Abstract
We have characterized monoclonal antibodies raised against the neuraminidase (NA) of a Sydney-like influenza virus (A/Memphis/31/98, H3N2) in a reassortant virus A/NWS/33HA-A/Mem/31/98NA (H1N2) and nine escape mutants selected by these monoclonal antibodies. Five of the antibodies use the same heavy chain VDJ genes and may not be independent. Another antibody, Mem5, uses the same VH and J genes with a different D gene and different isotype. Sequence changes in escape mutants selected by these antibodies occur in two loops of the NA, at amino acid 198, 199, 220, or 221. These amino acids are located on the opposite side of the NA monomer to the major epitopes found in N9 and early N2 NAs. Escape mutants with a change at 198 have reduced NA activity compared to the wild-type virus. Asp198 points toward the substrate binding pocket, and we had previously found that a site-directed mutation of this amino acid resulted in a loss of enzyme activity (M. R. Lentz, R. G. Webster, and G. M. Air, Biochemistry 26:5351-5358, 1987). Mutations at residue 199, 220, or 221 did not alter the NA activity significantly compared to that of wild-type NA. A 3.5-Å structure of Mem5 Fab complexed with the Mem/98 NA shows that the Mem5 antibody binds at the sites of escape mutation selected by the other antibodies.
Viruses exhibit different strategies to escape immune surveillance, and these escape mechanisms constitute significant hurdles in vaccine development. One of these strategies, genetic variation, involves selection of mutations in antibody epitopes that allow the virus to escape host immune defense because it can no longer be recognized and neutralized by specific antibodies.
Influenza A and B viruses have two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), both of which undergo extensive antigenic variation. Thus, influenza virus can efficiently escape from host antibodies through accumulation of mutations in these surface glycoproteins (antigenic drift) or by introduction of new subtypes of these glycoproteins by gene segment reassorting (antigenic shift). The HA attaches the virus to sialic acid and possibly other cell surface receptors (36) on the host cell, and upon endocytosis the HA triggers fusion between virus and host cell membranes. The NA is considered a receptor-destroying enzyme, permitting release of progeny virions that would otherwise aggregate due to the binding of HA to sialic acid on HAs of adjacent virions (24, 31). Antibodies against NA thus indirectly neutralize virus infectivity and protect animals against infection (44). By growing the virus in the presence of a monoclonal antibody, escape mutants, which usually result from single amino acid substitutions that rendered the mutant neutralization resistant, can be selected. Mapping these mutations provides information on the major neutralization epitopes, and knowledge of how the mutations are selected may lead to better understanding of antigenic drift and improved vaccine strategies (6).
We previously analyzed antigenic variation in N2 subtype NAs from H2N2 viruses (1, 21, 42, 43). We have even more detailed views of epitopes on N9 NA (3, 20, 27, 28, 40) since crystal structures of antibody Fab fragments bound to N9 NA have been obtained (25, 37, 38). Many years ago, Laver crystallized N2 NAs from viruses isolated between 1957 and 1967, but NAs of viruses isolated after about 1975 did not crystallize (16, 18). We have periodically screened more-recent N2 NAs for crystallization without success until we obtained diffractable crystals of NA from Sydney-like virus A/Memphis/31/98. To determine if the principles of escape seen in laboratory “antigenic drift” of avian N9 NA apply to antigenic drift of human viruses, we have begun an analysis of the antigenic structure of Mem/98 (N2) NA. We produced mouse monoclonal antibodies which were used to generate escape mutants. Sequence analysis of these antigenic variants showed that the mutations all map to the same region on the three-dimensional structure of NA, which is opposite the site recognized by most anti-N9 and anti-N2 NA antibodies.
MATERIALS AND METHODS
Viruses and cells.
The virus used in this study is A/Memphis/31/98 (H3N2). It was isolated in January 1998 from Robert Webster, and, although the virus was isolated in Memphis, Webster had just returned from Hong Kong and his infection may have originated in Hong Kong. The Mem/98 virus is antigenically similar to A/Sydney/5/97, which caused widespread worldwide epidemics in the 1997-1998 flu season and was used as the H3N2 vaccine strain for 1998-1999 and 1999-2000. We used the NA-minus virus NWS-Mvi (23, 45) to make a reassortant virus containing the HA of A/NWS/33 and NA of A/Mem/98, designated NWS-Mem/98. A high-growth virus with both HA and NA of A/Mem/98, designated Mem/98 (HG), was made by reassorting NWS-Mem/98 with the original Mem/98 virus in the presence of polyclonal antiserum against NWS HA.
Madin-Darby canine kidney (MDCK) cells were cultured in supplemented Dulbecco's modified Eagle medium (DMEM) (43). Viruses and variants were grown in MDCK cells in DMEM-F12 with ITS+ (Collaborative Biomedical Research) and trypsin (23). For large-scale production, the viruses were propagated in the allantoic cavities of 11-day old embryonated chicken eggs and purified by concentration (Amicon) and density gradient centrifugation through 10 to 40% sucrose (17).
Monoclonal antibodies.
