Abstract
Encapsidation of retroviral RNA involves specific interactions between viral proteins and cis-acting genomic RNA sequences. Human immunodeficiency virus type 1 (HIV-1) RNA encapsidation determinants appear to be more complex and dispersed than those of murine retroviruses. Feline lentiviral (feline immunodeficiency virus [FIV]) encapsidation has not been studied. To gain comparative insight into lentiviral encapsidation and to optimize FIV-based vectors, we used RNase protection assays of cellular and virion RNAs to determine packaging efficiencies of FIV deletion mutants, and we studied replicative phenotypes of mutant viruses. Unlike the case for other mammalian retroviruses, the sequences between the major splice donor (MSD) and the start codon of gag contribute negligibly to FIV encapsidation. Moreover, molecular clones having deletions in this region were replication competent. In contrast, sequences upstream of the MSD were important for encapsidation, and deletion of the U5 element markedly reduced genomic RNA packaging. The contribution of gag sequences to packaging was systematically investigated with subgenomic FIV vectors containing variable portions of the gag open reading frame, with all virion proteins supplied in trans. When no gag sequence was present, packaging was abolished and marker gene transduction was absent. Inclusion of the first 144 nucleotides (nt) of gag increased vector encapsidation to detectable levels, while inclusion of the first 311 nt increased it to nearly wild-type levels and resulted in high-titer FIV vectors. However, the identified proximal gag sequence is necessary but not sufficient, since viral mRNAs that contain all coding regions, with or without as much as 119 nt of adjacent upstream 5′ leader, were excluded from encapsidation. The results identify a mechanism whereby FIV can encapsidate its genomic mRNA in preference to subgenomic mRNAs.
Retroviral encapsidation is the process whereby two copies of viral genomic mRNA are incorporated into the assembling virion. Because both spliced (subgenomic) and unspliced (genomic) viral mRNAs exist in an infected cell and these in turn represent only a small fraction of the total cellular RNA, a mechanism to preferentially encapsidate the full-length viral genome is required.
Expression of only the Gag polyprotein (which in the presence of the viral protease is normally processed into virion structural proteins that include the matrix, capsid, and nucleocapsid proteins) is sufficient to direct formation of retroviral particles, and these particles are competent to encapsidate viral RNA (35, 36). These and other observations establish that Gag contains the protein determinants necessary for interacting with packaging signals in retroviral RNA (for reviews, see references 7, 16, 21, 22, 34, and 37). In mammalian retroviruses, the RNA sequences that participate in these specific interactions are located in a region that encompasses variable portions of the 5′ untranslated region and usually extend into the proximal gag open reading frame (ORF) (22). The functionally defined RNA elements are often termed E or psi elements, although the term psi has also been used simply to indicate the region between the major splice donor (MSD) and the gag ORF. Inclusion within E of intronic sequence downstream of the MSD (e.g., gag) allows discrimination between spliced and unspliced genomic mRNA. Moloney murine leukemia virus (MoMLV) encapsidation adheres most straightforwardly to this model: deletion of sequences between the MSD and the start codon of gag markedly attenuates encapsidation; attachment of this region plus a contiguous proximal segment of the gag ORF (psi′) to heterologous test RNAs confers nearly wild-type levels of encapsidation (1, 5).
In human immunodeficiency virus type 1 (HIV-1) and HIV-2, as in all mammalian retroviruses so far studied, deletion of the region between the MSD and the gag start codon impairs encapsidation and blocks productive replication (2, 20, 31). Four stable RNA stem-loops, termed SL1 through SL4, are located in a 115-nucleotide (nt) sequence that spans the HIV-1 MSD (which is located in SL2) and extends into the proximal gag gene (13). SL1, SL3, and SL4, but not SL2, appear to be required for efficient packaging (10, 13, 14, 26-28). SL1 also appears to be involved in initiating RNA dimerization, and dimerization and encapsidation may be linked processes (7, 14). SL3 contains a specific binding site for HIV-1 nucleocapsid protein (6, 8, 23). However, RNA mapping experiments suggest that additional HIV-simian immunodeficiency virus encapsidation determinants are present throughout the 5′ end of the mRNA, including U5 (15, 18, 27, 28, 39), and a continuous, discrete, transferable element analogous to that of MoMLV has not been defined. Packaging determinants extend into gag (13, 25) and may include more-downstream regions (9, 19). In contrast to the case for HIV-1, cotranslational encapsidation contributes to HIV-2 packaging specificity (18).
