Abstract
The baculovirus lef-12 (orf41) gene is required for transient expression of baculovirus late genes. To analyze the role of LEF-12 in the context of infected cells, two mutant viruses were constructed. Both mutants were viable in Trichoplusia ni High 5 and Spodoptera frugiperda Sf9 cells. Single-step growth curves, however, indicated that virus yields were reduced approximately fivefold in the absence of LEF-12. Pulse-labeling of infected cells revealed that LEF-12 mutant viruses entered the late phase and synthesized late proteins at levels equivalent to or only twofold lower than those of wild-type virus-infected cells. Western blot analyses confirmed that LEF-12 was not synthesized in cells infected with mutant virus. In wild-type virus-infected cells, LEF-12 was not detected until 18 h postinfection, and accumulation of LEF-12 peaked at 24 to 36 h postinfection. Primer extension mapping revealed that lef-12 mRNA was synthesized by 12 h postinfection and peaked between 18 and 24 h postinfection. Furthermore, synthesis of lef-12 mRNA and LEF-12 protein were inhibited by the addition of aphidicolin, indicating that lef-12 is expressed after DNA replication.
In the baculovirus system, most of the viral proteins involved in expression of late genes have been identified by transient-expression assays (25). In Spodoptera frugiperda cells, 19 viral proteins, called late expression factors (LEFs), are required for expression of a reporter gene cloned under the control of a late Autographa californica nuclear polyhedrovirus (AcNPV) promoter. Baculovirus late gene expression is absolutely dependent on viral DNA replication, and therefore these 19 genes include early transcription factors and DNA replication proteins as well as proteins directly involved in late transcription (21).
The identification of 19 LEFs is broadly applicable to all baculoviruses, although a few virus-specific and cell line-specific differences have been noted. For example, the apoptosis suppressor p35 can be replaced by inhibitor of apoptosis genes from other baculoviruses, and it is not required for expression in Trichoplusia ni cells because of the inherent resistance of this cell line to apoptosis (1, 10, 21, 29). In addition, three AcNPV genes (ie-2, lef-7, and lef-12) are essential or strongly stimulatory for transient expression of late genes in Spodoptera frugiperda cells but not in Trichoplusia ni cells (20, 22). Instead, transient late expression in T. ni requires HCF-1, which is nonessential for replication in S. frugiperda cells.
IE-2 transactivates early gene expression, stimulates DNA replication, and inhibits the host cell cycle in S. frugiperda cells (5, 6, 21, 24). IE-2 is expressed only transiently during the early phase of replication. Therefore, it seems likely that it is not directly involved in late gene expression but rather helps to establish viral replication factories. Little is known concerning the functions of LEF-7 and HCF-1, but they are also required for efficient transient DNA replication and therefore may also play an indirect role in late gene expression (9, 22).
LEF-12 is the only cell line-specific factor that does not affect transient DNA replication and thus was predicted to function directly at the stage of late transcription (25). The fact that LEF-12 is required for transient late gene expression in S. frugiperda cells but not in T. ni cells suggested to us that it might not be essential in the context of virus replication in T. ni. We decided to address this question through the construction of a mutant virus containing an interrupted lef-12 gene. We found that lef-12 was not essential for viral replication in either S. frugiperda or T. ni cells, although yields of virus were reduced in the mutant-infected cells. Analysis of lef-12 gene expression revealed that it was dependent on viral DNA replication, indicating that it should be classified as a late gene.
MATERIALS AND METHODS
Construction of lef-12 (orf41) mutant viruses.
To construct mutant viruses that do not express functional LEF-12, a fragment of DNA encoding the β-galactosidase gene was cloned into the lef-12 open reading frame. To do this, a 6-kb PstI-KpnI fragment, which represents the left half of the PstI-F fragment of the AcNPV genome and contains lef-12, was cloned into the PstI and SmaI sites of pUC8. The resulting clone, named PstF-PK, was digested with ApaI and then treated with T4 DNA polymerase in the presence of deoxynucleoside triphosphates to make blunt ends. The repaired DNA was then ligated with BglII linkers (GAAGATCTTC). This DNA was subsequently digested with BglII and then ligated to a 3.1-kb fragment encoding the β-galactosidase gene which had previously been purified from plasmid DP502 after digestion with BamHI. Colonies expressing the LEF-12-β-galactosidase fusion under the control of the lef-12 promoter were selected as blue colonies when grown in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). DNA sequence analysis was performed to confirm that the construction was correct, and the final clone was named pLEF12-βgal.
The vLEF12-βgal mutant virus was constructed by cotransfecting wild-type viral DNA and pLEF12-βgal into T. ni High Five (Invitrogen) tissue culture cells. Viruses that formed blue plaques when grown in the presence of X-Gal were picked, purified by plaque assay three additional times, and screened for the presence of the β-galactosidase gene within the lef-12 open reading frame by Southern blot analysis.
A second mutant virus was constructed in which the initiation codon for lef-12 was altered. To do this, QuickChange mutagenesis (Stratagene) was used to make substitutions within the methionine initiation codon of LEF-12 as well as the methionine at the third amino acid position. For ease in screening, the substitutions were planned so that a HindIII site was inserted at the 5′ end of the open reading frame in PstF-PK. The sequence ATGACTATG was changed to ATAAGCTTG, which alters the first three amino acids in the LEF-12 open reading frame from Met-Thr-Met to Ile-Ser-Leu. The mutant virus vLEF12-ΔATG was constructed by digesting vLEF12-βgal DNA with Bsu36I, which cuts viral DNA once within the β-galactosidase gene. Linearized viral DNA and pLEF12-ΔATG were cotransfected into S. frugiperda Sf9 cells, and white plaques were picked and amplified.
To screen for the presence of the LEF12-ΔATG mutation in recombinant viruses, the plaques were amplified by PCR with a primer named lef12.s3 (GTACCACCGATCTCGGCAC) that hybridized 322 nucleotides upstream of the start codon and a primer named lef12.as2 (AATCAACGTGATTGACGACGAG) that hybridized to the 3′ end of the lef-12 open reading frame. The PCR products were subsequently digested with HindIII and analyzed by agarose gel electrophoresis.
Production of LEF-12 antiserum.
To produce antiserum against LEF-12 in rabbits, the protein was cloned into a bacterial expression system. QuickChange mutagenesis (Stratagene) was used to construct an NdeI site at the ATG codon for lef-12. The resulting plasmid was digested with NdeI and EcoRV, and the resulting 570-bp fragment was cloned into the NdeI and BamHI (repaired with Klenow) sites of pET28a.
pET28a-lef12 was transformed into BL21(DE3)GroESL cells, and a 100-ml culture was grown in the presence of kanamycin and chloramphenicol at 30°C until the optical density at 280 nm reached 0.6. Expression of LEF-12 was induced by the addition of 0.5 mM isopropylthiogalactopyranoside, followed by incubation at 25°C overnight. Bacteria were resuspended in 5 ml of TEBG buffer (50 mM Tris [pH 7.5], 1 mM EDTA, 1 mM β-mercaptoethanol, and 10% glycerol). The resuspended cells were subsequently incubated in the presence of 5 μl of ReadyLyse (Epicentre Technologies). After 5 min at room temperature, the bacteria were lysed by the addition of 0.2% sodium deoxycholate. Nucleic acids were then digested with 5 μl of Omnicleave endonuclease (Epicentre Technologies) for 15 min at room temperature. Inclusion bodies were collected by centrifugation. Inclusion bodies were solubilized with 0.5% sodium dodecyl sulfate (SDS), and LEF-12 was purified by metal affinity chromatography on Talon (Clontech) in the presence of 0.1% SDS. Purified LEF-12 was subsequently dialyzed against Tris-buffered saline and injected into rabbits to produce polyclonal antibody by following a standard protocol (16).