Hybridoma cell lines producing antibodies to the N2 NA of A/Mem/31/98 were made as described previously (2, 41) at the Hybridoma Center, Oklahoma State University. The initial immunization was done with purified NWS-Mem/98 virus (10 μg/mouse) and was followed by two boosts with purified pronase-released NA (40 μg/mouse). The antibodies were initially screened by enzyme-linked immunosorbent assays (ELISA) using plates coated with NWS-Mem/98 virus, and specificity was determined with Mem/98 (HG), NWS-G70c (H1N9), and B/Hong Kong (HG) in ELISA and NA inhibition tests. Hybridomas producing antibodies to NA were cloned. Most antibodies were produced by growing the hybridomas to high density and then transferring them directly into Nutridoma (NS formulation; Boehringer Mannheim). For a few hybridomas, it was necessary to adapt them slowly to growth in Nutridoma to obtain good yields of antibody. A few antibodies were grown as ascites tumors and used without further purification.
Antibody purification.
A 5-ml column of recombinant protein A-Sepharose (Pharmacia) was washed with water and then equilibrated with 50 mM Tris buffer, pH 7.0. The Nutridoma supernatant (∼200 ml) was loaded onto the column and washed with 50 ml of 50 mM Tris, pH 7.0. Antibodies were eluted with 0.1 M citrate, pH 3.0; fractions of 6 ml were collected and immediately neutralized with 1.2 ml of 1 M Tris, pH 9.0. The column was regenerated with an additional 30 ml of citrate buffer. Total protein was determined by the Bradford method (protein assay; Bio-Rad Laboratories, Hercules, Calif.), and the purity was checked by sodium dodecyl sulfate-gel electrophoresis.
Serological and enzyme assays.
ELISA were carried out as described previously (14). NA and NA inhibition (NI) assays were performed with fetuin substrate (4, 29). The NA activity of mutant NAs was also measured with MUN (4-methylumbelliferyl-N-acetylneuraminic acid) (12, 32). HA assays were done at 4°C by using human red blood cells for Mem/98 (HG) and human or chicken red blood cells for NWS-Mem/98.
Sequencing of monoclonal antibodies.
For N-terminal protein sequencing, Edman degradation was carried out at the Molecular Biology Resource Facility, Oklahoma University Health Sciences Center, with a Perkin-Elmer, Applied Biosystems Procise model 492 protein sequencer equipped with on-line phenylthiohydantoin-amino acid analyzer and model 610A data system.
Most of the sequences were derived from mRNA by reverse transcription-PCR (RT-PCR). Total RNA was isolated from the hybridomas with Trizol (Life Technologies, Invitrogen Corporation, Carlsbad, Calif.) according to the manufacturer's instructions, and cDNA was made with the Superscript RT kit (Life Technologies) according to the recommended protocol. For RT and amplification of the heavy chain, we used primer γCH2, which primes from a sequence common to all mouse gamma 2 genes at the beginning of the CH2 domain back through the CH1 and V domains. For the reverse primer, we used a mixture of mouse VH primers (Pharmacia-Amersham Biosciences, Piscataway, N.J.). The PCR conditions were as follows: 30 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 1 min and a final extension at 72°C for 7 min.
The heavy chain V-CH1 domain product (∼700 bp) was purified by agarose gel electrophoresis and the V-D-J domain was sequenced by using an oligonucleotide (gamma) that primes at the beginning of the CH1 region. For antibodies that had not been isotyped, we used the γ-CH2 primer to obtain the CH1 domain sequence.
For a few antibodies that did not amplify with the mixed VH primers we designed primers based on the N-terminal protein sequence. These were used in combination with the γ-CH2 primer to reverse transcribe and amplify the heavy chain V-CH1 domains.
Selection of escape mutants.
Ascites fluid or purified antibody was used to select escape mutants (41). The selection and cloning were done in MDCK cells. An allantoic stock of NWS-Mem/98 was titrated on MDCK cells, and values of tissue culture infective units (TCIU) per microliter were calculated. To determine the approximate 100% growth inhibitory concentration (IC100) of each antibody, MDCK cells were infected with virus (10 TCIU) in the presence of 10-fold dilutions of the antibody starting with 10 μl. IC100 was taken as the midpoint between the last well that showed no cytopathic effect (CPE) or HA and the first well showing virus growth. For escape mutant selection, 10 or 100 TCIU of virus was titrated with purified antibody. Escape mutants were obtained from wells where the antibody concentration was one to two times the IC100. Supernatant from wells showing any trace of CPE was passaged again in the presence of the antibody and cloned at least three times by limiting dilution. Variants were confirmed by their similar levels of growth in the presence and absence of the antibody and lack of inhibition in NI tests. A few mutants were isolated by infecting the cells with 105 TCIU of NWS-Mem/98 in the presence of 50 μl of ascites fluid antibody; variants were selected and amplified in the same way.
Sequencing escape mutants.
The NA genes of all the escape mutants were sequenced completely to establish the amino acid sequence changes associated with the failure of the variants to bind the antibody used for their selection. Virus was pelleted from the MDCK supernatants, and RNA was extracted with Trizol (Life Technologies). RT was carried out by using the flu 12-mer primer (5′-AGCAAAAGCAGG), which is complementary to the 12 conserved nucleotides at the 3′ end of all influenza virus type A viral RNA segments, and a Superscript reverse transcription kit (Life Technologies). PCR amplification of the NA gene segment was performed by 25 rounds of PCR with N2 NA-specific terminal primers and Taq polymerase (Fisher). RT-PCR products excised after agarose gel electrophoresis were purified using a Wizard PCR purification system (Promega) and sequenced, using N2-specific primers (21, 33), with an ABI sequencer (Perkin-Elmer, Applied Biosystems) run by the DNA Sequencing Facility at Oklahoma Medical Research Foundation or by the Recombinant DNA/Protein Facility at Oklahoma State University.