In contrast to the extensive published analyses for HIV-1 and other primate lentiviruses, no investigations of encapsidation signals have been performed for any of the nonprimate (ungulate or feline group) lentiviruses. No information can be gained by direct sequence comparisons, as there is no significant nucleotide level homology between feline immunodeficiency virus (FIV) and HIV-1 other than at short, universally conserved retroviral elements such as the tRNA primer binding site (PBS) (12). To determine the requirements for encapsidation of this lentivirus and to enable optimization of FIV-based vectors, we systematically mapped FIV genome encapsidation determinants. We approached this problem by using multiple RNase protection probes to first directly determine relative virion RNA levels and then determine competitive encapsidation efficiencies versus that of the wild-type genome. We used these complementary assays to study a panel of FIV deletion mutants and a set of subgenomic vectors containing various increments of gag. In addition, we determined the replicative properties of viruses containing deletions in putative encapsidation domains. These experiments were facilitated by an expression system that permits production of high levels of FIV virions by transient transfection (32, 33).
MATERIALS AND METHODS
Plasmid construction.
The numbering of nucleotides is according to Talbott et al. (38). All mutants were confirmed by automated DNA sequencing. pCT5efs is a variant of wild-type FIV expression construct pCT5 (32, 33) with a frameshifting oligonucleotide insertion (29 bp) in the SU domain at nt 7176 (efs stands for envelope frameshift). pCT5Δpsi1 was generated from pCT5efs, using PCR to delete the region between the MSD and the gag start codon while simultaneously inserting an ApaI site. pCT5Δpsi2 was made by linearizing CT5Δpsi1 at the unique ApaI site, removing protruding 3′ends with T4 DNA polymerase and religating. pCF1Δenv is a previously described FIV vector packaging plasmid (33). pCF1efs was derived from pCF1Δenv by exchanging a fragment extending from the Bsu36I site at position 3573 to the PvuI site in the prokaryotic backbone with the corresponding fragment from pCT5efs. The same strategy was used to construct pCT5Δenv from pCT5efs. pCF16 was constructed by amplifying a fragment containing the human cytomegalovirus (CMV) immediate-early promoter and a β-globin splice donor from pCi (Promega) with oligonucleotides 5′-TATAACGCGTCAATATTGGCCATTAGCC-3′ and 5′-TATGGGCCCAGTTTCTATTGGTCTCC-3′; the PCR product was cleaved with MluI and ApaI and inserted between the same sites in pCT5Δpsi1.
pCT5SLm (loop mutant) was constructed by a three-part ligation. pCT5efs was digested with AlwNI, and protruding 3′ends were removed with T4 DNA polymerase. The fragment was restricted with either MluI (to isolate an 852-bp fragment) or Bsu36I (to isolate a 3,110-bp fragment). The fragments were subsequently ligated into the MluI-Bsu36I (8,318-bp) backbone of pCT5efs. The stem in CT5SmL (stem mutant) was mutated by inserting a double-stranded oligonucleotide (5′-CAGACCAAACAGTGAGTATCTCTAGTGAAGCGGACTCGAGCTCATAT-3′) restricted with SacI at the 3′end into CT5efs digested with MluI-Bsu36I (8318 bp) together with an 852-bp Mlu-AlwNI (blunt) fragment and a 3,069-bp SacI-Bsu36I fragment from CT5efs.
CE series constructs were generated by three-part ligations with (i) a 10,186-bp PvuI-PflFI fragment from CF1efs; (ii) a 1,495-bp PvuI-EcoRI PCR-generated fragment from pCT5efs, made with an EcoRI-tailed antisense PCR primer (5′-ATATGAATTCGAGGAGTCTCTTTGTTGAGG-3′) which introduces an EcoRI site after the 5′cap site; and (iii) EcoRI-PflFI fragments of 504 bp (to position 425) for pCEΔ209efs, 588 bp (to position 341) for pCEΔ125efs, and 650 bp (to position 279) for pCEΔ63efs.