LEF-12 transcriptional analysis.
Sf9 cells were gown in 100- or 500-ml spinner flasks and infected with wild-type virus at a multiplicity of infection (MOI) of 5. Cells were harvested at the indicated times postinfection, and RNAs were purified by guanidine-isothiocyanate extraction. RNAs prepared for the time course experiment in S. frugiperda cells were purified by ultracentrifugation in CsCl gradients as previously described (13). Additional RNAs were prepared by affinity chromatography according to the recommendations of the manufacturer (Eppendorf). The oligonucleotide lef12.gsp2 (GTTTCATCATTGTGGCGATC) was labeled at the 5′ end with T4 polynucleotide kinase and [γ-32P]ATP (27). Labeled primer was hybridized with 10 μg of total RNA isolated from infected cells at different times postinfection. Primer extension assays were performed as previously described (27). A sequencing reaction was performed with the same primer and Sequenase as recommended by the manufacturer (USB).
For reverse transcription (RT)-PCR analysis, RNAs were further purified by digestion with DNase I, followed by precipitation with 2.5 M LiCl. Reverse transcriptase reactions were performed with 1 μg of RNA, 20 pmol of LEF-12.pe2 primer (CCTGTTGCGTGCAATAGCCCTGCTG), and 1 μl of ImPromII reverse transcriptase (Promega) in a 20-μl reaction volume. Then 1 μl of the reverse transcription reaction was amplified in a 25-μl reaction mix with 2× PCR MasterMix (Promega) and 10 pmol each of lef12.gsp2 and lef12.5prime (GTGCTCCAACCGGGTG). RT-PCR products were separated on 2.5% agarose gels and photographed under UV light.
Analysis of viral protein expression.
Cells were infected with virus at an MOI of 5 and harvested at different times postinfection. Radiolabeling was performed in TNMF medium minus methionine and cysteine in the presence of 35S-Express (New England Nuclear) at 20 μCi/ml for 4 h. Soluble proteins were extracted by incubation with extraction buffer for 20 min on ice as previously described (12). Samples were used for immunoprecipitation by following a standard protocol (16). Unlabeled protein samples were prepared in the same manner and analyzed by immunoblot analysis (16).
Expression of polyhedrin was measured indirectly by viewing the formation of occlusion virus in the nuclei of infected cells. S. frugiperda cells were grown in Lab-Tek chamber slides and infected at an MOI of 5. At 36 and 48 h postinfection, cells were rinsed in phosphate-buffered saline and fixed in methanol at −20°C. Cells were examined under a Zeiss Axiophot microscope, and images were captured with a charge-coupled device camera. For each time point, six fields of cells at ×40 were scored for the presence of polyhedra.
RESULTS
Construction of vLEF12-βgal.
To determine whether lef-12 was essential for replication of AcNPV, we constructed a plasmid with the gene for β-galactosidase inserted into a unique ApaI site in the lef-12 open reading frame (Fig. 1A). The ApaI site was chosen for the site of insertion because it has previously been used to produce frameshift mutants that are not capable of stimulating late gene expression (18, 25). The chimeric plasmid was then transfected into T. ni (High 5) cells for in vivo recombination with wild-type viral DNA. The transfection and subsequent plaque purification were done in T. ni cells because we expected that LEF-12 might be essential in S. frugiperda but not in T. ni. Plaques with a blue phenotype in the presence of X-Gal were selected and amplified in T. ni cells.
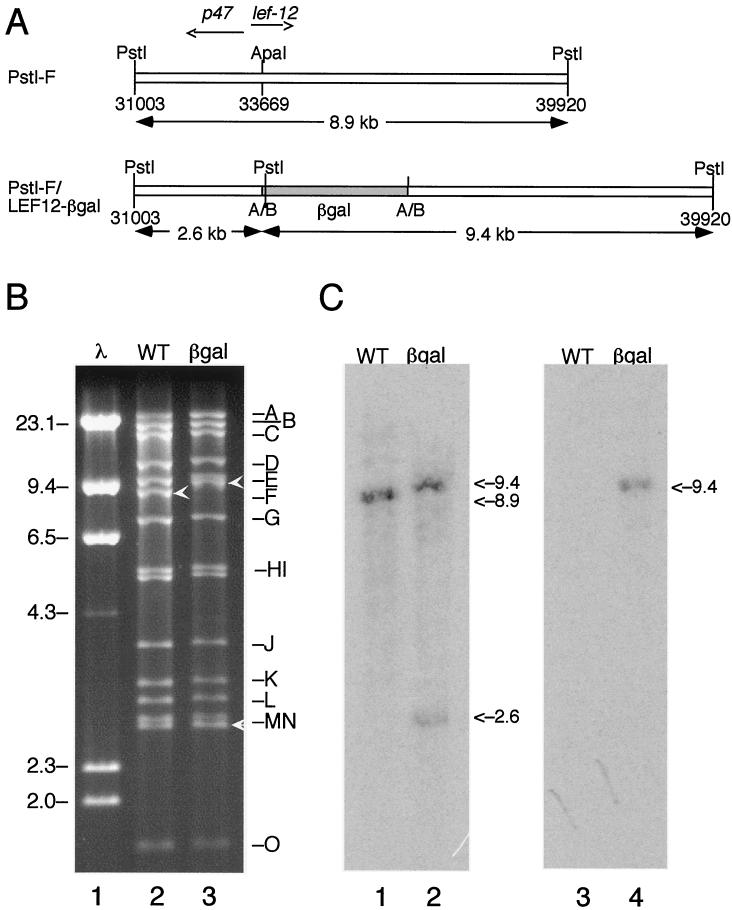
FIG. 1.
Construction of vLEF12-βgal. (A) Strategy. The recombinant virus vLEF12-βgal encodes the first 96 amino acids of LEF-12 fused in frame with β-galactosidase. This virus was constructed by in vivo recombination between wild-type viral DNA and a plasmid containing the β-galactosidase gene (shaded) inserted into an ApaI site within the open reading frame of lef-12. This virus was selected as a blue plaque recombinant when plated in the presence of X-Gal. The positions of the ApaI sites converted to BglII sites are indicated by A/B. The genome positions of the PstI and ApaI sites are indicated. (B) Restriction enzyme digest. DNA purified from wild-type virus (lane 2) and vLEF12-βgal virus (lane 3) was digested with PstI, and fragments were separated on a 0.8% agarose gel. Lambda DNA digested with HindIII was run in an adjacent lane as molecular size markers (lane 1). The locations of the genomic fragments are indicated on the right, and the sizes of the relevant lambda markers are indicated on the left (in kilobases). The PstI-F fragment in wild-type virus and the two PstI-F subfragments in the mutant virus are indicated by white arrowheads. (C) Southern blot. Duplicate sets of digests were transferred to nitrocellulose membranes and separately probed with uniformly radiolabeled PstI-F genomic DNA fragment (lanes 1 and 2) and β-galactosidase DNA (lanes 3 and 4).
The viral DNA was digested with PstI and separated on 0.8% agarose gel (Fig. 1B). Visualization of the ethidium bromide-stained gel revealed that the PstI-F band was missing in the mutant DNA. Digestion at the PstI site within the β-galactosidase gene produced two fragments containing portions of PstI-F. This was expected because cloning of the β-galactosidase gene into the lef-12 open reading frame resulted in the insertion of an additional PstI site immediately downstream of the insertion site. One of the new fragments, which represents the right portion of the PstI-F fragment fused to the β-galactosidase gene, has a predicted size of 9.4 kb, and it migrated more slowly than PstI-F at 8.9 kb. The other fragment, which comigrated with the PstI-N band at approximately 2.6 kb, represents the right portion of the PstI-F fragment.
