Abstract
The herpes simplex virus Us11 gene product inhibits activation of the cellular PKR kinase and associates with a limited number of unrelated viral and cellular RNA molecules via a carboxyl-terminal 68-amino-acid segment rich in arginine and proline. To characterize the determinants underlying the recognition of an RNA target by Us11, we employed an in vitro selection technique to isolate RNA ligands that bind Us11 with high affinity from a population of molecules containing an internal randomized segment. Binding of Us11 to these RNA ligands is specific and appears to occur preferentially on conformational isoforms that possess a higher-order structure. While the addition of unlabeled poly(I · C) reduced binding of Us11 to a selected radiolabeled RNA, single-stranded homopolymers were not effective competitors. Us11 directly associates with poly(I · C), and inclusion of an unlabeled selected RNA in the reaction reduces poly(I · C) binding, while single-stranded RNA homopolymers have no effect. Finally, Us11 binds to defined, double-stranded RNA (dsRNA) molecules that exhibit greater sequence complexity. Binding to these dsRNA perfect duplexes displays a striking dependence on length, as 39-bp or shorter duplexes do not bind efficiently. Furthermore, this interaction is specific for dsRNA as opposed to dsDNA, implying that the Us11 RNA binding domain can distinguish nucleic acid duplexes containing 2′ hydroxyl groups from those that do not. These results establish that Us11 is a dsRNA binding protein. The arginine- and proline-rich Us11 RNA binding domain is unrelated to known dsRNA binding elements and thus constitutes a unique recognition motif that interacts with dsRNA. The ability of Us11 to bind dsRNA may be important for inhibiting activation of the cellular PKR kinase in response to dsRNA.
Double-stranded (ds) or highly structured RNAs can be potent signaling molecules in eukaryotic cells (reviewed in references 16, 27, and 56). Indeed, it is believed that a major component involved in triggering the cellular antiviral response in infected cells is the accumulation of viral dsRNA (reviewed in references 27 and 36). This large fraction of dsRNA is thought to originate either as a by-product of the replication of RNA viruses or from the transcription of large DNA virus genomes in which many open reading frames are positioned on opposite DNA strands. The accumulation of dsRNA activates a battery of cellular enzymes important for establishing the antiviral state. One of these enzymes is the cellular PKR kinase. Upon activation by dsRNA or protein effectors, PKR dimerizes, and each subunit of the dimer phosphorylates the other. The phosphorylated, activated enzymes then can phosphorylate other substrates in trans, notably IKB and the alpha subunit of the translation initiation factor eIF2 (reviewed in reference 25). As phosphorylated eIF2 is unable to promote the initiation of protein synthesis, PKR activation by dsRNA could potentially prevent the production of polypeptides necessary for viral replication and effectively arrest the viral life cycle prior to its completion, thus containing the infection. To disarm the cellular response to dsRNA, many viruses encode functions that inhibit PKR activation (reviewed in reference 36). Adenovirus synthesizes copious amounts of VA RNAs, one of which binds PKR and prevents activation of the enzyme (reviewed in reference 36). Other viruses, such as influenza virus, vaccinia virus, and reovirus, encode proteins that prevent PKR activation and bind dsRNA (reviewed in references 22, 48, and 49). The interaction of these viral polypeptides with dsRNA is thought to sequester the dsRNA activator, preventing PKR activation and eIF2α phosphorylation.
Recently, multiple herpes simplex virus (HSV) gene products have been demonstrated to regulate eIF2α phosphorylation (6, 20, 31, 32). The γ34.5 protein is an important virulence factor that binds the cellular PP1α and mediates the dephosphorylation of phospho-eIF2 (9, 20). This overcomes the effect of PKR activation on translation by maintaining pools of active, unphosphorylated eIF2α. In the absence of the viral γ34.5 protein, PKR is activated, phosphorylated eIF2α accumulates, and late viral protein synthesis is blocked, precluding the completion of the viral life cycle (8, 10). Genetic studies have demonstrated that expression of the Us11 protein, normally produced late in the viral life cycle, early in infection prevents the premature cessation of protein synthesis observed in cells infected with γ34.5 mutants (31, 32). Furthermore, biochemical analysis has shown that Us11 can prevent the activation of PKR (6, 32, 37). Us11 is found in nucleoli and the cytosol of infected cells and is also packaged in the viral particle (43). A basic 68-amino-acid fragment from the carboxy terminus of the 21-kDa Us11 polypeptide associates with 60S ribosomal subunits and possesses an RNA binding activity (43, 46). The only RNA targets known to interact with Us11 are an in vitro-transcribed RNA complementary to the Us11 open reading frame; the elements responsive to the transactivators Rev and Rex encoded by human immunodeficiency virus and human T-cell leukemia virus; 60S rRNA; a segment derived from a region common to the HSV type 1 (HSV-1) UL12, -13, and -14 mRNAs; and a truncated version of the UL34 mRNA (1, 12, 40-42). The determinants governing the recognition of this diverse assortment of sequences by the Us11 polypeptide have not yet been expounded. Importantly, the Us11 RNA binding domain is also required to prevent PKR activation, possibly implicating RNA binding in this process (37).
The carboxyl-terminal 68 amino acids of Us11 contain 21 copies of the triplet Arg-X-Pro, where X is preferentially an uncharged polar or acidic amino acid. This repetitive motif is thought to fold into a type II poly-l-prolyl alpha helix (43, 46). The arginine- and proline-rich Us11 RNA binding domain does not exhibit any homology to known RNA binding motifs, although alpha-helical regions in other RNA binding proteins are known to be important for RNA binding. To gain insight into the types of RNA molecules that Us11 could potentially interact with, we isolated high-affinity RNA ligands that bind to the Us11 RNA binding domain by employing an in vitro selection procedure and characterized the interaction of Us11 with a variety of RNA substrates. We conclude that Us11 is a double-stranded RNA binding protein that recognizes structured RNA through a novel arginine- and proline-rich RNA binding element.
MATERIALS AND METHODS
Selection of high-affinity RNA ligands that bind Us11.