pCT5ΔRefs, pCT5ΔU5efs, and pCT5ΔRU5efs were created by using overlap extension PCR with CT5efs as a template. The 5′ outside primer was FIV11842 (5′-GCTTAGGGTTAGGCGTTTTGCGCTGC-3′), and the 3′outside primer was FIV556AS (5′-GTCACCAGATGTAATTTATCTGGG-3′). To generate CT5ΔRefs, the two primary PCRs were performed with the primers FIV11842 and FIVΔRAS (5′-ACTCGACAGGAACAAAGAGACTCCTCGAAGTTTC-3′), resulting in PCR product 1, and FIVΔRS (5′-GTCTCTTTGTTCCTGTCGAGTATCTGTGTAAT-3′) and FIV556AS, resulting in PCR product 2. The two primary products were combined and amplified with the two outside primers. The resulting fragment was restricted with MluI and SacI and inserted into the corresponding sites in pCT5efs. Similarly, pCT5ΔU5efs was generated with primers FIV11842 and FIVΔU5AS (5′-CGGGCGCCAACGTTCAATCTCAAATATTTATTGTATC-3′) for product 1 and primers FIVΔU5S (5′-GAGATTGAACGTTGGCGCCCGAACAGGGAC-3′) and FIV556AS for product 2. pCT5ΔRU5efs was made with primers FIV11842 and FIVΔRU5AS (5′-CGGGCGCCAACAACAAAGAGACTCCTCGAAGTTTC-3′) for product 1 and primers FIVΔRU5S (5′-GTCTCTTTGTTGTTGGCGCCCGAACAGGGAC-3′) and FIV556AS for product 2.
GiNWF-G311 is a FIV vector plasmid that contains, from 5′ to 3′, the hybrid U3-substituted promoter of pCT5, R, U5, the leader sequence, the first 311 bp of gag, the Rev response element (RRE) (nt 8537 to 8952 of the FIV 34TF10 genome), a sequence (FIV nt 4904 to 5191) containing the FIV central polypurine tract (cPPT) and the central termination sequence (CTS) (40), the human CMV immediate-early promoter, the enhanced green fluorescent protein (eGFP) gene, an internal ribosomal entry site, neoR, the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) (11), and the 3′ long terminal repeat (LTR). The cPPT-CTS combination is also referred to as the central DNA flap, as the strand initiations and terminations that occur at these loci result in a triple-stranded DNA flap structure at the completion of FIV reverse transcription (40).
pGiNWF-G311, was derived from pCT25, a previously described lacZ transfer vector (24), via a series of intermediates: pGiN (also called CT25.eGFP.ires.neo), pGiNW, pGiNWcPPT-CTS, and pGiNWF. To first construct pGiN, peGFP-1 (Clontech) was cleaved with NotI, treated with Klenow fragment, and cleaved with BamHI. The eGFP fragment was inserted into a murine retroviral vector between BamHI and HpaI, resulting in the sequence 5′-GCGGCCAACGAATTC-3′ at the 3′ junction. The modified eGFP insert was then excised with BamHI and EcoRI and inserted between the same sites in pCT25, thereby replacing lacZ with eGFP and generating pCT25.eGFP. A 1.5-kb Sal-Nhe fragment containing an internal ribosome entry site and neoR from pJZ308 (41) was then inserted into the EcoRI site pf pCT25.eGFP by blunt ligation, resulting in pGiN (pCT25.egfp.ires.neo). pGinW was constructed by blunt insertion of an EcoRV-XhoI fragment of pBluescriptIISK+WPRE-B11 (11) into the BspEI site of pGiN (producing a regenerated BspEI site that is Dam methylated). The central DNA flap was next inserted as a 279-nt PCR amplicon, using primers 5′-ATATTTCGAATCAAATCAAACTAATAAAGTATGTATTGTGAAACAACCTCCTTGGATAATGCC-3′ and 5′-ATATACCTCTTTTAGGTCTAGACTCTCATGTGTCTCCTAGG-3′. The longer sense PCR primer fuses the cPPT with the 3′ end of the FIV RRE while deleting an unnecessary FIV splice acceptor. After cleavage with BstBI and XbaI, the amplicon was inserted into BstBI- and XbaI-cleaved pGiNW, generating pGiNWcPPT-CTS. The last step removed the internal CMV promoter from pGinW while inserting the cPPT-CTS. To restore it, the BamHI-XhoI fragment of pGiNW was subcloned between the XhoI and BamHI sites in pCR2.1 (Invitrogen). An XbaI linker (GCTCTAGAGC) was blunted into the AflII site of this intermediate, and the 0.6-kb XbaI-XbaI fragment was then inserted into the Xba site of pGiNWcPPT-CTS, producing pGiNWF (also designated pGiNWF-G311 for comparison with the length of the gag segment in other plasmids in this work). The other GiNWF-G constructs were generated by PCR with the sense primer FIV11842 and one of the following SphI-tailed antisense primers: FIV627-SphIAS (5′-ATATGCATGCGTACGTTGCTGTAGAATCTCTCCTACC-3′) for GiNWF-G0, FIV771-SphIAS (5′-ATATGCATGCGTACGTCCTGTAGATACATTAGCC-3′) for GiNWF-G144, FIV1034-SphIAS (5′-ATATGCATGCGTACGGATATGCCTGTGGAGGGCC-3′) for GiNWF-G407, FIV1237-SphIAS (5′-ATATGCATGCGTACGCCAATATTTCTTTATCTGCAGCG-3′) for GiNWF-G610, or FIV1898-SphIAS (5′-ATATGCATGCGTACGCCCCGCCTTCCAGTTTCCCG-3′) for GiNWF-G1226. CTRZlb (32), which has a single nucleotide insertion at position 298 of gag, resulting in a frameshift, was used as a template. The PCR products were restricted with MluI and SphI and ligated to the 7,208-bp MluI-SphI backbone of GiNWF-G313. All plasmids were sequenced.