The correct assignment of the two restriction fragments was confirmed by Southern blot analysis (Fig. 1C). Probing of the digested DNAs from the mutant virus with the PstI-F fragment labeled both the 9.4-kb band and the 2.6-kb band and only PstI-F in the wild-type viral DNA. Radiolabeled β-galactosidase fragment hybridized only to the larger 9.4-kb band in the mutant viral DNA. As expected, the β-galactosidase probe did not hybridize to the wild-type viral DNA, and the PstI-F probe hybridized only to the PstI-F fragment. The particular viral isolate represented in Fig. 1 was named vLEF12-βgal.
The fact that we were able to rescue lef-12 mutant virus from T. ni cells indicated that this protein is not essential for replication of the virus in this cell line. To determine whether functional LEF-12 was required for replication in S. frugiperda Sf9 cells, the two cell lines were infected separately with the vLEF12-βgal and wild-type viruses at a multiplicity of infection of 2. Virus supernatants were harvested from both cell lines at 48 h postinfection, and viruses were counted on T. ni cells. These experiments revealed that mutant virus replicated in both cell lines, although the yields of mutant virus were 60 to 70% of those of wild-type virus; there were 1.4 × 108 ± 0.16 × 108 and 1.0 × 108 ± 0.08 × 108 wild-type viruses on T. ni and S. frugiperda cells, respectively, versus 8.2 × 107 ± 1.2 × 107 and 7.4 × 107 ± 1.0 × 107 mutant viruses, respectively (means ± standard error of the mean). This result indicates that LEF-12 is not essential for replication of AcNPV in either Sf9 or High 5 cells, although it may be required for optimal replication.
Construction of vLEF12-ΔATG.
The finding that viruses with an interrupted lef-12 gene were viable in S. frugiperda cells was somewhat surprising because lef-12 is required for transient late gene expression in that cell line (18, 25). We considered the possibility that the LEF-12-β-galactosidase fusion protein had residual LEF-12 activity, because the β-galactosidase portion was inserted after residue 91 of the 181-amino-acid protein. This seemed unlikely, because a frameshift mutant constructed at the ApaI site had previously been shown to be incapable of stimulating late gene expression (18, 25), but we believed that our results would be strengthened by construction of additional mutants.
The promoter for the adjacent p47 gene is contained within the lef-12 open reading frame, and therefore it did not seem reasonable to delete the N terminus of lef-12 or to insert the β-galactosidase gene closer to the N terminus. p47 is a component of the virus-encoded RNA polymerase and is essential for replication of the virus in infected cells and for transient expression of late genes (7, 15, 30). Therefore, we decided to alter the initiating methionine codon for lef-12. The N terminus of the lef-12 open reading frame encodes Met-Thr-Met. Since we were not sure which of the methionine codons was used for initiation, both were changed by site-directed mutagenesis, with concurrent insertion of a HindIII restriction enzyme site at the 5′ end of the open reading frame (Fig. 2A). This changed the first three codons to Ile-Ser-Leu, and therefore LEF-12 protein should not be produced in the absence of a methionine initiation codon. These substitutions did not affect the coding sequences for p47, although the N termini of lef-12 and p47 overlap by 2 nucleotides.
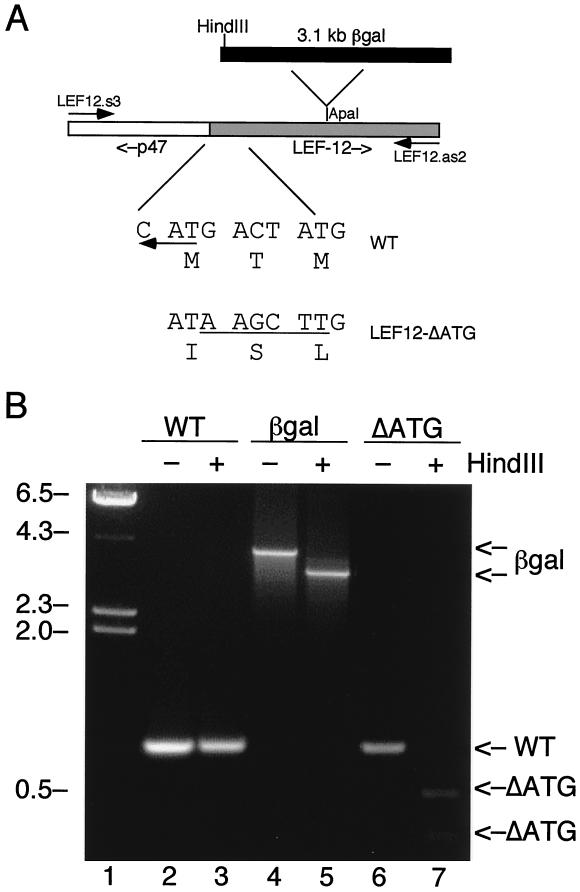
FIG. 2.
Construction of vLEF12-ΔATG. (A) Strategy. A set of PCR primers flanking the lef-12 gene were used to amplify DNA prepared from the wild-type, vLEF12-βgal, and vLEF12-ΔATG viruses. The diagram in the middle shows the orientation of the lef-12 gene and an upstream fragment of the p47 gene in the PCR product. The diagram at the top indicates the position of insertion of the β-galactosidase gene in LEF12-βgal DNA. The β-galactosidase gene is not drawn to scale. The sequences below the diagram show the nucleotide and predicted amino acid sequence of the 5′ ends of the lef-12 open reading frame in the wild-type and LEF12-ΔATG mutant viruses. The leftward-pointing arrow indicates the start codon for p47, which is encoded on the opposite strand from lef-12. (B) PCR analysis of wild-type (WT), LEF12-βgal (lane βgal), and vLEF12-ΔATG (lane ΔATG). Viral DNAs were amplified by PCR. The reaction mixes were split into two parts, and one part was digested with HindIII. Reaction products were separated on 2% agarose gels and stained with ethidium bromide. The positions of the LEF-12 PCR products and HindIII digestion products are indicated on the right. λ DNA digested with HindIII was run as molecular size markers (first lane), and the positions of the λ fragments are indicated on the left (in kilobases).
This virus, called vLEF12-ΔATG, was constructed by homologous recombination between a plasmid containing the desired mutation and vLEF12-βgal DNA linearized at the unique Bsu36I site within the β-galactosidase gene. Plaques that were colorless in the presence of X-Gal were picked and amplified. Viral DNA was screened by PCR and restriction digestion with HindIII (Fig. 2B). A primer set that was predicted to amplify a fragment of 865 bp containing the complete lef-12 open reading frame and 322 bp of upstream sequences was used. With wild-type viral DNA and vLEF12-ΔATG DNA, an 865-bp band was obtained, as expected. The migration of the wild-type DNA fragment was not altered after digestion with HindIII, as expected because there are no HindIII sites in this region of the AcNPV genome. But with vLEF12-ΔATG DNA, HindIII cut the 865-bp fragment into two smaller bands of approximately 320 and 540 bp, as expected due to the insertion of a HindIII site in the mutant. The same primer set amplified a 4-kb fragment on vLEF12-βgal DNA. Digestion of this PCR product with HindIII produced two fragments of 3.4 and 0.6 kb due to the presence of a HindIII site at the 5′ end of the β-galactosidase gene.