An oligonucleotide library that contains an internal randomized stretch of 40 nucleotides flanked by a 5′ invariant region that contains the T7 promoter and a 3′ invariant region was a gift from Fenyong Liu (University of California—Berkeley) and is described in reference 51. The library was amplified (25 cycles of 94°C for 1 min, 47°C for 1 min, and 72°C for 1 min; on the final cycle, a 10-min extension was performed) with Taq DNA polymerase using the primers JH1031 and JH1052 (51). Following purification of the PCR product on a 2% agarose gel, an in vitro RNA synthesis reaction was assembled with T7 RNA polymerase (NEB) using buffer supplied by the manufacturer supplemented with 0.5 mM nucleoside triphosphates, 20 U of RNasin (Promega), and 30 μCi of [α32P]CTP (3,000 Ci/mmol). The reaction was incubated at 37°C for 6 h; 3 U of RNase-free DNase (Promega) was then added, and the incubation continued for an additional 30 min. Samples were sequentially extracted with an equal volume of phenol followed by chloroform and precipitated with ethanol (EtOH) in the presence of 2 M NH4OAc. After the pellets were washed with 70% EtOH, they were air dried and resuspended in RNase-free distilled H2O (dH2O). One nanomole of RNA was incubated with 20 nmol of purified glutathione S-transferase (GST) protein prebound to glutothione-Sepharose beads (Pharmacia) equilibrated in 1× binding buffer (20 mM HEPES-KOH [pH 7.4], 50 mM NaCl, 1 mM EDTA, 5% glycerol) at 30°C for 15 min. The samples were periodically manually agitated to ensure that the beads did not settle. After the beads were pelleted by brief centrifugation, RNA in the supernatant was recovered and incubated with 20 nmol of a purified protein that contains the Us11 RNA binding domain fused to GST (GST Δ1-87) prebound to glutathione-Sepharose beads equilibrated in 1× binding buffer. The reaction mixtures were incubated at 30°C for 15 min and washed five times with binding buffer. Following the final wash, the pellets were suspended in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM EDTA, 1% sodium dodecyl sulfate, and 300 μg of proteinase K (Boehringer)/ml at 30°C for 30 min and manually agitated periodically. The beads were removed by centrifugation, and the supernatant was sequentially extracted with phenol followed by chloroform. Glycogen was added as a carrier to a final concentration of 0.1 mg/ml, and the RNA was precipitated with the addition of 2.5 volumes of EtOH. Following a 70% EtOH wash, the pellet was suspended in RNase-free dH2O; 200 pmol of the JH1052 primer was added, and the RNA was heated to 95°C to remove any secondary structure and then quickly cooled in an ice water bath. Avian myeloblastosis virus reverse transcriptase (Promega) (10 to 20 U) was added along with buffer supplied by the manufacturer supplemented with 0.5 mM deoxynucleoside triphosphates and 20 U of RNasin. After 1 h at 42°C, the cDNA was amplified by PCR (25 cycles as described above) using JH1031 and JH1052. The PCR product was gel purified, and the procedure was repeated with varying amounts of RNA and Us11 fusion protein until there was no further increase in RNA binding activity as evaluated by an electrophoretic mobility shift assay (12 cycles in all). In the second iteration, 10 pmol of RNA was incubated with 200 pmol of protein. In the third iteration, 2 pmol of RNA was incubated with 40 pmol of protein. In the fourth through sixth iterations, 1 pmol of RNA was incubated with 2 pmol of protein. All subsequent iterations contained 1 pmol of RNA and 0.2 pmol of protein.
To subclone the final population of PCR products, blunt ends were created by treating the products with T4 DNA polymerase (NEB) according to the manufacturer's specifications. After the polymerase was heat inactivated, the PCR products were digested with an excess of HindIII for 3 h at 37°C. The large fragments were purified on agarose gels and ligated into pUC19 that had been previously digested with SmaI and HindIII, and the resulting plasmid molecules were transformed into Escherichia coli DH5α. DNA sequencing was performed with a Sequenase kit (U.S. Biochemicals) according to the manufacturer's instructions.
RNA binding assays.
The RNA binding reaction mixtures contained 50,000 cpm of the selected sequence in 1× binding buffer supplemented with 1 mM dithiothreitol, 100 μg of E. coli RNA (Ambion)/ml, and various quantities of purified GST or GST Δ1-87. The reaction mixtures were assembled on ice and subsequently incubated at 30°C for 10 min. In experiments with excess cold unlabeled competitor, protein was added last to the reaction mixtures. At the conclusion of the binding incubation, the reaction mixtures were loaded onto a 5% native polyacrylamide gel (19:1 acrylamide-bisacrylamide) that had been prerun for 1 h at room temperature in 0.5× Tris-borate-EDTA (TBE). Bromophenol blue was loaded in adjacent lanes to serve as a marker to monitor the progress of the electrophoresis. The gels were fixed in 10% methanol-10% acetic acid, dried on Whatman paper, and exposed to Kodak XAR film. The gels were also exposed in a PhosphorImager cassette (Molecular Dynamics), and the intensities of the bands were measured using the Imagequant software package. The reaction mixtures for nitrocellulose filter binding contained 1 mg of yeast RNA (Boehringer)/ml in place of the E. coli RNA but were otherwise prepared and incubated in an identical fashion. At the conclusion of the incubation, the reaction mixtures were applied to a manifold (Bio-Rad) containing a 0.45-μm-pore-size nitrocellulose sheet (Protran; Schleicher & Schuell) that had been preequilibrated in 1× binding buffer. Each sample was washed three times with 0.2 ml of 1× binding buffer. The filters were air dried and exposed to Kodak XAR film. Radioactivity present in the excised spots was quantified by Cerenkov counting.
Preparation of RNA binding substrates.
Plasmid clones containing selected inserts were linearized with KpnI and subsequently treated with 100 μg of proteinase K/ml in 0.5% sodium dodecyl sulfate-1 mM EDTA for 30 min at 50°C. Samples were extracted twice with phenol and once with chloroform, precipitated with EtOH, and resuspended in RNase-free H2O. Ten micrograms of DNA was incubated with 50 U of T7 RNA polymerase (NEB) in buffer supplied by the manufacturer supplemented with 20 U of RNasin and 0.5 mM ATP, UTP, and GTP. The reaction mixtures also contained 0.012 mM CTP and 25 μCi of [α-32P]CTP (3,000 Ci/mmol). After 1 h at 37°C, RNase-free DNase (Promega) was added and the incubation continued for an additional 30 min. The reaction was extracted with phenol followed by chloroform and then precipitated with EtOH in the presence of NH4OAc. After the pellet was washed with 70% EtOH, the RNA was resuspended in RNase-free dH2O.
To isolate RNA conformational isoforms for RNA binding studies, radiolabeled RNA synthesis reaction mixtures were fractionated on 5% native polyacrylamide gels, and the different bands were excised. After overnight elution at 4°C in 2 M NH4OAc, the RNA was precipitated with EtOH, the pellet was washed with 70% EtOH, and the RNA was resuspended in RNase-free H2O.
Large quantities of unlabeled RNA for competition experiments were synthesized using a T7 Megashort script kit (Ambion) according to the manufacturer's instructions. To remove potential dsRNA contaminants from labeled or unlabeled RNA preparations, a modified version of the protocol described by Pe'ery and Mathews (35) was employed. The RNA was first gel purified on an 8% polyacrylamide-7 M urea gel run in 0.5× TBE. Cold RNA was visualized under UV light following staining with ethidium bromide. The excised band was then layered onto a 10% native gel run in 0.5× TBE. The band removed from this nondenaturing gel was eluted overnight at 4°C in 10 mM Tris-HCl (pH 7.6)-1 mM EDTA. The RNA was concentrated by extraction with n-butanol, extracted with phenol followed by chloroform, precipitated with EtOH, and resuspended in RNase-free H2O.
To prepare labeled dsRNAs of different lengths, the plasmid pBSK II(+) was digested with either EagI, BamHI, EcoRI, HindIII, SalI, XhoI, HaeIII, or KpnI, and RNA products were synthesized in vitro with T7 RNA polymerase in the presence of [α-32P]CTP. Each RNA preparation was subsequently annealed to a 245-nucleotide RNA synthesized by T3 RNA polymerase from PvuII-digested pBSK II(+). Single-stranded regions were removed by treatment with RNase A and RNase T1, and the RNA duplexes were purified in nondenaturing polyacrylamide gels as described previously (29). Each filter binding reaction contained 5,000 cpm of labeled substrate. For competition experiments, an 81-bp dsDNA derived from the same sequence as the 81-bp RNA duplex was synthesized by PCR and gel purified.
Poly(I · C) was 5′-end labeled with T4 polynucleotide kinase (NEB) according to the manufacturer's instructions. Unincorporated nucleotides were removed on a Biogel P30 spin column (Bio-Rad). Each filter binding reaction contained 50,000 cpm. All nucleotide polymers for RNA binding and competition experiments were from Pharmacia.