Plasmids used as templates for production of riboprobes were constructed as follows. For the noncompetitive experiments, riboprobes pSPT-CT5PB and pSPT-EGFP were used. pSPT-CT5PB was generated by cloning a BalI-PstI fragment from positions 659 to 938 (gag) of CT5efs into pSPT18 (Roche) digested with PstI and SmaI. The resulting plasmid was linearized with PvuII, and the protected RNA species is 279 nt long. pSPT-EGFP was created by cloning a 163-bp-long BsrFI fragment of pEGFP-N1 into pSPT18 digested with XmaI. The plasmid was linearized with EcoRI, and the protected RNA species is 163 nt long. For the competitive experiments, the following riboprobes were made. pSPT-CT5SR was constructed by cloning an RsaI-SacI fragment from positions 172 to 504 of CT5efs into pSPT19 digested with SacI and HindII. The plasmid was linearized with HindIII, and the protected RNA species are 293 nt for CT5efs, 230 nt for CEΔ63efs, and 168 nt for CEΔ125efs. pSPT-CT5MR was generated by cloning an RsaI-MscI fragment from positions 172 to 659 of CT5efs into SmaI-digested pSPT18. The plasmid was linearized with EcoRI. The protected RNA species are 447 nt for CT5efs, 373 nt for CT5ΔRefs, and 307 nt for CT5ΔU5efs and CT5ΔRU5efs. pSPT-CF1MM was constructed by cloning an McrI-MslI fragment from positions 460 to 741 of CF1 into SmaI-digested pSPT18. The plasmid was linearized with EcoRI, and the protected RNA species are 281 nt for CF1Δenv and 233 nt for CT5efs. pSPT-FLAP was created by cloning a BstBI-HindIII fragment from positions 1328 to 1686 of pGiNWF-G311 into pSPT18 digested with HindIII and AccI. The plasmid was linearized with EcoRI, and the protected RNA species are 358 nt for all GiNWF-G clones and 296 nt for CT5efs.
Cell culture and transfections.
293T cells and CrFK.CXCR4 cells, which are Crandell feline kidney cells that stably express CXCR4 (32), were cultured in Dulbecco's modified Eagle medium with 10% fetal calf serum. Transient transfections were performed by the calcium phosphate coprecipitation method. In noncompetitive assays, 15 μg of the test plasmid were cotransfected with 1 μg of pEGFP-NI. In competitive assays, 7.5 μg of pCT5efs (wild type) and 7.5 μg of mutant plasmid were cotransfected.
RNA isolation.
At 42 h after transfection, cellular RNA was isolated with Trizol (Invitrogen) according to the manufacturer's protocol. Viral particles were purified from the medium by removing cell debris by centrifugation at 2,000 rpm in a Sorvall tabletop centrifuge for 10 min and filtering the clarified supernatant through a 0.45-μm-pore-size filter. The virus was then pelleted through a 20% sucrose cushion for 2 h at 25,000 rpm in an SW28 rotor. The viral pellet was resuspended in TNE buffer (100 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA), and aliquots were stored at −80°C. Viral particle production was measured by a reverse transcriptase (RT) assay (see below). Viral RNA was isolated by disrupting the particles in sodium dodecyl sulfate (SDS) (1%)-proteinase K (100 μg/ml) at 37°C for 30 min in the presence of 10 μg of yeast RNA. After two extractions with phenol (pH 4.5)-chloroform, RNA was ethanol precipitated in the presence of 0.3 M sodium acetate. Both cellular and viral RNAs were treated with 10 U of RNase-free DNase I in a buffer containing 20 mM Tris (pH 8.0), 10 mM MgCl2, and 40 U of RNase inhibitor (Roche) at 37°C for 30 min. The reaction was terminated by adding an equal volume of a 2× stop buffer (0.4% SDS, 0.6 M sodium acetate, 20 mM EDTA). The RNA was extracted with phenol-chloroform and precipitated with ethanol. The concentration of cellular RNA was determined by measuring the spectrophotometric absorption at 260 nm.