A virus with a wild-type copy of lef-12 was constructed from vLEF12-βgal to use as a revertant control. This was constructed by homologous recombination of pPstI-F and vLEF12-βgal DNA linearized at the unique BsuI site. Plaques that were colorless in the presence of X-Gal were picked and screened by PCR and HindIII digestion as described above. A plaque with the same size of PCR fragment as wild-type virus was named vLEF12-R (data not shown).
Single-step growth curves of mutant virus.
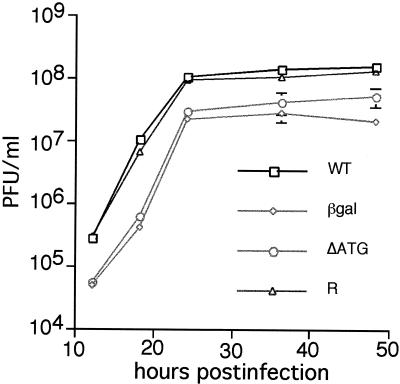
To examine the kinetics of viral infection, all four viruses were propagated and titrated in Sf9 cells, and then Sf9 cells were separately infected with wild-type and mutant viruses at an MOI of 5 (Fig. 3). Infected cells were harvested at 12, 18, 24, 36, and 48 h postinfection, and virus in the supernatant was counted on Sf9 cells. As shown in Fig. 3, the rates of virus production were equivalent in cells infected with the wild-type and vLEF12-R viruses. The titers for the two mutant viruses were three- to fivefold lower at all times postinfection than those for the wild-type virus. This indicates that the mutant viruses were viable, although impaired in virus production.
FIG. 3.
Single-step growth curves. S. frugiperda cells were infected with wild-type (WT), vLEF12-βgal (βgal), vLEF12-ΔATG (ΔATG), or vLEF12-R (R) virus at an MOI of 5. Cells were harvested at the indicated times, and viruses in the supernatants were counted on S. frugiperda cells. Titers represent means of duplicate determinations of triplicate infections. Standard deviations were plotted for every point, although most of the error bars are smaller than the data points.
Late gene expression in the absence of LEF-12.
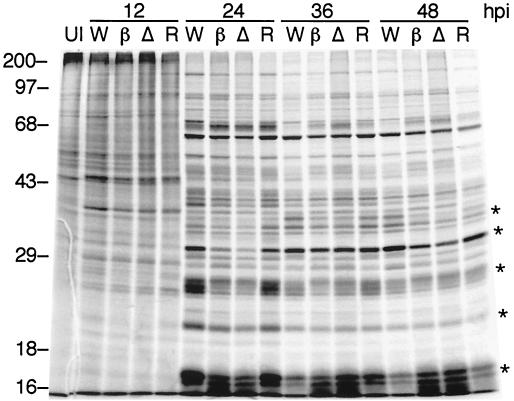
Most of the viral structural proteins are encoded by late genes. Therefore, the titration experiments suggested that cells infected with the lef-12 mutant virus entered the late phase and synthesized late proteins with normal or nearly normal kinetics. To directly address this issue, radiolabeling experiments were performed. Sf9 cells were separately infected with mutant or wild-type virus and pulse-labeled for 4 h prior to harvest at 12, 24, 36, or 48 h postinfection. Analysis of SDS-polyacrylamide gels revealed that the patterns of late protein synthesis were similar, though not identical, in wild-type and mutant virus-infected cells (Fig. 4). At 24 h, several highly expressed late proteins were synthesized at lower levels in mutant-infected cells than in the cells infected with the wild-type or the reconstructed wild-type virus. These differences were less obvious at 36 and 48 h.
FIG. 4.
Kinetics of late protein synthesis. S. frugiperda cells were infected with wild-type virus (lanes W), vLEF12-βgal (lanes β), vLEF12-ΔATG (lanes Δ), or vLEF12-R (lanes R). Cells were pulse-labeled with a mixture of [35S]methionine and [35S]cysteine for 4 h prior to the time of harvest (indicated at the top of the gel; hpi, hours postinfection). Total proteins were resolved on 12% acrylamide-SDS gels. Uninfected cells are shown in lane UI. The migration of prestained molecular size markers is shown on the left. The sizes of the markers are indicated in kilodaltons.
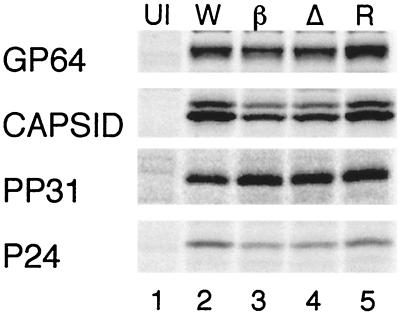
To specifically examine the synthesis of particular proteins, radioimmunoprecipitation experiments were performed with antibodies raised against several highly expressed late proteins (Fig. 5). Significant decreases in expression were observed for P39 (ORF89) capsid protein and P24 (ORF129) capsid-associated protein (Table 1). Both of these genes are transcribed exclusively during the late phase (2). On the other hand, PP31 (ORF36) and GP64 (ORF128) were expressed at wild-type levels in mutant-infected cells. pp31 and gp64 are regulated by tandem early and late promoters (4, 14). At 24 h postinfection, most of the pp31 and gp64 mRNAs originate from their respective late promoters, although early transcripts are still present and are relatively stable throughout the late phase of infection. The presence of early transcripts may account for the higher levels of expression of PP31 and GP64 as opposed to the strictly late proteins, P39 and P24.
FIG. 5.
Radioimmunoprecipitation of viral proteins. Cells were labeled and harvested at 24 h postinfection as described in the legend to Fig. 4. Individual panels show precipitation of the specific viral proteins indicated on the left.
TABLE 1.
Radioimmunoprecipitation of viral late proteinsa
| Protein | % of wild-type radioactivity
|
||
|---|---|---|---|
| vLEF12-βgal | vLEF12-ΔATG | Revertant | |
| gp64 | 93 ± 4 | 96 ± 3 | 106 ± 6 |
| p39/capsid | 46 ± 15 | 52 ± 5 | 89 ± 5 |
| pp31 | 107 ± 11 | 86 ± 12 | 97 ± 11 |
| p24 | 58 ± 8 | 60 ± 8 | 88 ± 19 |
S. frugiperda cells were separately infected with four different viruses and pulse-labeled with [35S]methionine from 20 to 24 h postinfection. Viral proteins were immunoprecipitated and separated by SDS-polyacrylamide gel electrophoresis. Gels were quantitated by PhosphorImager analysis, using the object average of the uninfected sample to subtract background. Values are expressed as percentages of the wild-type value. The values shown here represent means and standard deviations of triplicate determinations.
Production of occluded virus in mutant-infected cells.