RESULTS
Isolation of high-affinity RNA ligands that bind to Us11.
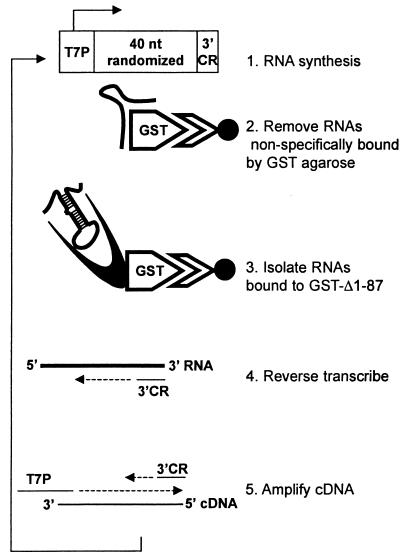
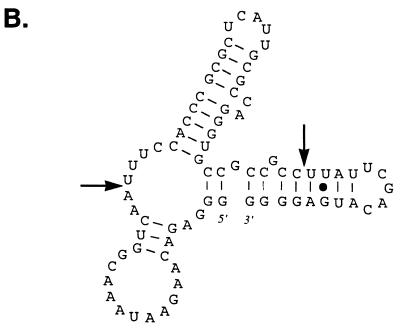
To identify RNA molecules capable of binding with high affinity to the Us11 protein, we employed an in vitro selection technique that has been used previously to successfully isolate RNA ligands that bind to a variety of relatively simple and complex molecular structures (2, 4, 11, 13, 45, 51, 55). This methodology allowed us to amplify specific RNA ligands from a combinatorial library consisting of molecules with a randomized central region. Our strategy is outlined in Fig. 1. Briefly, a library of in vitro-synthesized RNA containing a 40-nucleotide randomized internal region was first incubated with purified GST bound to glutathione-agarose beads to remove molecules that nonspecifically interact with this polypeptide and the supporting solid matrix. The unbound RNAs were subsequently mixed with a purified hybrid protein comprising the carboxyl-terminal 87 amino acids of Us11 fused to GST (GST Δ1-87). This segment of Us11 contains a proline- and arginine-rich segment known to bind RNA and inhibit activation of the cellular PKR kinase. RNA molecules retained in this complex were collected on glutathione-agarose beads and comprised the first cycle of enriched RNA ligands that bound to the Us11 RNA binding domain. These RNAs were then reverse transcribed, the population was amplified by PCR, and the process was repeated. After 12 iterations of this procedure, individual clones were isolated and sequenced from the PCR-amplified population. Sequence analysis of 300 clones resulted in the isolation of nine unique sequences within the randomized component (Fig. 2A). Although no substantial primary sequence homology was detected in the selected clones, their predicted secondary structures contained some common features. Multiple short duplexed segments were observed, along with sequences predicted to remain single stranded. Notable among these was a stem-loop region followed by a CA or CCA nucleotide bulge present in the selected sequences in eight of nine different clones (Fig. 2B).
FIG. 1.
Selection scheme for isolating RNA ligands that associate with the Us11 RNA binding domain. An oligonucleotide pool that contains a randomized central region situated between a T7 promoter (T7P) and a 3′ constant region (3′ CR) was amplified with specific primers. Labeled RNA synthesized from this population was first incubated with purified GST protein immobilized on Sepharose beads to remove RNA molecules that interact nonspecifically with GST. Unbound RNA was then incubated with a purified protein that contains the Us11 RNA binding domain fused to GST (GST Δ1-87). The bound RNA was recovered and reverse transcribed with a primer homologous to the 3′ CR, and the cDNA was amplified with a primer designed to regenerate an intact T7P sequence along with the 3′ CR primer. This process was repeated multiple times to enrich for RNA molecules that preferentially associate with Us11. nt, nucleotide.
FIG. 2.
Sequence alignment of selected RNA ligands that bind to the Us11 RNA binding domain. (A) Sequences corresponding to the 5′ invariant region primer (left) and the 3′ constant region primer (right) are boxed. (B) RNA secondary structure for L12-262 predicted using the m-fold algorithm (30, 58). The selected segment, located between the two arrows, folds into a stem-loop structure followed by a CA bulge.
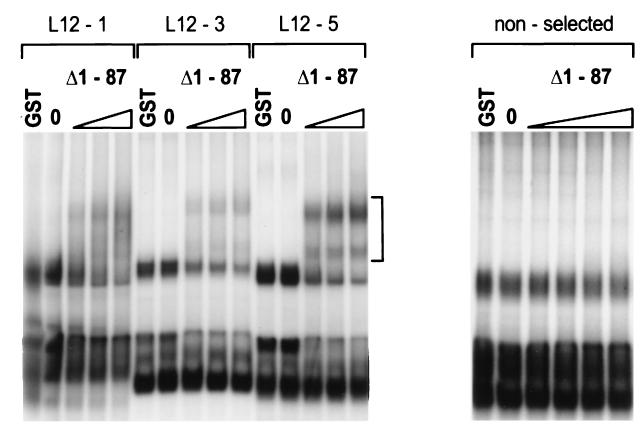
The ability of RNA produced from these clones to complex with the Us11 RNA binding domain was evaluated in an electrophoretic mobility shift assay. Three representative examples are presented in Fig. 3. In the absence of exogenous protein, all of the selected RNAs migrate as multiple bands in native polyacrylamide gels, suggestive of alternative RNA conformations. This pattern is unaltered by the addition of 1 μg of purified GST. Addition of as little as 30 ng of purified GST Δ1-87 to the binding reaction mixture results in a decrease in the amount of certain unbound conformers and the appearance of retarded complexes that represent bound ligand. The intensities of these retarded complexes are greatest in reactions programmed with L12-5 RNA, intermediate in reactions containing L12-1 RNA, and weakest in the group represented by L12-3. It is difficult to reconcile these variations with distinct binding affinities, as these clones must bind with sufficient avidity to survive 12 iterations of the selection procedure. The observed differences in the extent of complex formation could be attributed to variability in the formation of active conformational isoforms capable of binding Us11 among the different ligands. RNA prepared from molecules in the initial unselected population did not interact with Us11 to the same extent (Fig. 3 and data not shown). The levels of binding of GST Δ1-87 to the selected RNA sequences in the absence and presence (0.1 to 5 mM) of MgCl2 were indistinguishable. In addition, the pattern of RNA conformational isoforms observed following electrophoresis in a nondenaturing polyacrylamide gel was unaltered in reactions assembled either in the presence or the absence of Mg2+ (not shown). This is consistent with reports demonstrating that Mg2+ is not required for simple Watson-Crick pairing (5, 54) but can contribute to some tertiary interactions (18). Finally, purified Us11 containing an amino-terminal His tag also bound to the selected sequences, ruling out the possibility that the GST segment of the fusion protein is required for binding to the selected ligands (not shown).
FIG. 3.
Binding of Us11 to selected RNA ligands. Labeled RNA prepared from a representative member of either the selected (L12-5, L12-1, and L12-3) or the initial unselected population was incubated without any protein (0), with 1 μg of GST, or with increasing amounts of purified GST Δ1-87 (left to right, 30, 100, and 300 ng; nonselected RNA, 10, 30, 100, and 300 ng). Protein-RNA complexes were resolved by electrophoresis in native polyacrylamide gels, and the fixed dried gel was exposed to Kodak XAR film. The vertical bracket indicates the region containing protein-RNA complexes.