Sequence analysis was carried out by using Wisconsin Package, version 9.0, software from the Genetics Computer Group (Madison, Wis.), Vector NTI Suite (InforMax), and Multalin (8).
To identify antibody gene families and germline genes, we used the mouse germline database of Instituto de Biotecnologia, Universidad Nacional Autónoma de México, immunoglobulin (Ig) germline gene and IgBLAST databases of the National Center for Biotechnology Information, and the Kabat Ig sequence database (13).
Crystallization and molecular replacement characterization of the Mem/98 NA-Mem5 Fab complex.
Purified pronase-released NA heads from NWS-Mem/98 were complexed with Mem1, Mem4, or Mem5 Fab generated by insoluble papain (19), and the complex was purified by size exclusion fast performance liquid chromatography (FPLC) and screened for crystallization. Crystals of the Mem5 Fab-NA complex grew in hanging drops over 2 M ammonium sulfate-100 mM Tris, pH 7.5. Diffraction data were collected in house with an RU-H3R X-ray generator with Osmic confocal optics and an R-Axis IV imaging plate detector. The structure was solved by molecular replacement by using the program AMORE (26), with the A/Tokyo/67 N2 NA (PDB file 1NN2) and the Fv domain of Fab from the N9-NC41 complex structure (1NCA) as independent search models.
RESULTS
Characterization of monoclonal antibodies.
A panel of more than 20 antibodies that reacted with the N2 NA of Sydney-like Memphis/98 virus was obtained. These were further characterized by NI assay, which showed that the panel included antibodies that inhibited NA enzyme activity, antibodies specific for NA but noninhibitory, and antibodies cross-reactive with type A and type B viruses that did not inhibit NA activity, presumably directed to the sulfated carbohydrate “host antigen” on the egg-grown virus glycoproteins. None of the antibodies inhibited hemagglutination. Most of the antibodies are isotype IgG2b as determined by an Ouchterlony double-diffusion assay or heavy chain constant region sequencing. Properties of the antibodies used in further studies are shown in Table 1.
TABLE 1.
Biological properties of monoclonal antibodies against NWS-Mem/98
| Monoclonal antibody | Isotypea | NIb | IC100c | Binding to B/HK (HG) | Variant selectiond |
|---|---|---|---|---|---|
| Mem1 | IgG2b | 0 | No inhibition | + | ND |
| Mem2 | IgG2b | ++ | 0.79 μg | − | + |
| Mem3 | IgG2b | ++ | 0.98 μg | − | + |
| Mem4 | IgG2b | ++ | 0.83 μg | − | + |
| Mem5 | IgG2a | ++ | 0.22 μg | − | 0 |
| Mem6 | + | >20 μle | − | 0 | |
| Mem7 | + | — | − | ND | |
| Mem8 | IgG2b | ++ | 1.79 μg | − | + |
| Mem9 | IgG2b | ++ | 5.13 μg | − | 0 |
Determined by Ouchterlony double diffusion or from the CH1 sequence. Blank, isotype not known.
NI assays were done with fetuin as the substrate. ++, antibody inhibits NA activity 70 to 100%; +, inhibition of <50%; 0, no inhibition.
IC100 for inhibition of virus growth was calculated as described in Materials and Methods. ND, not done.
+, we were able to select escape mutants with this antibody; 0, we could not select escape mutants; —, selection not done.
Mem6 assays were done with ascites fluid.
Characterization of escape mutants.
Seven antibodies (Mem2 to Mem6, Mem8, and Mem9) were used to select escape mutants. We obtained nine escape mutants from four antibodies (Mem2, -3, -4, and -8). We did not obtain variants from Mem5, although it inhibits NA activity, from Mem6, which only partially inhibits NA activity, or from Mem9, which bound with low affinity as indicated by the NI curve and inhibition of virus growth (Table 1).
The variants were cloned at least three times by limiting dilution to ensure a single population and were grown for two passages in the absence of antibody to check the stability of the mutation. Mostly the mutations were stable and the variants grew well when challenged again with the antibody, but the mutants with changes at amino acids 220 and 221 sometimes reverted, as indicated by regain of NI activity by the antibody.
Sequence changes in escape mutants.
The NA genes of the variants were completely sequenced. Single nucleotide changes, giving single amino acid changes at 198, 199, 220, or 221, were found in most of the variants (Table 2). Double changes were found in two variants: E199V with K221E in M8vg1 and D198V with T69A in M4vsua1. The deduced amino acid changes in the escape mutants are shown in Table 2.
TABLE 2.