RT-PCR and sequencing of mutant viruses.
Virions (CT5Δpsi1 and CT5Δpsi2) isolated from day 15 supernatants of infected cultures were pelleted through a 20% sucrose cushion, RNA was isolated as described above, and reverse transcription was performed with 5 μl of RNA and 200 U of MoMLV RT (Gibco-BRL) in 50 mM Tris (pH 8.3)-75 mM KCl-3 mM MgCl2-10 mM dithiothreitol-0.5 mM deoxynucleoside triphosphates and primer FIV999AS (TGCTTCTTTCATAGATGGCC) for 60 min at 37°C. RT was left out for control reactions. Subsequently, PCR was performed with primers FIV214Eco (ATATGAATTCGAGGAGTCTCTTTGTTGAGG) and FIV929AS (ATATGACACCGTCATATTTAAAAGTCCTGC) and Taq polymerase. PCR fragments were cloned into pCRII by TA cloning (Invitrogen). Five amplicons were sequenced for each virus.
RPA.
32P-labeled riboprobes were synthesized by in vitro transcription of linearized plasmids, using SP6 RNA polymerase (Roche), [α-32P]rUTP (800 Ci/mmol) and 5 μM unlabeled rUTP. Riboprobes were purified from 6% polyacrylamide-8 M urea gels prior to use. The RNase protection assay (RPA) was performed with a commercially available kit (Ambion) according to the manufacturer's protocol. Five micrograms of cellular RNA or different amounts of viral RNA supplemented with 2 μg of yeast RNA were mixed with 5 × 105 cpm of 32P-labeled probe in 10 μl of hybridization buffer and incubated at 42°C for 12 h. Unhybridized regions were then digested with 0.37 U of RNase A and 15 U of RNase T1 for 30 min at 37°C. Protected fragments were precipitated, resuspended in RNA loading buffer, and separated on 6% polyacrylamide-8 M urea gels. For size determination, [α-32P]dCTP-labeled fragments of pBR322 digested with MspI were run in parallel. Protected RNA species were quantified by PhosphorImager analysis (Molecular Dynamics).
RT assay.
Isotopic RT assays were performed as described previously (32), with modifications to permit high-throughput processing. Five microliters of the RT reaction mix was transferred to a DE81 paper-lined, 96-well Whatman plate. The plate was dried and then washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 95% ethanol, using an automated 96-well FilterMate Harvester (Packard). After drying at 37°C for 15 min, scintillation cocktail was added and the plate was analyzed in a TopCount scintillation counter (Packard).
Western blots.
Transiently transfected 293T cells or viral particles were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) containing protease inhibitors (Complete Mini; Roche). Lysates were combined with SDS sample buffer containing β-mercaptoethanol, electrophoresed through an SDS-10% polyacrylamide gel, and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The membranes were blocked in 0.2% I-block (Tropix)-0.1% Tween and then probed first with Petaluma serum (1:1,000), a FIV-infected-cat serum. The secondary antibody was a peroxidase-conjugated goat anti-feline immunoglobulin G (1:1,000; ICN). Bands were visualized with enhanced chemiluminescence detection reagents (Amersham Pharmacia).
RESULTS
Sequences between the MSD and the start codon of gag contribute negligibly to FIV encapsidation.