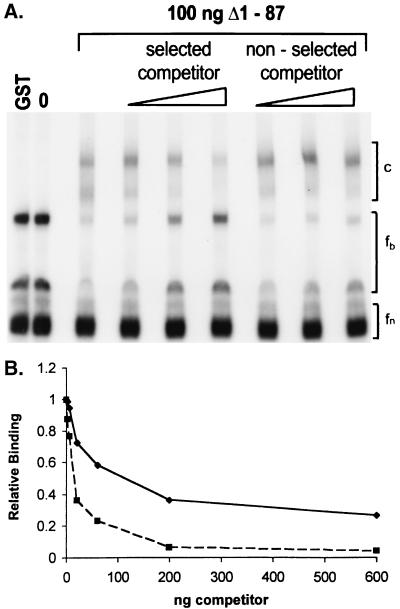
To further address the specificity of ligand binding, competition experiments were performed with a labeled RNA ligand at a single concentration of GST Δ1-87 protein in the presence of excess unlabeled specific or nonspecific challenger RNA molecules. This RNA was purified through a series of native and denaturing gels to remove any potential small double-stranded RNA contaminants that might affect RNA binding (35). Figure 4A demonstrates that the addition of increasing amounts of unlabeled L12-5 RNA substantially reduces the amount of bound retarded complex while simultaneously increasing the amount of some of the unbound material (fb). Quantifying these data on a PhosphorImager revealed an 80% decrease in the bound complex at 100 pmol of L12-5 competitor along with a 2.5-fold increase in the unbound form. A fraction of the unbound RNA (fn) is apparently unable to bind protein and is not in rapid equilibrium with the unbound form, which can produce protein-RNA complexes. Including 100 pmol of unlabeled RNA isolated from a member of the initial unselected population, prepared in an identical fashion, resulted in only a 28% decrease in the amount of RNA bound by the Us11 protein. Quantitative data were obtained by using a nitrocellulose filter binding assay along with highly purified RNA templates. Incubation of the labeled high-affinity L12-5 RNA ligand with increasing amounts of purified GST Δ1-87 leads to the retention of greater quantities of radioactivity on the nitrocellulose filter (not shown). At a fixed concentration of GST Δ1-87 protein, addition of increasing amounts of the unlabeled specific L12-5 competitor is more effective than identical quantities of a nonspecific competitor at reducing binding to the labeled probe (Fig. 4B). Furthermore, the concentration of L12-5 ligand required to achieve a 50% reduction in binding to the labeled probe is approximately 10-fold less than the concentration of nonspecific competitor necessary to generate an equivalent reduction in binding. These results demonstrate that the RNA ligands isolated by the in vitro selection procedure bind specifically and with high affinity to the Us11 RNA binding domain.
FIG. 4.
Selected RNA ligands preferentially associate with US11. (A) Purified GST Δ1-87 (100 ng) was incubated with labeled L12-5 RNA in the presence and absence of increasing amounts of unlabeled L12-5 RNA or RNA from the unselected initial population (left to right, 10, 30, and 100 pmol). Labeled L12-5 RNAs incubated without any protein (0) and with 1 μg of purified GST are also shown. Protein-RNA complexes were resolved as described in the legend to Fig. 2. GST Δ1-87 RNA-protein complexes migrate in the bracketed region labeled c. Free, unbound isoforms capable of binding Us11 are within the bracketed region labeled fb, while nonbinding isoforms are labeled fn. (B) Purified GST Δ1-87 (10 ng) was incubated with labeled L12-5 RNA in the presence or absence of increasing amounts of specific (L12-5 [▪] or nonspecific (RNA from initial unselected population [♦]) competitor. The amount of radioactivity in each sample retained on a nitrocellulose filter was quantified. RNA binding in the absence of competitor was normalized to 1.0.
Preferential association of Us11 with specific RNA conformational isoforms.
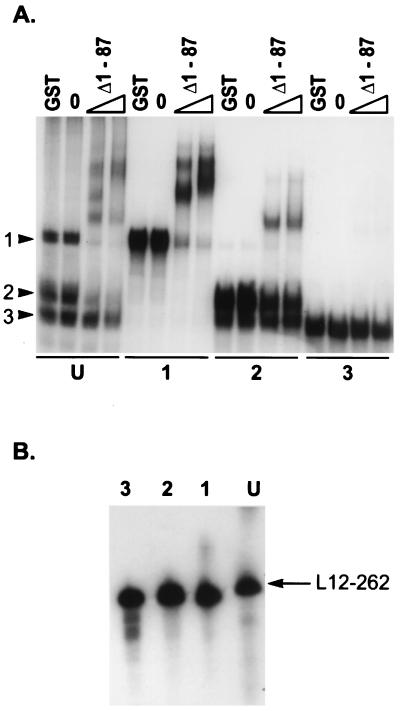
All of the RNA molecules enriched for by the in vitro selection procedure migrate as multiple species on native polyacrylamide gels (Fig. 5A). Furthermore, addition of the Us11 RNA binding protein to the reaction mixtures results in the appearance of a defined set of multiple retarded bands following electrophoresis in native polyacrylamide gels. These bands could represent the assembly of multimeric protein structures on the RNA ligands or characteristic mobility shifts of Us11 associating with a specific RNA conformational isoform. To ascertain the nature of each species and to analyze the relative ability of each species to bind to Us11, three unbound RNA species were purified on native polyacrylamide gels and designated class 1, class 2, or class 3. All three of these purified species comigrated on denaturing polyacrylamide gels, indicating that they may indeed represent different folded conformational isoforms of a single primary sequence of defined length (Fig. 5B). In addition, the gel-purified species 1 and 3 each retained its characteristic mobility when subjected to subsequent electrophoresis on a native polyacrylamide gel, indicating that these structures are stable and do not regenerate a heterogenous population of conformational isoforms following purification (Fig. 5A). While the class 2 preparation does not contain significant amounts of the class 1 isoform, some class 3 RNA is present. Due to the proximity of the class 2 and 3 bands in native gels, we are unable to determine whether the class 2 preparation is contaminated with some class 3 molecules or whether some of the purified class 2 RNA is capable of adopting the mobility characteristic of the class 3 isoform. Figure 5A demonstrates that 100 and 300 ng of purified GST Δ1-87 generates a heterogenous mixture of several retarded complexes following incubation with the unfractionated L12-262 ligand. Interestingly, only the class 1 and class 2 preparations exhibit significant binding to the Us11 fusion protein. The class 3 conformational isoform fails to demonstrate any detectable binding, suggesting that the class 2 RNA is responsible for the binding activity observed in the class 2 preparation (Fig. 5A). The retarded complexes generated by purified class 1 isoforms migrate more slowly than the retarded complexes observed with class 2 RNA. In addition, the complexes observed in the class 1 and class 2 binding reactions appear to account for all of the retarded complexes observed in the unfractionated starting material. There are, however, multiple retarded bands visible in the class 1 and class 2 binding reactions, consistent with the possible assembly of Us11 multimers on these templates or with a Us11-induced structural change in the RNA ligand. Similar results have been obtained with the other selected RNAs (data not shown). Together, these observations suggest that, in addition to defined sequence determinants, there are significant conformational features of the RNA ligand that are important for recognition by the Us11 RNA binding domain.
FIG. 5.