Biological activitiesa of escape mutants
| Virus | Antibody used for selection | Sequence change(s) | NA activity/800 HAUc with:
|
Relative NA/HA activityb (%) with:
|
||
|---|---|---|---|---|---|---|
| Fetuin (A549) | MUN (fluorescence) | Fetuin | MUN | |||
| NWS-Mem/98 | None (wild type) | 7.5 ± 1.0 | 1,490 ± 46 | 100 | 100 | |
| M2vsu1 | Mem2 | K221N | 4.0 ± 0.3 | 1,825 ± 41 | 53 ± 4 | 123 ± 3 |
| M3vd1 | Mem3 | E199G | 6.4 ± 0.6 | 1,192 ± 83 | 85 ± 7 | 80 ± 6 |
| M3vg2 | Mem3 | E199V | 4.0 ± 0.4 | 751 ± 32 | 53 ± 5 | 50 ± 2 |
| M4vs3 | Mem4 | K220N | 6.7 ± 0.7 | 1,723 ± 144 | 89 ± 9 | 116 ± 10 |
| M4vsu1 | Mem4 | D198V | 0.6 ± 0.9 | 112 ± 36 | 8 ± 11 | 8 ± 2 |
| M4vsu2 | Mem4 | D198V | 0.6 ± 0.7 | 93 ± 29 | 7 ± 10 | 6 ± 2 |
| M4vsua1 | Mem4 | T69A, D198V | 0.1 ± 0.3 | 111 ± 68 | 2 ± 4 | 8 ± 5 |
| M8vg1 | Mem8 | K221E, E199V | 5.8 ± 3.0 | 1,254 ± 381 | 77 ± 40 | 84 ± 26 |
| M8vsu1 | Mem8 | D198V | 1.0 ± 2.0 | 118 ± 4 | 13 ± 26 | 5 ± 1 |
Values shown are averages of two to four observations ± standard deviations.
High standard deviation for the D198V mutants is due to the very low NA activity.
HAU, hemagglutinating units.
Biological activities of escape mutants.
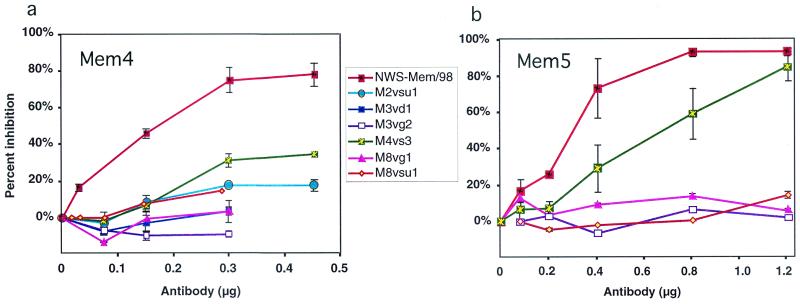
The escape mutants were tested for NA and HA activity. Amino acid substitutions at position 199, 220, or 221 resulted in an NA:HA activity ratio that was at least 50% of the ratio for the wild type, whereas the variants with a mutation at 198 averaged less than 10% of wild-type NA activity, whether measured with a high-molecular-weight substrate (fetuin) or a small substrate (MUN) (Table 2). All escape mutants were resistant to inhibition by Mem2, Mem3, Mem4, Mem5, or Mem8, indicating that the epitopes overlap. Inhibition curves for Mem4 and Mem5 are shown in Fig. 1.
FIG. 1.
Antibody inhibition curves of NWS-Mem/98 and the escape mutants. Purified viruses were titrated with antibodies Mem4 (a) and Mem5 (b).
Cross-reactivity between antibodies and escape mutants.
Antigenic sites can be operationally defined by the ability of sequence changes in one group of variants to prevent the binding of a corresponding group of monoclonal antibodies but, at the same time, to have no effect on the binding of other monoclonal antibodies which recognize different antigenic sites on the same molecule (11). The cross-reactivity in NI assays of the monoclonal antibodies and the variants selected by them (Fig. 1) showed that Mem2, Mem3, Mem4, Mem5, and Mem8 behaved very similarly in all the assays, and we wondered if they were all the same. To resolve this, we sequenced the antibodies.
Sequencing of monoclonal antibodies.
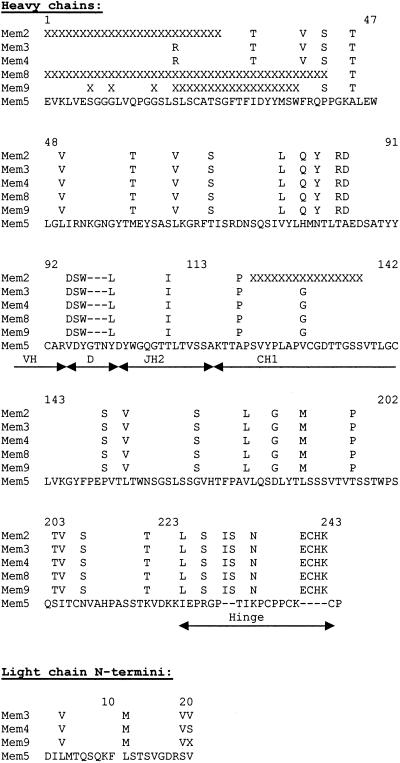
Heavy chain V-CH1 domain genes were amplified by RT-PCR. A clear band was observed close to the expected ∼700 bp, and we obtained good sequences for V regions from Mem2, Mem3, Mem4, Mem8, and Mem9 by using the gamma primer. A discrepancy between nucleotide and protein sequences at amino acid 5 can be attributed to the primer sequences (30). Mem5 did not amplify with mixed variable-region primers from Pharmacia, so we constructed a reverse primer based on the N-terminal heavy chain sequence deduced from the heavy-plus-light chain protein sequence and an oligonucleotide (IgG CH1, which can prime on all mouse IgGs) at the beginning of the CH1 domain. Using these primers, we successfully amplified and sequenced the V-D-J domains. We have not succeeded in amplifying the VH domain of Mem1, and the N terminus appears to be blocked.