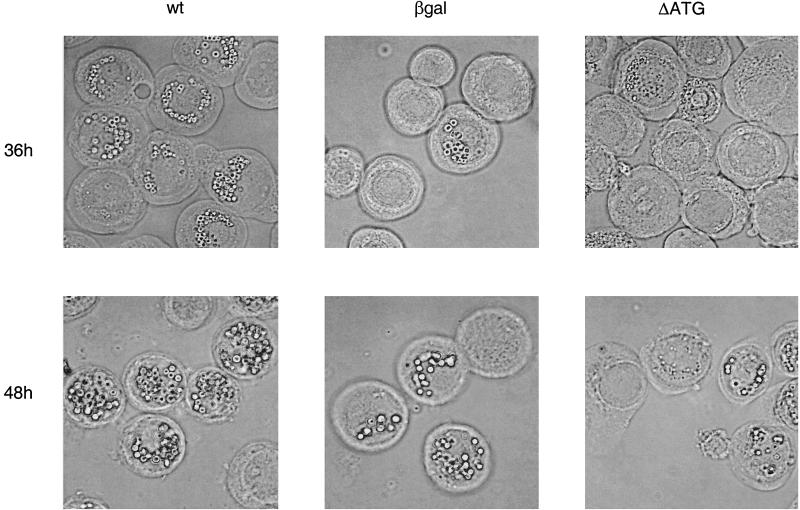
To monitor production of occluded virus, which is dependent upon expression of very late genes, infected cells were examined under a light microscope (Fig. 6). At 36 h postinfection, 75% of cells infected with wild-type virus were filled with polyhedra. In mutant virus-infected cells, only 33% of the cells contained polyhedra, and those contained fewer polyhedra per cell than wild-type-infected cells. In most of the cells lacking polyhedra, the virogenic stroma was clearly evident. This indicates that the cells were infected but were progressing through the infection cycle more slowly than the wild-type-infected cells. At 48 h postinfection, 86% of cells infected with the vLEF12-βgal virus contained polyhedra, 90% of cells infected with vLEF12-ΔATG contained polyhedra, and 96% of the wild-type virus-infected cells contained polyhedra. The mutant-infected cells had fewer polyhedra per cell even at this late time, although most cells contained numerous polyhedra, which made it difficult to quantitate the number of polyhedra per cell. Cells infected with the reconstructed (revertant) wild-type virus had approximately the same percentage of cells with polyhedra as the wild-type-infected cells at 36 and 48 h postinfection (data not shown).
FIG. 6.
Very late protein synthesis in LEF-12 mutant virus. Cells were infected in 60-mm dishes with wild-type (wt), vLEF12-βgal (βgal), or vLEF12-ΔATG (ΔATG). At 36 and 48 h postinfection, cells were harvested, concentrated, spotted onto glass slides, and photographed with a Zeiss Axiophot video camera system.
Immunodetection of LEF-12 expression in infected cells.
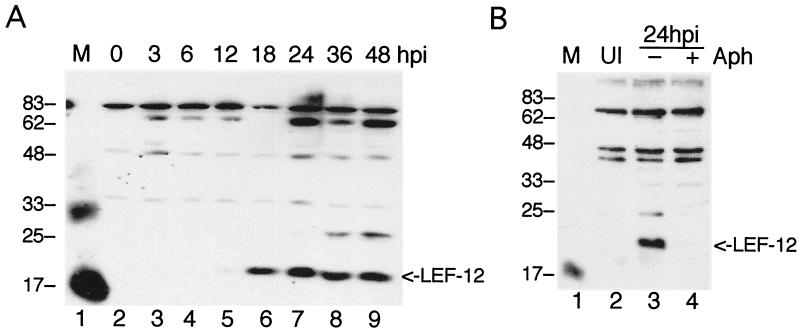
To confirm that the mutant viruses were not able to direct synthesis of LEF-12, antiserum was raised in rabbits against protein produced in bacteria. This antiserum was first used to examine the profile of LEF-12 synthesis in cells infected with wild-type virus. Infected cells were harvested at different times postinfection, and proteins were separated on 12% acrylamide-SDS gels (Fig. 7A). A very weak LEF-12 signal was detected at 12 h postinfection, and this increased in intensity by 18 h postinfection. Accumulation of LEF-12 peaked at 24 h postinfection and then declined somewhat at later times.
FIG. 7.
Western blot analysis of LEF-12. (A) Time course of LEF-12 protein synthesis. Extracts of soluble proteins from S. frugiperda cells infected with wild-type virus were harvested at the indicated times (hours) postinfection, separated on 12% gels, transferred to nitrocellulose membranes, and probed with rabbit antibody raised against LEF-12. (B) Expression of LEF-12 requires viral DNA synthesis. Cells were infected with wild-type virus in the presence of aphidicolin (Aph, lane 4) or an equal concentration of DMSO (lane 3) and harvested at 24 h postinfection (hpi). Molecular size markers were run in lane M of each panel, and the positions and sizes of the markers are indicated on the left (in kilodaltons). Antisera from two different rabbits were used in this figure, and therefore some of the background bands are different. Both antisera cross-reacted with the 17,000-molecular-weight marker (lysozyme); only one reacted with the 33,000-molecular-weight marker (triosephosphate isomerase). The position of LEF-12 is indicated on the right. The blot was developed with SuperSignal West Femto substrate (Pierce). UI, uninfected cells.
These kinetics of expression suggested that lef-12 was expressed during the late phase of infection. To directly test whether LEF-12 expression was dependent on viral DNA replication, cells were infected and then grown for 24 h in the presence of 5 μg of aphidicolin per ml (Fig. 7B). LEF-12 expression was detected in control cells, which were exposed to the solvent dimethyl sulfoxide (DMSO) only, but there was no LEF-12 signal in the aphidicolin-treated cells. Aphidicolin is an inhibitor of the virus-encoded DNA polymerase, and therefore this result indicates that LEF-12 synthesis is dependent on viral DNA replication.
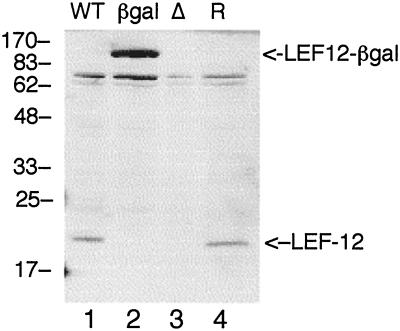
Immunoblot analysis with LEF-12 antiserum was performed to confirm that mutant viruses did not synthesize LEF-12 protein (Fig. 8). Soluble extracts were prepared from the mutant virus-infected cells at 24 h postinfection, at which time synthesis of LEF-12 protein was maximal in the wild-type virus infection. A large protein with a molecular weight of approximately 100,000 was detected in the cells infected with vLEF12-βgal. This is the expected position for the fusion protein of β-galactosidase with LEF-12. The intensity of this band was much stronger than that of the wild-type LEF-12 protein, suggesting that the fusion protein was more stable than native LEF-12. No LEF-12 signal was detected in cells infected with the LEF12-ΔATG mutant. There was also no indication that shorter versions of LEF-12 were produced in cells infected with the vLEF12-ΔATG mutant.
FIG. 8.
Analysis of LEF-12 expression in mutant virus-infected cells. S. frugiperda cells were infected with wild-type (WT), vLEF12-βgal (βgal), vLEF12-ΔATG (Δ), or vLEF12-R (R) virus and harvested at 24 h postinfection. LEF-12 was detected by immunoblot analysis as described in the legend to Fig. 7, except that the blot was developed with the ECL kit (Amersham). The positions and sizes of molecular size markers are indicated on the left (in kilodaltons). The positions of LEF-12 and the LEF-12-β-galactosidase fusion are indicated on the right.
Truncated LEF-12 could potentially arise from initiation of translation at downstream methionine codons. The lef-12 open reading frame has two in-frame methionines, 22 and 23 residues downstream of the native start site. Initiation at either methionine is predicted to encode proteins with molecular weights of 18,615 and 18,484, respectively. If produced, these proteins would migrate slightly slower than the 17,000-molecular-weight marker, yet they were not observed. The signal from vLEF12-R was equal in intensity to that of wild-type virus, confirming that the lack of LEF-12 synthesis was directly related to the mutations that were introduced.
Primer extension analysis of LEF-12.