Association of Us11 with RNA conformational isoforms. (A) Labeled RNA (L12-262) was fractionated by electrophoresis in native polyacrylamide gels, and three species (class 1, class 2, and class 3) were isolated. Each gel-purified RNA species, along with the unfractionated starting material (U), was incubated in the absence of added protein (0) or in the presence of either 1 μg of purified GST, 100 ng of GST Δ1-87, or 300 ng of GST Δ1-87. Protein-RNA complexes were resolved as described in the legend to Fig. 2. The arrowheads numbered 1, 2, and 3 to the left refer to the mobilities of the three isoforms. The numbers below the panel refer to the gel-purified isoform used in the binding reaction, and U represents the unfractionated starting material. (B) Each purified RNA species, along with the unfractionated starting material, was subjected to electrophoresis in a denaturing polyacrylamide gel. The fixed dried gel was subsequently exposed to Kodak XAR film.
The Us11 RNA binding domain recognizes double-stranded RNA.
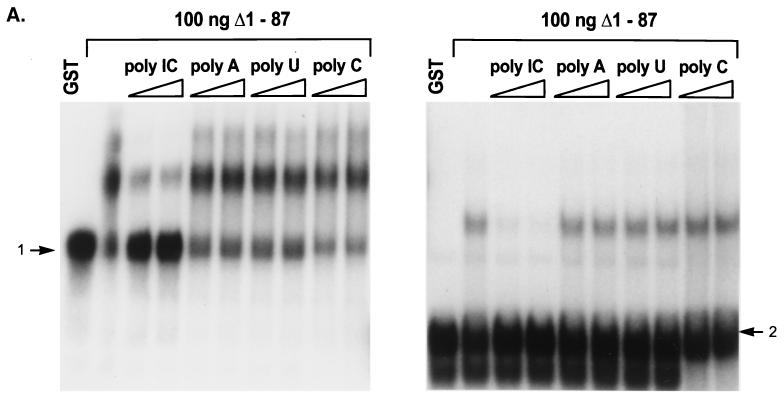
To address the role of potential structural and sequence elements in RNA recognition by the Us11 RNA binding domain, we evaluated the abilities of a variety of synthetic RNA polymers to compete with the selected ligand for binding. In the experiment presented in Fig. 6A, Us11 binding to the gel-purified, radiolabeled conformational isoforms class 1 and class 2 was examined in the presence of two concentrations of poly(C), poly(U), poly(A), or poly(I · C). In all cases, significant effects on RNA ligand binding were observed only in the presence of poly(I · C), a synthetic double-stranded RNA molecule. Poly(C), a single-stranded homopolymer containing a nucleotide component of poly(I · C), along with poly(U) and poly(A), had no measurable effect on RNA binding. In a nitrocellulose filter binding assay, unlabeled poly(I · C) was at least as effective as unlabeled L12-5 in reducing binding to the labeled L12-5 probe (Fig. 6B). The simplest explanation of this observation is that Us11 has a high affinity for double-stranded RNA.
FIG. 6.
Recognition of selected RNA sequences by Us11 in the presence of defined single-stranded and double-stranded RNA polymers. (A) Two species of labeled selected RNA L12-262 (left, class 1; right, class 2) were incubated with 100 ng of GST Δ1-87 in the absence or presence of two amounts of either poly(A), poly(U), poly(C), or poly(I · C) (from left to right, 33 and 100 μg/ml). Protein-RNA complexes were resolved as described in the legend to Fig. 2. Each RNA species was also incubated with 1 μg of GST. Unbound material from each class is indicated with an arrow. (B) Purified GST Δ1-87 (10 ng) was incubated with labeled L12-5 in the presence of increasing amounts of unlabeled poly(I · C) (▴) or L12-5 (▪). The amount of radioactivity retained on a nitrocellulose filter for each sample was quantified. RNA binding in the absence of competitor was normalized to 1.0.
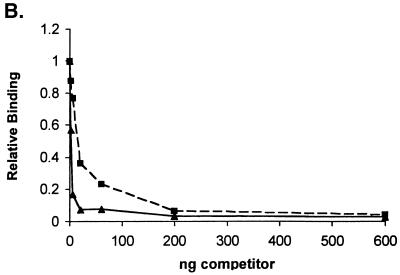
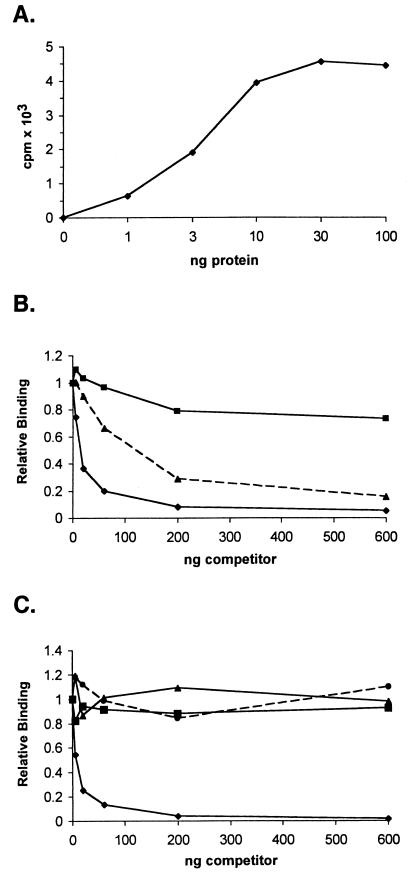
To directly examine the ability of Us11 to bind double-stranded RNA, radiolabeled poly(I · C) was utilized as a substrate in a nitrocellulose filter binding assay. Following incubation of labeled poly(I · C) with purified GST Δ1-87, the binding reaction mixtures were filtered through nitrocellulose on a manifold, and the amount of radioactivity retained on the filter was quantified by liquid scintillation counting. As little as 1 ng of purified GST Δ1-87 bound measurable amounts of poly(I · C) (Fig. 7A). The binding to radiolabeled poly(I · C) was effectively competed by increasing amounts of unlabeled poly(I · C) (Fig. 7B). Importantly, highly purified, unlabeled L12-5 selected ligand was also a potent competitor of binding to poly(I · C), while a representative RNA from the initial unselected population was only slightly active as a competitor (Fig. 7B). The unlabeled single-stranded RNA homopolymers poly(C), poly(U), and poly(A) were not effective competitors when Us11 binding to labeled poly(I · C) was evaluated by nitrocellulose filter binding (Fig. 7C). This directly demonstrates that Us11 is a double-stranded RNA binding protein and that the RNA ligand isolated by in vitro selection is a potent competitor in an assay that measures binding to double-stranded RNA.
FIG. 7.
Us11 RNA binding domain directly binds double-stranded RNA. (A) Labeled poly(I · C) was incubated with increasing amounts of purified GST Δ1-87, and the amount of radioactivity retained on a nitrocellulose filter for each sample was determined. (B) Ten nanograms of GST Δ1-87 was incubated in the presence of labeled poly(I · C) in the absence of competitor or with increasing amounts of an unlabeled RNA selected to bind Us11 (L12-5 [▴]), an unlabeled RNA from the initial unselected population (▪), or unlabeled poly(I · C) (♦). The amount of radioactivity retained on a nitrocellulose filter for each sample was determined. RNA binding in the absence of competitor was normalized to 1.0. (C) Binding to labeled poly(I · C) was as described for panel B with the modification that the reaction mixtures contained increasing amounts of either unlabeled poly(A) (▪), poly(C) (•), poly(U) (▴), or poly(I · C) (♦).