Mem3 and Mem4 have identical heavy chain V, D, and J sequences with identical junctions and are both IgG2b. Almost-complete data indicate that Mem2, Mem8, and Mem9 sequences are identical to these. Mem5 is derived from the same germline VH gene but has a different D sequence, and the C domain is IgG2a. N-terminal protein sequencing for 20 cycles showed that the N termini were not blocked and mixed light chain and heavy chain sequences were obtained for Mem3, Mem4, Mem5, and Mem9, so by subtraction we could see that the N-terminal VL sequences of Mem3, Mem4, and Mem9 are also identical. The sequences are shown in Fig. 2.
FIG. 2.
Sequences of the heavy chains of monoclonal antibodies (with Kabat numbering). The heavy chain (VDJ) amino acid sequences are assembled from N-terminal protein sequence and cDNA sequences. The N-terminal sequences of the light chains were obtained by Edman degradation and subtraction of the heavy chain. X, not determined.
Crystallization and molecular replacement characterization of NA-Mem5 Fab complex.
Crystals of purified pronase-released NA heads from NWS-Mem/98 complexed with Mem5 Fab were grown as described in Materials and Methods. Washed crystals, dissolved and run on gel electrophoresis, showed NA and Fab (light and heavy chain) bands, indicating that the crystals were indeed of the complex. The crystals diffracted to 3.1 Å but did not survive freezing, and they decayed in the X-ray beam at room temperature. Nevertheless, a 93.7% complete 3.5-Å data set was collected (Rsym = 11.2%, overall I/σ [I] = 10.0, and I/σ [I] for outer shell = 2.8, where Rsym is the residual index of the symmetric data and I is the intensity). The space group is P422, and unit cell dimensions are as follows: a = b = 159.6 Å, c = 104 Å, with α = β = γ = 90.00°. We attempted to solve the structure by molecular replacement by using the program AMORE (26), with the N2 NA (PDB file 1NN2) and the Fab portion of the N9-NC41 complex structure (1NCA) as independent search models. We located the NA with reasonable statistics (correlation coefficient, 55.9%; R factor, 38.8%) but not the Fab. We then searched with just the Fv domain from 1NCA and located it (correlation coefficient, 59.3%; R factor, 35.0%). Finally, we searched separately for the C domain of the Fab, but it was not located by AMORE, suggesting that the elbow angle is mobile, as it was in the N9NA-NC10 complex crystal structure (25), or that the packing is not well ordered. A refinement of the structure and inclusion of side chains of Mem98 NA complexed with Fv of antibody Mem5 await better-quality crystals, but the 3.5-Å molecular replacement structure shows the Fv domain sitting over the sites of escape mutations (see Fig. 4b).
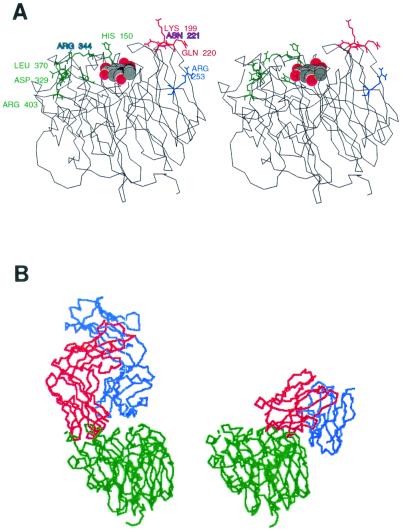
FIG. 4.
(A) Stereo view of the location of escape mutations on the three-dimensional structure of N2 (A/Tokyo/67) NA (39). The color coding is the same as in Fig. 3. Sialic acid in the active site is shown as a space-filling model. (B) Comparison of crystal structures of NC41 Fab-N9 NA (left) and Mem5 Fab complexed with Mem/98 NA as solved by molecular replacement at 3.5 Å by using Tokyo/67 NA and the NC41 Fv domain (37) as starting models (right). The NA (green) is in the same orientation in both complexes. The antibody heavy chains are red, and light chains are blue.
DISCUSSION
Structure determination of antibody-antigen complexes by X-ray crystallography remains the only definitive way to learn how an antibody recognizes and binds to the surface of a foreign protein and thus to define the antigenic determinant. However, identification of mutations in escape mutants, which do not bind the selecting monoclonal antibody, has proven to be a useful method to map at least the center of an epitope. Whenever the escape mutant analysis has been tested by comparison with the X-ray crystal structure of the complex, the amino acids that change in escape mutants have been found to be in contact with the antibody (15, 25, 27, 35, 37, 38). It appears that global conformational change due to a single amino acid substitution cannot be transmitted across a protein without disturbing the function, and so for a viable escape mutant the effects of the mutation are strictly local.
At least in tissue culture, antibodies will neutralize influenza virus infection only if they inhibit the activity of the protein to which they bind. MDCK cells lack Fc receptors and so have no opsonizing activity. Thus, in our panel of antibodies against the Mem/98 NA, antibodies Mem2, -3, -4, -5, and -8 inhibit NA activity and neutralize virus infection. Other antibodies, such as Mem9 weakly inhibit NA activity and reduce infection, but not enough for selection of escape mutants (3, 40). Mem6 binds to N2 NA but only weakly inhibits NA activity. It does not bind to other subtypes of NA; presumably it recognizes an epitope which is remote from the biologically active center. Antibody Mem1 binds to NA but does not inhibit NA activity and does not inhibit infection. Mem1 is highly cross-reactive (even with type B virus) and may be directed to the sulfated carbohydrate host antigen on the egg-grown virus glycoproteins.