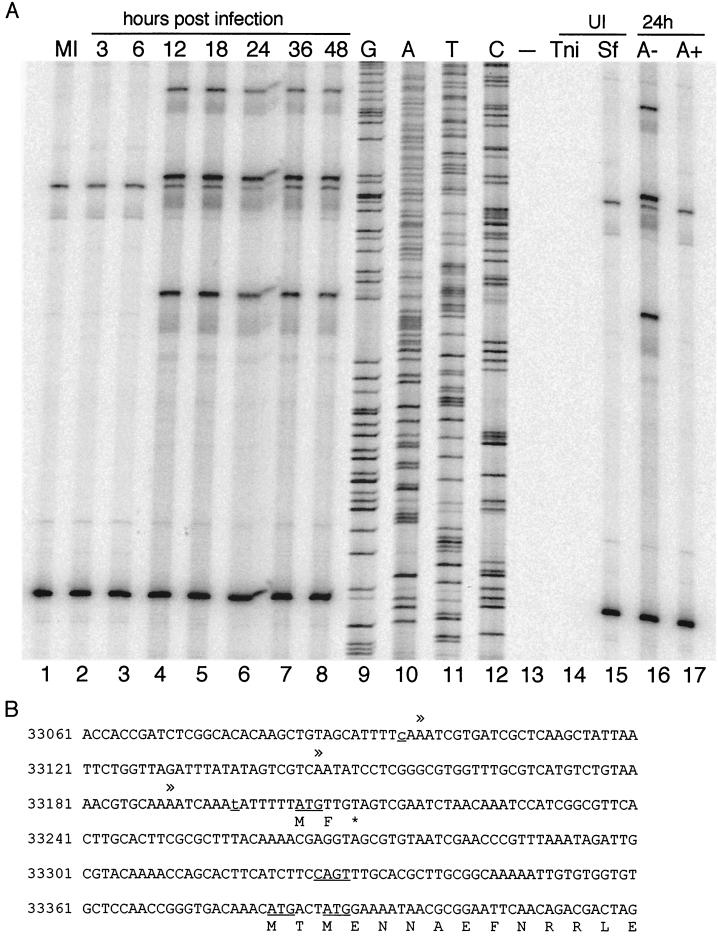
The AcNPV lef-12 gene was predicted to be expressed as an early gene due to the presence of an early initiator element and the absence of late transcription motifs in the promoter-proximal region. To identify the start site and also to confirm the kinetics of lef-12 transcription, primer extension analysis was performed (Fig. 9A). The primer used in this experiment was lef12.gsp2, which corresponds to positions 33453 to 33434 of the AcNPV map. Two extension products were observed with RNAs harvested during the early phase of infection and also in the mock-infected cells. The intensity of each band was constant throughout the time course, indicating that RNA levels were constant. The fact that these bands were detected in the mock-infected cells suggested that they arose from a host RNA that is homologous to the lef-12 primer.
FIG. 9.
Primer extension analysis of LEF-12 transcription. (A) Primer extension reactions. Total cellular RNA was isolated from wild-type AcNPV-infected S. frugiperda cells at the indicated times postinfection (lanes 1 to 8). The 5′ ends of LEF-12 transcripts were mapped by primer extension analysis with lef12.gsp2. The sequencing ladder was generated with the same primer (lanes 9 to 12). The samples in lanes 14 to 17 were harvested at 24 h after mock infection of T. ni cells (Tni, lane 14) or S. frugiperda (Sf) cells (lane 15) or infection in the presence of aphidicolin (5 μg/ml) (A+, lane 17) or an equal concentration of DMSO (A−, lane 16). Lane 13 shows the products of a primer extension reaction conducted in the absence of added RNA. The sequence ladder is antisense relative to the sequence of the lef-12 promoter shown in panel B. (B) Sequence of the lef-12 promoter. The transcriptional start sites mapped by primer extension are indicated by double arrowheads above the sequence. The N-terminal sequence of LEF-12 is indicated by the single-letter amino acid code. The ATG codon for a short minicistron downstream of the 5′-proximal start site is underlined, as are the two ATG codons at the N terminus of the lef-12 open reading frame. Two nucleotide substitutions noted for the E2 isolate of AcNPV compared to the published AcNPV sequence are indicated by lowercase underlined residues. The 5′-most substitution is a silent mutation in the p47 open reading frame, and the other results in a substitution of isoleucine for methionine at codon 62 in the p47 open reading frame. The Bombyx mori nuclear polyhedrovirus p47 gene encodes isoleucine at position 62, and therefore it is likely to be a functional mutation.
A similar pattern of host RNAs was obtained with another lef-12 primer, called lef12.pe2, which corresponds to nucleotides 33590 to 33483. In this case, three different primer extension products were observed, also with equal intensities at each time point (data not shown). Extension products were not produced with uninfected T. ni cell RNA as template (Fig. 9A, lane 14) or with tRNA as template (Fig. 9A, lane 13). The RNAs used for primer extension reactions shown in lanes 12 to 17 were purified by affinity chromatography, while the time course RNAs (Fig. 9A, lanes 2 to 8) were purified by CsCl centrifugation. In addition, older preparations of RNA, which have been used successfully in the past for primer extension mapping of other viral genes, also gave identical results (data not shown) (15, 26). Together, these controls suggest that the primer extension products obtained with uninfected RNAs suggest the presence of an S. frugiperda gene that is homologous to the lef-12 primers and not contamination of RNA preparations.
Virus-specific RNAs initiating upstream of the lef-12 open reading frame were not detected until 12 h postinfection. Extended products of three different lengths were observed, and the intensities of each were relatively constant from 12 to 48 h postinfection. This timing of transcription was consistent with classification of lef-12 as a late gene, and this was further confirmed by analysis of a culture infected in the presence of 5 μg of aphidicolin per ml and harvested at 24 h postinfection.
The virus-specific extension products were compared with a sequencing ladder primed with the same oligonucleotide (Fig. 9B, lanes 9 to 12). This analysis revealed that the early CAGT initiator element, at map positions 33326 to 33329, was not used for transcription. Rather, the results suggest that RNA synthesis initiated at three distinct sites further upstream, at map positions 33095, 33146, and 33190. This predicts that the 5′ leader of lef-12 mRNAs is relatively long for a baculovirus gene and also indicates the presence of a short minicistron upstream of the lef-12 open reading frame. Primer extension analyses performed with lef12.pe2 as the primer also produced three virus-specific extension products with identical kinetics of expression (data not shown). These transcripts mapped to the same position on the viral genome as the transcripts mapped with lef12.gsp2.
The primer extension reactions indicate that lef-12 RNAs are transcribed during the late phase but do not correspond to canonical late transcription start sites. To confirm that RNAs initiated at these locations and were not produced by RNA splicing, 5′ rapid amplification of cDNA ends (RACE) was conducted with RNA harvested at 24 h postinfection. Sequencing of 10 5′ RACE clones identified cDNAs with 5′ ends corresponding to the promoter-proximal start site at position 33190. In addition, one clone with a 5′ end mapping upstream of the p47 open reading frame was identified, as well as several shorter clones that may represent incomplete extension products.
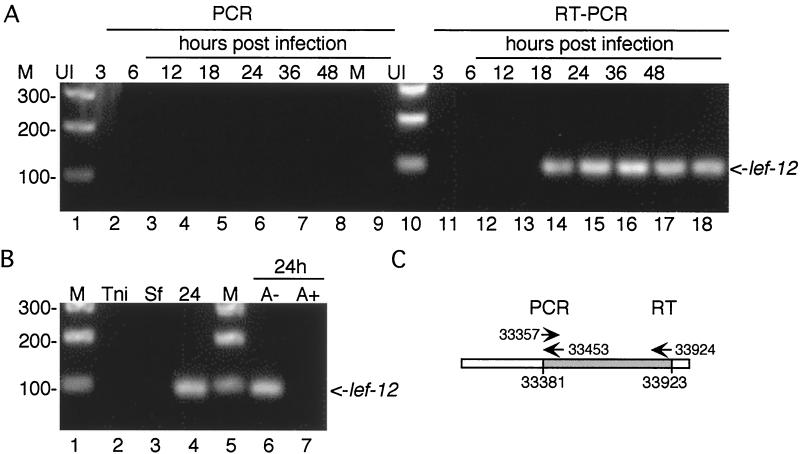
RT-PCR analysis of LEF-12 transcripts.