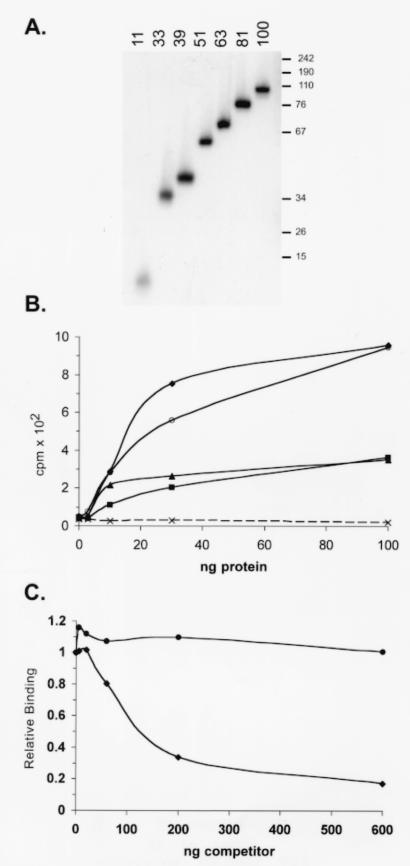
As poly(I · C) preparations consist of two complementary homopolymers and vary significantly in length, we next evaluated the ability of Us11 to bind dsRNA molecules of defined lengths that possess greater sequence complexity. Labeled dsRNA molecules derived from the E. coli cloning vector pBS and ranging in size from 11 to 100 bp were prepared in vitro and incubated with increasing amounts of purified GST Δ1-87 (Fig. 8A). RNA duplexes of 81 bp and above bound GST Δ1-87 with similar efficiencies, while binding to smaller RNA duplexes was reduced (Fig. 8B). Importantly, binding to RNA duplexes of 39 bp or shorter was not detected, demonstrating that the length of a perfectly duplexed RNA target can have a substantial impact on binding to Us11. Purified full-length Us11 that contained an amino-terminal tag composed of six histidine residues also bound efficiently to defined dsRNA molecules, exhibiting an apparent Kd of approximately 12.6 nM for the 81-bp substrate in a filter binding assay (not shown).
FIG. 8.
The Us11 RNA binding domain exhibits length-dependent binding to dsRNA and prefers RNA duplexes to DNA duplexes. (A) Labeled RNA duplexes of defined lengths were prepared as described in Materials and Methods. A sample of each gel-purified dsRNA was resolved by electrophoresis in a nondenaturing polyacrylamide gel. An exposure of the fixed, dried gel is shown. The numbers on the top refer to the length in base pairs of the purified RNA duplex. The numbers to the right indicate the migration of dsDNA markers. (B) Labeled dsRNA of defined length was incubated with increasing amounts of purified GST Δ1-87 and subsequently filtered through a nitrocellulose membrane. Radioactivity retained on the membrane was quantified. ♦, 100-bp dsRNA; ○, 81-bp dsRNA; ▴, 63-bp dsRNA; ▪, 51-bp dsRNA; (×) 39-, 33-, or 11-bp dsRNA. (C) Ten nanograms of GST Δ1-87 was incubated with a labeled 81-bp dsRNA substrate in the presence of increasing amounts of unlabeled dsRNA (♦) or dsDNA (•). Both nucleic acid duplexes were 81 bp in length and were derived from the same sequence.
To determine if Us11 binding to a nucleic acid duplex was specific for RNA, the binding of GST Δ1-87 to an 81-bp dsDNA fragment and its binding to an 81-bp dsRNA duplex with an identical sequence were compared. Increasing amounts of unlabeled dsRNA or dsDNA were added to reaction mixtures that contained a fixed amount of radiolabeled dsRNA. Following the addition of GST Δ1-87, the reaction mixtures were incubated for 10 min at 30°C, RNA-protein complexes were collected on nitrocellulose filters, and the fractions of radioactivity retained on the filters were quantified by counting in liquid scintillant. Figure 8C demonstrates that unlabeled dsRNA is significantly more effective than dsDNA at reducing the binding of GST Δ1-87 to the labeled dsRNA probe. This demonstrates that the Us11 RNA binding domain preferentially binds dsRNA duplexes of 51 bp or longer and can effectively discriminate between dsRNA and dsDNA.
DISCUSSION
Us11 binds RNA through a novel 68-amino-acid domain that contains a repetitive arginine- and proline-rich motif (43, 46). As the Us11 RNA binding domain is not homologous to any known RNA recognition motif and the few RNAs known to interact with Us11 are unrelated at the level of primary sequence, we proceeded to characterize features of RNA molecules that associate with Us11. After isolating RNA ligands that bound Us11 from a combinatorial library consisting of molecules that contained a randomized segment, we demonstrated that Us11 bound specifically to these selected molecules and preferentially associated with discrete conformational isoforms. While the addition of unlabeled poly(I · C) reduced the binding of Us11 to a radiolabeled selected RNA, single-stranded homopolymers were not effective competitors. Our study establishes that Us11 binds directly to dsRNA polynucleotides and stably associates with perfect RNA duplexes of 51 bp or longer; furthermore, Us11 preferentially associates with nucleic acid duplexes that contain 2′ hydroxyl groups, since dsDNA duplexes of identical sequence and length do not reduce binding to dsRNA. This is the first demonstration that the Us11 RNA binding domain can recognize dsRNA.
While earlier studies reported that the Us11 polypeptide associates with a synthetic RNA probe complementary to the 5′ end of the Us11 mRNA and an internal segment of the Us12 mRNA, they did not adequately address the RNA determinants involved in nucleic acid recognition (40). Thus, their observed reduction in Us11 binding to RNA probes with 80-nucleotide deletions at their 3′ ends may not reflect binding to a specific site or sequence but rather the global alteration of an RNA structure required for binding that involves sequences from disparate parts of the molecule. Similarly, the changes in the distribution of RNA conformational isoforms they described does not establish that Us11 is a conformation-specific RNA binding protein, as the experiments were all performed with crude extracts prepared from HSV-1-infected cells. Other activities in the extract in conjunction with Us11 could be responsible for modifying the structure of the RNA probe. Finally, the ability of Us11 to associate with the different RNA conformational isoforms was never examined, as the conformers were in rapid equilibrium and could not be isolated. Our study, in contrast, was performed with both purified protein and RNA conformational isoforms; furthermore, we have established that Us11 associates with dsRNA and structured RNA as opposed to single-stranded polynucleotides or dsDNA.