Four Mem/98 NA-specific monoclonal antibodies (Mem2, -3, -4, and -8) inhibited NA activity in the fetuin assay, inhibited virus replication, and selected variants under in vitro conditions. Sequence changes in escape mutants selected with these antibodies all mapped in the same area of the three-dimensional structure of NA, and all mutants had similar cross-reactivities with the other three antibodies and also with Mem9. Our suspicion that these might be the same antibody was confirmed by sequencing VH-DH-JH and the N-terminal 20 amino acids of the VH chain. The hybridomas were prepared from a single mouse, and, since those producing Mem2, -3, -4, -8, -9 grew very vigorously, it is likely that they were amplified before being cloned out rather than that their identity reflected a bias in favor of the use of this VDJ combination. The Mem5 sequence is related but not identical, so we have at least two independent antibodies recognizing the same region of NA. The other antibodies have different properties; they bind to NA but inhibit weakly or not at all. The VH sequences in the Mem2 group and Mem5 are from the group 7 (S107/T15) family pBV19B4 germline gene (34) joined to JH2 via two different D sequences.
Changes in escape mutants.
The sequence changes in the escape mutants indicated that the antibodies characterized (the Mem2 group and Mem5) recognize epitopes on the top corner of the NA subunit (Fig. 3 and 4), adjacent to the active site, but opposite to the 329-to-403 area, which contained almost all the epitopes of monoclonal antibodies against earlier N2 NAs (1, 42, 43) and also N9 NA (3, 20, 40).
FIG. 3.
Location of escape mutations in recent and early N2 NAs. The amino acid positions where changes have been selected by monoclonal antibodies (mAb) are indicated by the colored bars: green, X-7(F1) (RI/5+/57) escape mutants; blue, Tokyo/67 escape mutants, including escape mutants of Tokyo/67 selected by antibodies made against A/Jap/305/57 (H2N2) and A/Texas/77 (H3N2); red, Mem/31/98 (Sydney-like) escape mutants. Data for X-7(F1) and Tokyo/67 escape mutants are from references 1, 18, 21, 42, and 43).
Single amino acid sequence changes at positions 198, 199, 220, and 221 were sufficient to effectively abolish antibody binding. Two mutants had two changes, at 199 and 221 (M8vg1) and at 198 and 69 (M4vsua1). The change at amino acid 69 is in the NA stalk domain and is unlikely to affect the antigenic properties of the NA (9). When sequences of human N2 NAs from 1957 to 1998 are compared, regions of high variability can be seen. As previously noted, the stalk region is highly variable but has not been associated with the binding of neutralizing antibodies (2, 5, 9). The variable regions in the NA “head” domain are amino acids 140 to 155, 328 to 370, and to a lesser extent 381 to 403. There are, however, other changeable positions, including amino acids 199, 220, and 221, where we find escape mutations. The 328-to-370 region has dominated variation in human H2N2 and H3N2 isolates from 1957 to 1998, accounting for more than 50% of all amino acid changes. Escape mutant analysis of early viruses containing N2 NA (A/RI/5+/57 and A/Tokyo/3/67) showed that almost all changes were in the 329-to-370 region, the four exceptions being at 403, 150, 221, and 253 (1, 18, 21, 42, 43). Figure 3 summarizes the distribution of changes in Mem/98 escape mutants compared to previous studies and shows the dominance of the 327-to-403 region in escape mutations of earlier strains, which we have not seen with Mem/98.
When escape mutant changes are plotted on the three-dimensional structure of N2 NA (7), all the changes except that at position 253 are located on the upper surface of the molecule, surrounding the active-site pocket (Fig. 4a), in accord with the observation that escape mutants can only be selected by antibodies that inhibit NA activity. To confirm that the antibody binds at the site of escape mutation, we have begun a crystallographic study of these antibodies bound to Mem/98 NA. We have solved the crystal structure of the Mem5 Fab-Mem/98 NA complex at 3.5-Å resolution by molecular replacement with the Tokyo/67 NA (38) and NC41 Fab (37) as starting models, and our structure shows Mem5 binding to loops containing the escape mutations, amino acids 198, 199, 220, and 221, selected by our other antibodies. We did not succeed in selecting escape mutants with Mem5, but escape mutants selected by the Mem2 antibody group were not inhibited, or were weakly inhibited, by Mem5 (Fig. 1b), indicating that it binds to the same region of NA. The structure is not yet refined; we are working on obtaining crystals that diffract to higher resolution, but the comparison of antibody binding sites is clear and is shown in Fig. 4b.
Fanning et al. (10) have analyzed “phylogenetically informative amino acid positions” (PIPs) and “phylogenetically important regions” (PIRs; many PIPs near one another) in influenza virus A NAs of subtypes N1 and N2. They found 12 PIRs on N2. One PIR is amino acids 197 to 199, comprising two PIPs at 197 and 199. Residue 198 was excluded since it is conserved in all N2 sequences although not in all NAs. We see changes at 198 and 199. Our analyses (Table 2) show that substitution at 198 significantly reduced NA activity (∼7.5% of the wild-type activity). Indeed we previously found that a site-directed mutation of Asp198 resulted in loss of enzyme activity (22). Mutations at 199, 220, or 221 had no significant effect on NA activity.
Our analysis of epitopes on a recent human N2 NA suggests that new antigenic variants may arise from the loops containing amino acids 198 to 221. Epitopes in this region were represented, but not dominant, in antibodies against the NA of H2N2 viruses isolated from 1957 to 1967.