To further confirm that the primer extension products obtained with RNAs from uninfected and mock-infected S. frugiperda cells were really due to host RNAs and not viral contamination, RT-PCR analyses were performed. The rationale was that we should be able to identify a 5′ primer that hybridized with viral DNA but not with the homologous host gene, thereby generating a specific RT-PCR product with infected cells but not uninfected cells. To this end, RNAs from the time course experiment shown in Fig. 9 were first extended with lef12.pe2 and reverse transcriptase and then amplified by PCR with lef12.gsp2 and lef12.5prime (Fig. 10). Agarose gel analysis of the RT-PCR products revealed the presence of the predicted 95-bp product during the late phase of viral infection. No bands were observed in reactions primed with RNAs prepared from uninfected T. ni cells or with uninfected or mock-infected S. frugiperda cells.
FIG. 10.
RT-PCR analysis of LEF-12 expression. (A) Time course analysis. The same RNA preparations used for primer extension in Fig. 9 were subjected to RT-PCR. In lanes 2 to 9 and 11 to 18, cells were harvested at the indicated times postinfection. Lanes 2 to 9 are controls showing PCR products in the absence of reverse transcription. (B) Mock-infected and aphidicolin-treated cells. RT-PCR of RNAs prepared from mock-infected T. ni (lane 2), mock-infected S. frugiperda cells (lane 3), infected S. frugiperda cells at 24 h postinfection (lane 4), and infected S. frugiperda cells at 24 h postinfection in the presence of DMSO (lane 6) and 24 h postinfection in the presence of aphidicolin (lane 7). (C) Diagram of 1,000 bp surrounding the lef-12 open reading frame, showing the primers used for RT-PCR. The 5′ and 3′ ends of the open reading frame are shown below the line, and the 5′ ends of the primers are shown above.
RNAs prepared during the early phase of infection were also not capable of directing the synthesis of a specific RT-PCR product, as were cells infected in the presence of aphidicolin. Lanes 2 to 9 served as controls to verify that PCR amplification was dependent upon reverse transcription, indicating that the RNA preparations were not contaminated with viral DNA. These results also serve to confirm that lef-12 is not transcribed during the early phase of viral infection or in the presence of aphidicolin. The highly sensitive RT-PCR should have detected even very small amounts of transcript that are below the level of detection by primer extension.
DISCUSSION
The results presented here indicate that the lef-12 gene of AcNPV is not essential for replication of virus in tissue culture cells. This finding was unexpected because it has been reported that LEF-12 is required for gene expression from a late viral promoter in transient assays (18, 25). One possibility that we considered was that the β-galactosidase insertion construct retained partial activity. LEF-12 has 181 amino acids, and the N-terminal 96 amino acids should be synthesized and fused to β-galactosidase. The insertion was made at an ApaI site, which had previously been used to construct lef-12 frameshift mutants that lacked activity (18, 25). This would argue that the N-terminal 96 amino acids were not functional in the absence of the C-terminal portion of the protein. But there may be an essential difference between these two types of mutations (β-galactosidase insertion versus premature termination) with respect to the stability of the proteins. We observed that the LEF-12-β-galactosidase fusion protein was stable and accumulated to higher levels than authentic LEF-12. The fate of the LEF-12 truncation was not analyzed, and it may have been degraded (18, 25).
Therefore, an additional mutant was made in which the initiating methionine residues were removed, which should prevent the synthesis of LEF-12 protein in the mutant virus vLEF12-ΔATG. Both of these mutants replicated in tissue culture cells, with only a modest decrease in overall yields of virus. Furthermore, analysis of viral protein synthesis revealed that the mutants produced late proteins at normal or 50% reduced levels, indicating that late protein synthesis was not significantly impaired. Viruses were also occluded in the nucleus, although at a decreased rate compared to the wild type.
Together, these mutants provide compelling evidence that the lef-12 gene is not essential for replication of AcNPV or for expression of late genes. This suggests that another viral protein may be functionally redundant with LEF-12, and therefore it is not required in the context of the whole genome. This hypothesis is consistent with previous results showing that LEF-12 was stimulatory but not essential for late gene expression when the LEFs were supplied on large genomic fragments (18, 25). It was only when the individual LEFs were supplied on separate plasmids, each containing only a single open reading frame, that LEF-12 was absolutely required.
Another possibility is that the requirement for LEF-12 is limited to certain late genes, like vp39 capsid, which was the late promoter used in both previous transient-expression screens (18, 25). We examined the effects of LEF-12 on the expression of four late genes. Only expression of VP39 capsid and P24 was significantly lower in the mutant-infected cells than in the wild-type virus-infected cells. The other two proteins, PP31 and GP64, were expressed at wild-type levels in the absence of LEF-12. PP31 is an abundant viral protein that forms the nuclear matrix, and GP64 is the major membrane protein of budded virus. Both pp31 and gp64 are expressed from tandem early and late promoters (4, 14). It has previously been shown that at 24 h postinfection, when the samples were harvested, the late RNAs for pp31 and gp64 are far more abundant than the early messages. Therefore, synthesis of PP31 and GP64 should primarily reflect levels of late mRNAs for these genes, although the early transcripts are presumably active and likely contribute to protein synthesis. Both vp39 and p24 are expressed solely from late promoter motifs (2, 3, 28, 31), and it is possible that they were more severely affected because late transcription is influenced by LEF-12, while early transcription is independent of LEF-12. Further understanding of the mechanism of LEF-12 function should help to define its role in the expression of viral late genes.
The yields of infectious virus were 20 to 40% of the level obtained with wild-type and revertant viruses. This is roughly equivalent to the decrease in synthesis of the capsid and P24 proteins. A more dramatic effect on the rate of virus occlusion in the nucleus was noted. A decrease in both the number of cells with polyhedra and the number of polyhedra per cell was evident at 36 h postinfection. At 48 h postinfection, the number of mutant-infected cells with polyhedra was only slightly less than the number of wild-type-infected cells with polyhedra, although the number of polyhedra per cell was still lower. Occlusion of virus particles into polyhedra is the final step in virus infection and thus is sensitive to any mutations that decrease the rate of viral protein synthesis as well as the rate of virus assembly.
Immunoblot analysis of LEF-12 expression revealed that protein did not accumulate to detectable levels until 18 h postinfection. Furthermore, LEF-12 was not detected when cells were infected in the presence of aphidicolin, an inhibitor of viral DNA synthesis. Together, these results suggest that lef-12 should be classified as a late gene. This conclusion conflicts with earlier predictions that lef-12 would be expressed as an early gene due to the presence of a CAGT initiator element in the proximal upstream region (25). These authors also noted that the nearest late transcription motif (TAAG) is 711 bp upstream of the lef-12 initiation codon. Transcripts initiating at this point are unlikely to direct synthesis of LEF-12 because there are numerous AUG codons between this TAAG motif and the lef-12 initiation codon. Northern blot analyses of the p47/lef-12 region in Choristoneura fumiferana also suggested that lef-12 was expressed as an early gene in that virus, although aphidicolin and primer extension mapping experiments were not done to confirm the timing of expression or the start site (17).