Numerous proteins that bind dsRNA contain a 65- to 75-amino-acid motif (dsRNA binding motif [dsRBM]) that either folds into or is predicted to adopt a tertiary structure where two alpha helices flank a three-stranded antiparallel beta sheet (α-β-β-β-α). This domain comprises a dsRNA recognition motif (reviewed in reference 15). Multiple copies of this motif are found in many dsRNA binding proteins, although a single copy, as in the case of the vaccinia virus E3L protein, is sufficient. Recognition of dsRNA, however, is not limited to proteins that contain the dsRBM structure. Alternative dsRNA binding structures that have no homology to each other or to the dsRBM have been observed in several viral proteins. Multiple basic residues that do not appear to fold into an independent domain or assume a structure on their own are distributed along the side of the sigma 3 polypeptide encoded by reovirus. Recent structural studies propose that sigma 3 binds to RNA as a dimer via a positively charged surface patch on one side of the structure. This charged region is formed by the association of sigma 3 monomers and spans the interface between the two subunits (34). Binding to dsRNA by the influenza virus NS-1 protein also involves dimerization. In this case, structural studies revealed that the dimer contains six helices that contribute to multimerization and RNA binding (52). Although the NS1 protein lacks a canonical dsRBM, it can also associate with a cellular protein, Staufen, that contains this motif (14). In addition to its dsRNA binding activity, NS-1 also recognizes poly(A) stretches and U6 snRNA (19, 38, 39). We have now established that the RNA binding domain in the HSV-1 Us11 protein constitutes yet another class of elements that can bind dsRNA. While the structure of the Us11 RNA binding domain has not been determined, it is predicted to adopt the conformation of a type 2 polyprolyl alpha helix built up of a series of repeats of the amino acid triplet Arg-X-Pro (43, 46).
Copious amounts of Us11 are synthesized at late times in the infectious cycle. HSV-1-infected cells have been demonstrated to contain a large amount of dsRNA, and dsRNA is a potent activator of cellular enzymes poised to execute an antiviral response (23). The dsRNA binding domain of Us11 may therefore function stoichiometrically to sequester viral dsRNA and prevent it from engaging cellular dsRNA binding proteins such as PKR. Along these lines, the minimum lengths for RNA duplexes to efficiently activate PKR and to bind to Us11 are strikingly similar (29). RNA-dependent and -independent interactions involving Us11 and PKR may also reduce levels of activated PKR (reference 7 and unpublished observations). A related mechanism invoking both dsRNA binding and a physical association with PKR has been proposed for the vaccinia virus dsRNA binding protein, E3L (44).
Although dsRNA is thought to accumulate in HSV-1-infected cells, the activities of the Us11 protein are not essential for viral replication in cultured cells or pathogenesis in mice, since Us11 is one of two known HSV-1 proteins capable of modulating the cellular antiviral response (21, 28, 33). Should viral dsRNA activate the cellular PKR kinase and lead to the accumulation of phosphorylated eIF2α, the viral γ34.5 gene product, in association with the cellular protein phosphatase 1α, can mediate the dephosphorylation of phospho-eIF2α and prevent the premature cessation of protein synthesis that would otherwise occur (20). Us11 is also packaged in the virus particle and therefore is delivered into the cell at times that precede the expression of viral immediate-early genes (42). While the role of Us11 in the virion is not understood, RNAs have not been found packaged in a Us11-dependent manner (47). The smaller quantities of Us11 delivered by the virus particles could suffice to sequester the reduced amounts of dsRNA formed early in the infection. In addition, Us11 has been found in nucleoli and associated with ribosomes. As PKR has also been detected in polysomes and nucleoli, similar subcellular localization of Us11 may be important in preventing PKR activation (24, 53, 57).
The selected RNA ligands that bound Us11 all formed a series of conformational isoforms. Several of these isoforms were stable upon gel purification, and Us11 binding activity was limited to those isoforms that possessed increased structure. We do not know the precise nature of these putative structural determinants, as they may be either intramolecular, intermolecular, or a combination of these alternatives. While Us11 prefers to bind perfect RNA duplexes of 51 bp or longer, the selected RNA molecules are predicted to contain duplex regions substantially shorter than 51 bp. Yet the selected RNAs associate with Us11, and a 39-bp dsRNA perfect duplex does not. As both of these templates are composed of approximately 80 nucleotides, this suggests that the selected RNAs contain binding determinants that are distinct from perfectly base paired regions. These determinants could depend solely on structure, primary sequence, or a combination of both. Along these lines, eight of the nine selected sequences are predicted to contain a CA or CCA bulge in the stem of a dsRNA segment, and this element could conceivably be important for binding to Us11. All of the RNAs reported by others to associate with Us11 are quite large, ranging from 200 to 600 nucleotides in length (1, 12, 40, 41, 43, 46). While these studies convincingly demonstrate the binding of Us11 to specific RNA molecules, they fall short of establishing sequence-specific binding. In fact, the interaction of Us11 with RNA observed in published reports could be explained either entirely or in part by recognition of RNA structure. Only the Rev- and Rex-responsive elements, both of which bind Us11, have been determined to contain extensive regions of secondary structure, although the involvement of their structured segments in binding to Us11 has not been explored (3, 17, 26, 50). The remaining lengthy RNA molecules reported to bind Us11, some of which have a high GC content characteristic of herpesvirus mRNAs, would invariably possess a large degree of predicted secondary structure, making it difficult to distinguish recognition of structural determinants from recognition of sequence determinants. Indeed, the relative contributions of sequence and structure are not evident and are worthy of future investigation. Is there, for example, a role for specific RNA sequence recognition by Us11, or is the role of primary sequence simply to create a secondary structure that is effectively recognized by the Us11 RNA binding domain? Similarly, although simple RNA homopolymeric sequences did not compete effectively for Us11 in binding experiments with labeled poly(I · C), this does not conclusively prove that Us11 could not recognize discreet unstructured RNA molecules of greater sequence complexity. Our isolation of a small, defined RNA sequence that binds with high affinity to Us11 will make it possible to approach these questions experimentally. Finally, Us11 might execute several different functions, depending on the nature of its RNA ligand. Complexes with dsRNA may function to prevent PKR activation, while other unidentified sequence-specific targets may be important for additional activities that remain to be discovered.
Acknowledgments
We are grateful to Fenyong Liu for generously providing the combinatorial library. We thank Lisa Parker and Tsafi Pe'ery for advice concerning the RNA binding procedures, Joel Belasco and Matt Mulvey for their critical comments on the manuscript, and Joel Belasco for numerous discussions concerning RNA structure.
This work was supported by a grant from the NIH to I.M.