Acknowledgments
We thank Robert Webster, St. Jude Children's Research Hospital, Memphis, Tenn., for very kindly providing the virus A/Memphis/31/98; Francisca A. Neethling and Brandi Simmons, Hybridoma Center, Dept. of Entomology and Plant Pathology, Oklahoma State University, for hybridoma production; Sheryl Christofferson at the DNA Sequencing Facility, Oklahoma Medical Research Foundation, and Janet Rogers, Recombinant DNA/Protein Facility at Oklahoma State University, for DNA sequencing; and Kenneth Jackson and Casey Batson at the Molecular Biology Resource Facility, OUHSC, for protein sequencing. L. David Porter selected the mutant M3vd1.
This work was supported by a merit grant (AI-19084) from the National Institute of Allergy and Infectious Diseases, NIH.
REFERENCES
- 1.Air, G. M., M. C. Els, L. E. Brown, W. G. Laver, and R. G. Webster. 1985. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology 145:237-248. [DOI] [PubMed] [Google Scholar]
- 2.Air, G. M., W. G. Laver, M. Luo, S. J. Stray, G. Legrone, and R. G. Webster. 1990. Antigenic, sequence and crystal variation in influenza B neuraminidase. Virology 177:578-587. [DOI] [PubMed] [Google Scholar]
- 3.Air, G. M., W. G. Laver, and R. G. Webster. 1990. Mechanism of antigenic variation in an individual epitope on influenza virus N9 neuraminidase. J. Virol. 64:5797-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aymard-Henry, M., M. T. Coleman, W. R. Dowdle, W. G. Laver, G. C. Schild, and R. G. Webster. 1973. Influenza virus neuraminidase and neuraminidase inhibition test procedures. Bull. W. H. O. 48:199-202. [PMC free article] [PubMed] [Google Scholar]
- 5.Blok, J., and G. M. Air. 1982. Variation in the membrane-insertion and “stalk” sequences in eight subtypes of influenza type A virus neuraminidase. Biochemistry 21:4001-4007. [DOI] [PubMed] [Google Scholar]
- 6.Bush, R. M., C. A. Bender, K. Subbarao, N. J. Cox, and W. M. Fitch. 1999. Predicting the evolution of human influenza A. Science 286:1921-1925. [DOI] [PubMed] [Google Scholar]
- 7.Colman, P. M., J. N. Varghese, and W. G. Laver. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303:41-44. [DOI] [PubMed] [Google Scholar]
- 8.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Els, M. C., G. M. Air, K. G. Murti, R. G. Webster, and W. G. Laver. 1985. An 18 amino acid deletion in an influenza neuraminidase. Virology 142:241-247. [DOI] [PubMed] [Google Scholar]
- 10.Fanning, T. G., A. H. Reid, and J. K. Taubenberger. 2000. Influenza A virus neuraminidase: regions of the protein potentially involved in virus-host interactions. Virology 276:417-423. [DOI] [PubMed] [Google Scholar]
- 11.Gerhard, W., J. Yewdell, M. Frankel, and R. G. Webster. 1981. Antigenic structure of influenza virus hemagglutinin defined by hybridoma antibodies. Nature 290:713-717. [DOI] [PubMed] [Google Scholar]
- 12.Ghate, A. A., and G. M. Air. 1998. Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design. Eur. J. Biochem. 258:320-331. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, G., and T. T. Wu. 2001. Kabat database and its applications: future directions. Nucleic Acids Res. 29:205-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kida, H., L. E. Brown, and R. G. Webster. 1982. Biological activity of monoclonal antibodies to operationally defined antigenic regions on the hemagglutinin molecule of A/Seal/Massachusetts/1/80 (H7N7) influenza virus. Virology 122:38-47. [DOI] [PubMed] [Google Scholar]
- 15.Knossow, M., R. S. Daniels, A. R. Douglas, J. J. Skehel, and D. C. Wiley. 1984. Three-dimensional structure of an antigenic mutant of the influenza virus hemagglutinin. Nature 311:678-680. [DOI] [PubMed] [Google Scholar]
- 16.Laver, W. G. 1978. Crystallization and peptide maps of neuraminidase “heads” from H2N2 and H3N2 influenza virus strains. Virology 86:78-87. [DOI] [PubMed] [Google Scholar]
- 17.Laver, W. G. 1969. Purification of influenza virus, p. 82-86. In K. Habel and N. P. Salzman (ed.), Fundamental techniques in virology. Academic Press, New York, N.Y.
