glycoprotein seems to have a function either in targeting the viral glycoproteins into the cell surface raft microdomains or in stabilizing the glycoprotein on the cell surface (6, 14, 20, 24, 29).
We found that removal of the palmitoylated site in the MuLV Env protein did not significantly affect the syncytium formation activity of Env; the palmitoylation-deficient Env protein was still functional in fusion activity, although the kinetics of syncytium formation by the mutant Env was slower and the size of the multinucleated cells was smaller than that observed with the wild-type Env. Nonetheless, it is possible that the interaction between the MuLV Env protein and its cell receptor mCAT-1, which is highly concentrated in rafts on the cell surface (12), could promote clustering of the Env protein, which may play a role in initiating syncytium formation between the cells infected with the mutant MuLV and the target XC cells. It has been reported that mutation of the three palmitoylated cysteine residues on the TM domain and cytoplasmic tail of the HA protein of influenza virus strain A/Aichi/2/68 (H3), which caused reduced association of HA with lipid rafts, did not affect the ability of HA to form syncytia and had no effect on the pH of the conformational change required for fusion activity (32). In contrast, deacylation of the HA protein of influenza virus A/USSR/77 (H1) dramatically decreased fusion pore formation and syncytium formation activity (27), indicating the different roles of acylation in different viral envelope proteins. Dispersion of the lipid rafts by cholesterol depletion with methyl-β-cyclodextrin has been found to inhibit syncytium formation induced by human T-cell leukemia virus type 1 (18). We observed that after removal of methyl-β-cyclodextrin from the medium, cells expressing MuLV Env protein were still capable of inducing cell-to-cell fusion, even in the presence of the cholesterol biosynthesis inhibitors lovastatin and mevalonate. However, no fusion was observed when cells were continuously exposed to methyl-β-cyclodextrin or when the target XC cells were pretreated with cyclodextrin (data not shown). Since lipid rafts play important roles in many biological processes and the integrity of the cell plasma membrane is required for efficient membrane fusion, disruption of the lipid rafts may affect the distribution of other essential components involved in membrane fusion or affect the lipid composition of the cellular plasma membrane, which is required for fusion pore formation.
Acknowledgments
We thank Tanya Cassingham for assistance in preparing the manuscript.
This study was supported by grant CA 18611 from the National Institutes of Health. A. Weidmann was supported by a fellowship from the Bundesministerium für Bildung und Forschung (BMBF-LPD 9901/8-29).
REFERENCES
- 1.Ali, A., and D. P. Nayak. 2000. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology 276:289-303. [DOI] [PubMed] [Google Scholar]
- 2.Aloia, R. C., F. C. Jensen, C. C. Curtain, P. W. Mobley, and L. M. Gordon. 1988. Lipid composition and fluidity of the human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 85:900-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloia, R. C., H. Tian, and F. C. Jensen. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA 90:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D. A., and E. London. 1998. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164:103-114. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 7.Gray, K. D., and M. J. Roth. 1993. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J. Virol. 67:3489-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooper, N. M. 1999. Detergent-insoluble glycosphingolipid/cholesterol-rich membrane domains, lipid rafts and caveolae. Mol. Membr. Biol. 16:145-156. [DOI] [PubMed] [Google Scholar]
- 10.Li, M., C. Yang, and R. W. Compans. 2001. Mutations in the cytoplasmic tail of murine leukemia virus envelope protein suppress fusion inhibition by R peptide. J. Virol. 75:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, X., and J. Silver. 2000. Ecotropic murine leukemia virus receptor is physically associated with caveolin and membrane rafts. Virology 276:251-258. [DOI] [PubMed] [Google Scholar]
- 13.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melkonian, K. A., A. G. Ostermeyer, J. Z. Chen, M. G. Roth, and D. A. Brown. 1999. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 274:3910-3917. [DOI] [PubMed] [Google Scholar]
- 15.Moffett, S., D. A. Brown, and M. E. Linder. 2000. Lipid-dependent targeting of G proteins into rafts. J. Biol. Chem. 275:2191-2198. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee, S., and F. R. Maxfield. 2000. Role of membrane organization and membrane domains in endocytic lipid trafficking. Traffic 1:203-211. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niyogi, K., and J. E. Hildreth. 2001. Characterization of new syncytium-inhibiting monoclonal antibodies implicates lipid rafts in human T-cell leukemia virus type 1 syncytium formation. J. Virol. 75:7351-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsenbauer, C., S. R. Dubay, and E. Hunter. 2000. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol. Cell. Biol. 20:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochsenbauer-Jambor, C., D. C. Miller, C. R. Roberts, S. S. Rhee, and E. Hunter. 2001. Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J. Virol. 75:11544-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickl, W. F., F. X. Pimentel-Muinos, and B. Seed. 2001. Lipid rafts and pseudotyping. J. Virol. 75:7175-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 25.Roth, M. G., R. V. Srinivas, and R. W. Compans. 1983. Basolateral maturation of retroviruses in polarized epithelial cells. J. Virol. 45:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakai, T., R. Ohuchi, and M. Ohuchi. 2002. Fatty acids on the A/USSR/77 influenza virus hemagglutinin facilitate the transition from hemifusion to fusion pore formation. J. Virol. 76:4603-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 29.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 30.Simons, K., and E. Ikonen. 2000. How cells handle cholesterol. Science 290:1721-1726. [DOI] [PubMed] [Google Scholar]
- 31.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 32.Steinhauer, D. A., S. A. Wharton, D. C. Wiley, and J. J. Skehel. 1991. Deacylation of the hemagglutinin of influenza A/Aichi/2/68 has no effect on membrane fusion properties. Virology 184:445-448. [DOI] [PubMed] [Google Scholar]
- 33.Stephens, E. B., R. W. Compans, P. Earl, and B. Moss. 1986. Surface expression of viral glycoproteins is polarized in epithelial cells infected with recombinant vaccinia viral vectors. EMBO J. 5:237-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, A., K. D. Gray, and M. J. Roth. 1997. Analysis of mutations within the cytoplasmic domain of the Moloney murine leukemia virus transmembrane protein. Virology 227:305-313. [DOI] [PubMed] [Google Scholar]
- 35.Vincent, S., D. Gerlier, and S. N. Manie. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, J. K., E. Kiyokawa, E. Verdin, and D. Trono. 2000. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc. Natl. Acad. Sci. USA 97:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witte, O. N., and D. F. Wirth. 1979. Structure of the murine leukemia virus envelope glycoprotein precursor. J. Virol. 29:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55Gag in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, C., and R. W. Compans. 1997. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J. Virol. 71:8490-8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, C., and R. W. Compans. 1996. Palmitoylation of the murine leukemia virus envelope glycoprotein transmembrane subunits. Virology 221:87-97. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]