REFERENCES
- 1.Attrill, H. L., S. A. Cumming, J. B. Clements, and S. V. Graham. 2002. The herpes simplex virus type 1 US11 protein binds the coterminal UL12, UL13, and UL14 RNAs and regulates UL13 expression in vivo. J. Virol. 76:8090-8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, D. P., M. L. Zapp, M. R. Green, and J. W. Szostak. 1991. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell 67:529-536. [DOI] [PubMed] [Google Scholar]
- 3.Baskerville, S., M. Zapp, and A. D. Ellington. 1995. High-resolution mapping of the human T-cell leukemia virus type 1 Rex-binding element by in vitro selection. J. Virol. 69:7559-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D., J. Brown, C. Kang, L. Gold, and P. Allen. 1997. Single-stranded RNA recognition by the bacteriophage T4 translational repressor, regA. J. Biol. Chem. 272:14969-14974. [DOI] [PubMed] [Google Scholar]
- 5.Bukard, M. E., T. Xia, and D. H. Turner. 2001. Thermodynamics of RNA internal loops with a guanosine-guanosine pair adjacent to another noncanonical pair. Biochemistry 40:2478-2483. [DOI] [PubMed] [Google Scholar]
- 6.Cassady, K., M. Gross, and B. Roizman. 1998. The herpes simplex virus Us11 protein effectively compensates for the gamma1(34.5) gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassady, K. A., and M. Gross. 2002. The herpes simplex virus type1. Us11 protein interacts with protein kinase R in infected cells and requires a 30 amino acid sequence adjacent to a kinase substrate domain. J. Virol. 76:2029-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou, J., and B. Roizman. 1992. The γ34.5 gene of Herpes Simplex Virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 1 34.5, a gene non-essential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 10.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of a M(r) 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with gamma 1 34.5-mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrad, R. C., L. Giver, Y. Tian, and A. D. Ellington. 1996. In vitro selection of nucleic acid aptamers that bind proteins. Methods Enzymol. 267:336-367. [DOI] [PubMed] [Google Scholar]
- 12.Diaz, J. J., M. D. Dodon, N. Schaerer-Uthurralt, D. Simonin, K. Kindbeiter, L. Gazzolo, and J. J. Madjar. 1996. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature (London) 379:273-277. [DOI] [PubMed] [Google Scholar]
- 13.Doudna, J. A., T. R. Cech, and B. A. Sullenger. 1995. Selection of an RNA molecule that mimics a major autoantigenic epitope of the human insulin receptor. Proc. Natl. Acad. Sci. USA 92:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falcon, A. M., P. Fortes, R. M. Marion, A. Beloso, and J. Ortin. 1999. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 27:2241-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fierro-Monti, I., and M. B. Mathews. 2000. Proteins binding to duplex RNA: one motif, multiple functions. Trends Biochem. Sci. 25:241-246. [DOI] [PubMed] [Google Scholar]
- 16.Gerber, A. P., and W. Keller. 2001. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci. 26:376-384. [DOI] [PubMed] [Google Scholar]
- 17.Gosser, Y., T. Hermann, A. Majumdar, W. Hu, R. Frederick, F. Jiang, W. Xu, and D. J. Patel. 2001. Peptide-triggered conformational switch in HIV-1 RRE RNA complexes. Nat. Struct. Biol. 8:146-150. [DOI] [PubMed] [Google Scholar]
- 18.Hanna, R., and J. Doudna. 2000. Metal ions in ribozyme folding and catalysis. Curr. Opin. Chem. Biol. 4:166-170. [DOI] [PubMed] [Google Scholar]
- 19.Hatada, E., and R. Fukada. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 73:3325-3329. [DOI] [PubMed] [Google Scholar]
- 20.He, B., M. Gross, and B. Roizman. 1997. The gamma (1) 34.5 protein of herpes simplex virus complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi, K., R. Fawl, R. J. Roller, and B. Roizman. 1993. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J. Virol. 67:2123-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs, B. L. 2000. Translational control in poxvirus-infected cells, p. 951-972. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Jacquemont, B., and B. Roizman. 1975. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J. Virol. 15:707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jimenez-Garcia, L. F., S. R. Green, M. B. Mathews, and D. L. Spector. 1993. Organization of the double-stranded RNA activated protein kinase DAI and virus associated VA RNAI in adenovirus-2 infected HeLa cells. J. Cell Sci. 106:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman, R. J. 2000. Double-stranded RNA-activated protein kinase PKR, p. 503-528. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Kjems, J., M. Brown, D. D. Chang, and P. A. Sharp. 1991. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc. Natl. Acad. Sci. USA 88:683-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 28.Longnecker, R., and B. Roizman. 1986. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction sequences, an origin of DNA synthesis, and several genes, including those specifying glycoprotein E and the alpha 47 gene. J. Virol. 58:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manche, L., S. R. Green, C. Schmedt, and M. B. Mathews. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 15:358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathews, D. H., J. Sabina, M. Zucker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 31.Mohr, I. J., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 32.Mulvey, M., J. Poppers, A. Ladd, and I. Mohr. 1999. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J. Virol. 73:3375-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishiyama, Y., R. Kurachi, T. Daikoku, and K. Umene. 1993. The Us 9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology 194:419-423. [DOI] [PubMed] [Google Scholar]
- 34.Olland, A. M., J. Jane-Valbuena, L. A. Schiff, M. L. Nibert, and S. C. Harrison. 2001. Structure of the reovirus outer capsid and dsRNA binding protein sigma 3 at 1.8A resolution. EMBO J. 20:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pe'ery, T., and M. B. Mathews. 1997. Synthesis and purification of single-stranded RNA for use in experiments with PKR and in cell-free translation systems. Methods 11:371-381. [DOI] [PubMed] [Google Scholar]
- 36.Pe'ery, T., and M. B. Mathews. 2000. Viral translational strategies and host defense mechanisms, p. 371-424. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 40.Roller, R. J., and B. Roizman. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roller, R. J., and B. Roizman. 1991. Herpes simplex virus 1 RNA binding protein Us11 negatively regulates the accumulation of a truncated viral mRNA. J. Virol. 65:5873-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein Us11 is a virion component and associates with 60S ribosomal subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roller, R. J., L. L. Monk, D. Stuart, and B. Roizman. 1996. Structure and function in the herpes simplex virus 1 RNA-binding protein Us11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J. Virol. 70:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano, P. R., F. Zhang, S. L. Tan, M. T. Garcia-Barrio, M. G. Katze, T. E. Dever, and A. G. Hinnebusch. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18:7304-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sassanfar, M., and J. W. Szostak. 1993. An RNA motif that binds ATP. Nature (London) 364:550-553. [DOI] [PubMed] [Google Scholar]
- 46.Schaerer-Uthurralt, N., M. Erard, K. Kindbeiter, J. J. Madjar, and J. J. Diaz. 1998. Distinct domains in herpes simplex virus type 1 Us11 protein mediate post transcriptional transactivation of human T-lymphotropic virus type 1 envelope glycoprotein gene expression and specific binding to the Rex responsive element. J. Gen. Virol. 79:1593-1602. [DOI] [PubMed] [Google Scholar]
- 47.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shatkin, A. J. 2000. Reovirus translational control, p. 915-932. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Tan, S.-L., M. Gale, Jr., and M. G. Katze. 2000. Translational reprogramming during influenza virus infection, p. 933-950. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Toyoshima, H., M. Itoh, J. Inoue, M. Seiki, F. Takaku, and M. Yoshida. 1990. Secondary structure of the human T-cell leukemia virus type 1 Rex-responsive element is essential for Rex regulation of RNA processing and transport of unspliced RNAs. J. Virol. 64:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J., H. Jiang, and F. Liu. 2000. In vitro selection of novel RNA ligands that bind human cytomegalovirus and block viral infection. RNA 6:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of basic amino acids. RNA 5:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, S., K. U. Kumar, and R. J. Kaufman. 1998. Identification and requirement of three ribosome binding domains in dsRNA-dependent protein kinase (PKR). Biochemistry 37:13816-13826. [DOI] [PubMed] [Google Scholar]
- 54.Xia, T., J. Santa Lucia, Jr., M. E. Burkard, R. Kierzek, S. J. Schroeder, X. Jiao, C. Cox, and D. H. Turner. 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37:14719-14735. [DOI] [PubMed] [Google Scholar]
- 55.Xu, W., and A. D. Ellington. 1996. Anti-peptide aptamers recognize amino acid sequence and bind a protein epitope. Proc. Natl. Acad. Sci. USA 93:7475-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamore, P. D. 2001. RNA interference: listening to the sound of silence. Nat. Struct. Biol. 8:746-750. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, S., P. R. Romano, and R. C. Wek. 1997. Ribosome targeting of PKR is mediated by two double-stranded RNA-binding domains and facilitates in vivo phosphorylation of eukaryotic initiation factor-2. J. Biol. Chem. 272:14434-14441. [DOI] [PubMed] [Google Scholar]
- 58.Zucker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. NATO ASI series. Kluwer Academic Publishers, Dordrecht, The Netherlands.