- 18.Laver, W. G., G. M. Air, R. G. Webster, and L. J. Markoff. 1982. Amino acid sequence changes in antigenic variants of type A influenza virus N2 neuraminidase. Virology 122:450-460. [DOI] [PubMed] [Google Scholar]
- 19.Laver, W. G., R. G. Webster, and P. M. Colman. 1987. Crystals of antibodies complexed with influenza virus neuraminidase show isosteric binding of antibody to wild-type and variant antigens. Virology 156:181-184. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. T., and G. M. Air. 2002. Contacts between influenza N9 neuraminidase and monoclonal antibody NC10. Virology 300:256-268. [DOI] [PubMed]
- 21.Lentz, M. R., G. M. Air, W. G. Laver, and R. G. Webster. 1984. Sequence of the neuraminidase gene of influenza virus A/Tokyo/3/67 and previously uncharacterized monoclonal variants. Virology 135:257-265. [DOI] [PubMed] [Google Scholar]
- 22.Lentz, M. R., R. G. Webster, and G. M. Air. 1987. Site-directed mutation of the active site of influenza neuraminidase and implications for the catalytic mechanism. Biochemistry 26:5351-5358. [DOI] [PubMed] [Google Scholar]
- 23.Liu, C., and G. M. Air. 1993. Selection and characterization of a neuraminidase-minus mutant of influenza virus and its rescue by cloned neuraminidase genes. Virology 194:403-407. [DOI] [PubMed] [Google Scholar]
- 24.Liu, C., M. C. Eichelberger, R. W. Compans, and G. M. Air. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 69:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malby, R. L., W. R. Tulip, V. R. Harley, J. L. McKimm-Breschkin, W. G. Laver, R. G. Webster, and P. M. Colman. 1994. The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody. Structure 2:733-746. [DOI] [PubMed] [Google Scholar]
- 26.Navaza, J. 1994. An automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 27.Nuss, J., P. Whitaker, and G. Air. 1993. Identification of critical contact residues in the NC41 epitope of a subtype N9 influenza virus neuraminidase. Proteins Struct. Funct. Genet. 15:121-132. [DOI] [PubMed] [Google Scholar]
- 28.Nuss, J. M., and G. M. Air. 1994. Defining the requirements for an antibody epitope on influenza virus neuraminidase: how tolerant are protein epitopes? J. Mol. Biol. 235:747-759. [DOI] [PubMed] [Google Scholar]
- 29.Nuss, J. M., and G. M. Air. 1991. Transfer of the hemagglutinin activity of influenza virus neuraminidase subtype N9 into an N2 background. Virology 183:496-504. [DOI] [PubMed] [Google Scholar]
- 30.Orlandi, R., D. H. Gussow, P. T. Jones, and G. Winter. 1989. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:3833-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature-sensitive influenza virus mutants defective in neuraminidase. Virology 61:397-410. [DOI] [PubMed] [Google Scholar]
- 32.Potier, M., L. Mameli, M. Belisle, L. Dallaire, and S. B. Melancon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl)-α-D-N-acetylneuraminate substrate. Anal. Biochem. 94:287-296. [DOI] [PubMed] [Google Scholar]
- 33.Ritchie, L. R., R. G. Webster, W. G. Laver, and G. M. Air. 1987. Heterogeneity of neuraminidase genetic information in an H1N2 reassortant influenza virus [X-7(F1)]. Arch. Virol. 96:303-308. [DOI] [PubMed] [Google Scholar]
- 34.Siu, G., E. A. Springer, H. V. Huang, L. E. Hood, and S. T. Crews. 1987. Structure of the T15 VH gene subfamily: identification of immunoglobulin gene promoter homologies. J. Immunol. 138:4466-4471. [PubMed] [Google Scholar]
- 35.Smith, T. J., N. H. Olson, R. H. Cheng, E. S. Chase, and T. S. Baker. 1993. Structure of a human rhinovirus-bivalently bound antibody complex: implications for viral neutralization and antibody flexibility. Proc. Natl. Acad. Sci. USA 90:7015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stray, S. J., R. D. Cummings, and G. M. Air. 2000. Influenza virus infection of desialylated cells. Glycobiology 10:649-658. [DOI] [PubMed] [Google Scholar]
- 37.Tulip, W. R., J. N. Varghese, W. G. Laver, R. G. Webster, and P. M. Colman. 1992. Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J. Mol. Biol. 227:122-148. [DOI] [PubMed] [Google Scholar]
- 38.Tulip, W. R., J. N. Varghese, R. G. Webster, W. G. Laver, and P. M. Colman. 1992. Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface. J. Mol. Biol. 227:149-159. [DOI] [PubMed] [Google Scholar]
- 39.Varghese, J. N., J. L. McKimm-Breschkin, J. B. Caldwell, A. A. Kortt, and P. M. Colman. 1992. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins Struct. Funct. Genet. 14:327-332. [DOI] [PubMed] [Google Scholar]
- 40.Webster, R. G., G. M. Air, D. W. Metzger, P. M. Colman, J. N. Varghese, A. T. Baker, and W. G. Laver. 1987. Antigenic structure and variation in an influenza N9 neuraminidase. J. Virol. 61:2910-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster, R. G., and M. T. Berton. 1981. Analysis of antigenic drift in the hemagglutinin molecule of influenza B virus with monoclonal antibodies. J. Gen. Virol. 54:243-251. [DOI] [PubMed] [Google Scholar]
- 42.Webster, R. G., L. E. Brown, and W. G. Laver. 1984. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology 135:30-42. [DOI] [PubMed] [Google Scholar]
- 43.Webster, R. G., V. S. Hinshaw, and W. G. Laver. 1982. Selection and analysis of antigenic variants of the neuraminidase of N2 influenza viruses with monoclonal antibodies. Virology 117:93-104. [DOI] [PubMed] [Google Scholar]
- 44.Webster, R. G., P. A. Reay, and W. G. Laver. 1988. Protection against lethal influenza with neuraminidase. Virology 164:230-237. [DOI] [PubMed] [Google Scholar]
- 45.Yang, P., A. Bansal, C. Liu, and G. M. Air. 1997. Hemagglutinin specificity and neuraminidase coding capacity of neuraminidase-deficient influenza viruses. Virology 229:155-165. [DOI] [PubMed] [Google Scholar]