To directly analyze the kinetics of lef-12 transcription in AcNPV-infected cells, we performed primer extension assays. Transcripts extending through the lef-12 open reading frame were not detected until 12 h postinfection. The levels of transcript were constant through 48 h postinfection. The timing is consistent with the transcription pattern of other viral late genes. Furthermore, we found that the early CAGT motif upstream of lef-12 was not used for transcription initiation and that lef-12 was not transcribed in the presence of aphidicolin. These observations confirm the protein data and indicate that lef-12 should be classified as a late gene.
Questions still remain concerning the mechanism of lef-12 transcription. The transcription initiation sites do not correspond to late promoter motifs. The sequence A/GTAAG is believed to be the major determinant for transcription initiation by the virus-encoded RNA polymerase and has been found at the 5′ ends of most viral late RNAs. While the noncanonical start site of lef-12 is unexpected, it may help to explain the observations of Li et al. (18), who found that a region approximately 2.5 kb downstream of lef-12 was required for optimal activity. This downstream region was apparently not due to the presence of an open reading frame, but rather appeared to be a nucleotide sequence that increased expression of lef-12. Activation of expression by very distal sequences has not been reported previously for any baculovirus genes. These observations of Li et al., as well as the transcriptional mapping presented here, indicate that the regulation of lef-12 expression differs from that of other previously described baculovirus late genes.
Eukaryotic ribosomes usually initiate translation at the 5′-proximal AUG. LEF-12 mRNAs are predicted to contain a short 5′ open reading frame. A number of baculovirus clones with minicistrons upstream of open reading frames have been identified. The minicistrons apparently have a regulatory function and affect the efficiency of translation initiation (8).
The finding that lef-12 is expressed as a late gene is consistent with our conclusion that lef-12 is not essential for expression of late genes. Baculovirus DNA is infectious, and thus, it is obvious that late genes cannot be essential for their own expression. Most of the other transcription LEFs have been analyzed for their expression patterns, and their transcripts were detected by 3 to 6 h postinfection and protein by 4 to 8 h postinfection (7, 11, 15, 19, 23). Yet expression of two highly expressed late genes was lower in the mutant-infected cells, indicating that LEF-12 does have an effect on late gene expression. The exact mechanism of its action and whether it affects transcription or translation remain to be determined.
Acknowledgments
This research was supported by a grant from the National Science Foundation (MCB 0110925).
We thank April Peterson and Kristin Garcia, who were supported by REU supplements, for assistance during the early phases of this research.
REFERENCES
- 1.Ahrens, C. H., and G. F. Rohrmann. 1995. Replication of Orgyia pseudotsugata baculovirus DNA: lef-2 and ie-1 are essential and ie-2, p34, and Op-iap are stimulatory genes. Virology 212:650-662. [DOI] [PubMed] [Google Scholar]
- 2.Blissard, G. W., R. L. Quant-Russell, G. F. Rohrmann, and G. S. Beaudreau. 1989. Nucleotide sequence, transcriptional mapping, and temporal expression of the gene encoding p39, a major structural protein of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata. Virology 168:354-362. [DOI] [PubMed] [Google Scholar]
- 3.Blissard, G. W., and G. F. Rohrmann. 1991. Baculovirus gp64 gene expression: analysis of sequences modulating early transcription and transactivation by IE1. J. Virol. 65:5820-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170:537-555. [DOI] [PubMed] [Google Scholar]
- 5.Carson, D. D., L. A. Guarino, and M. D. Summers. 1988. Functional mapping of an AcNPV immediately early gene which augments expression of the IE-1 trans-activated 39K gene. Virology 162:444-451. [DOI] [PubMed] [Google Scholar]
- 6.Carson, D. D., M. D. Summers, and L. A. Guarino. 1991. Transient expression of the Autographa californica nuclear polyhedrosis virus immediate-early gene IE-N is regulated by three viral elements. J. Virol. 65:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carstens, E. B., A. L. Lu, and H. L. Chan. 1993. Sequence, transcriptional mapping, and overexpression of p47, a baculovirus gene regulating late gene expression. J. Virol. 67:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, M. J., and G. W. Blissard. 1997. Baculovirus gp64 gene expression: negative regulation by a minicistron. J. Virol. 71:7448-7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. J., and S. M. Thiem. 1997. Differential infectivity of two Autographa californica nucleopolyhedrovirus mutants on three permissive cell lines is the result of lef-7 deletion. Virology 227:88-95. [DOI] [PubMed] [Google Scholar]
- 10.Clem, R. J., M. Fechheimer, and L. K. Miller. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388-1390. [DOI] [PubMed] [Google Scholar]
- 11.Durantel, D., G. Croizier, M. Ravallec, and M. Lopez-Ferber. 1998. Temporal expression of the AcMNPV lef-4 gene and subcellular localization of the protein. Virology 241:276-284. [DOI] [PubMed] [Google Scholar]
- 12.Guarino, L. A., W. Dong, B. Xu, D. R. Broussard, R. W. Davis, and D. L. Jarvis. 1992. Baculovirus phosphoprotein pp31 is associated with virogenic stroma. J. Virol. 66:7113-7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino, L. A., and M. Smith. 1992. Regulation of delayed-early gene transcription by dual TATA boxes. J. Virol. 66:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarino, L. A., and M. D. Summers. 1986. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J. Virol. 57:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Lapointe, R., D. W. Back, Q. Ding, and E. B. Carstens. 2000. Identification and molecular characterization of the Choristoneura fumiferana multicapsid nucleopolyhedrovirus genomic region encoding the regulatory genes pkip, p47, lef-12, and gta. Virology 271:109-121. [DOI] [PubMed] [Google Scholar]
- 18.Li, L., S. H. Harwood, and G. F. Rohrmann. 1999. Identification of additional genes that influence baculovirus late gene expression. Virology 255:9-19. [DOI] [PubMed] [Google Scholar]
- 19.Lin, G., J. M. Slack, and G. W. Blissard. 2001. Expression and localization of LEF-11 in Autographa californica nucleopolyhedrovirus-infected Sf9 cells. J. Gen. Virol. 82:2289-2294. [DOI] [PubMed] [Google Scholar]
- 20.Lu, A., and L. K. Miller. 1995. Differential requirements for baculovirus late expression factor genes in two cell lines. J. Virol. 69:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, A., and L. K. Miller. 1996. Species-specific effects of the hcf-1 gene on baculovirus virulence. J. Virol. 70:5123-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Passarelli, A. L., and L. K. Miller. 1994. Identification and transcriptional regulation of the baculovirus lef-6 gene. J. Virol. 68:4458-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prikhod'ko, E. A., A. Lu, J. A. Wilson, and L. K. Miller. 1999. In vivo and in vitro analysis of baculovirus ie-2 mutants. J. Virol. 73:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reilly, L. M., and L. A. Guarino. 1994. The pk-1 gene of Autographa californica multinucleocapsid nuclear polyhedrosis virus encodes a protein kinase. J. Gen. Virol. 75:2999-3006. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Thiem, S. M., and L. K. Miller. 1989. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 63:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todd, J. W., A. L. Passarelli, A. Lu, and L. K. Miller. 1996. Factors regulating baculovirus late and very late gene expression in transient-expression assays. J. Virol. 70:2307-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todd, J. W., A. L. Passarelli, and L. K. Miller. 1995. Eighteen baculovirus genes, including lef-11, p35,39K, and p47, support late gene expression. J. Virol. 69:968-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolgamot, G. M., C. H. Gross, R. L. Russell, and G. F. Rohrmann. 1993. Immunocytochemical characterization of p24, a baculovirus capsid-associated protein. J. Gen. Virol. 74:103-107. [DOI] [PubMed] [Google Scholar]