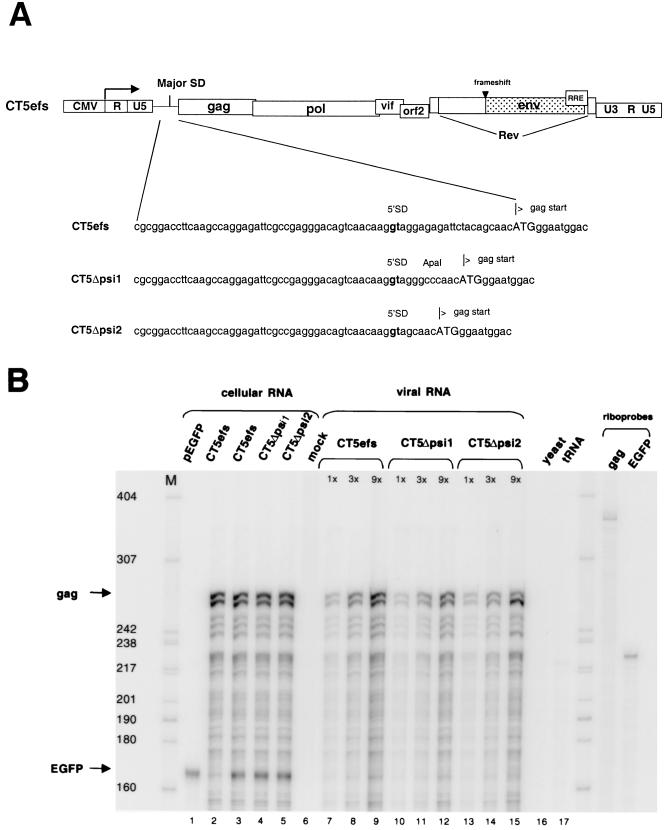
Deletions between the MSD and gag gene of FIV were studied first, because such deletions are known to markedly impair encapsidation of the genomic mRNAs of human lentiviruses as well as simpler mammalian retroviruses (1, 2, 20, 31). However, this sequence is shorter in FIV than, for example, in HIV-1 (20 versus 44 nt) and is much shorter than in MoMLV (351 nt). To express FIV genomes in cells that are amenable to efficient transfection and high-level virion production (e.g., 293T cells), we derived expression constructs for the present work from pCT5, a plasmid that produces infectious, wild-type FIV in both feline and nonfeline cells (Fig. 1A) (32). However, enabling of heterologous FIV expression results in marked syncytium formation in all human cell lines, which is due to interaction between the FIV envelope glycoprotein and the ubiquitous human CXCR4 chemokine receptor (32). Therefore, for the present work we derived a pCT5 variant with a frameshifting oligonucleotide insertion (29 bp) in the SU domain (pCT5efs). Avoiding any deletions in this way permitted unbiased analysis of the entire FIV genomic mRNA for encapsidation determinants while eliminating Env-induced cell lysis that could cause copurification of virion RNA with cell fragments containing intracellular viral RNAs. Two deletions between the MSD and gag were made in pCT5efs, and the resulting constructs were designated CT5Δpsi1 and CT5Δpsi2 (Fig. 1A).
FIG. 1.
Sequences between the MSD and the start codon of gag contribute negligibly to encapsidation. (A) Schematic representation of CT5efs, a provirus with a frameshifting (29-nt) oligonucleotide insertion in the SU domain of env. In CT5Δpsi1 and CT5Δpsi2,sequences between the MSD and the gag start codon are deleted. (B) 293T cells were transiently transfected with CT5efs, CT5Δpsi1, or CT5Δpsi2 together with pEGFP-N1. Cellular RNA and viral RNA were subjected to an RPA with antisense riboprobes pSPT-CT5PB and pSPT-EGFP. Protected fragments are 279 and 163 nt long, respectively. RNAs from cells transfected with either pEGFP or CT5efs alone are shown in lanes 1 and 2, respectively. Viral RNAs were analyzed in three different amounts (1×, 3×, and 9×).
Encapsidation efficiencies of genomic RNAs from the three constructs (CT5efs, CT5Δpsi1, and CT5Δpsi2) were first analyzed in a noncompetitive assay, in which wild-type and mutant RNAs were expressed in separate transfections, and intracellular and virion RNAs were quantified (Fig. 1B). The viral constructs were transiently transfected into 293T cells together with an eGFP expression plasmid, pEGFP-N1. eGFP, quantified with a separate riboprobe, served as a measure for transfection efficiency and also as an abundant intracellular control mRNA whose exclusion from virion pellet samples could verify their purity as bona fide virion RNAs.
Cellular RNA and RNA from virion particles pelleted through sucrose cushions were isolated. Virion RNA was extracted from equal amounts of viral particles, as determined by RT assay. Both of these RNAs were then subjected to an RPA with a 32P-labeled riboprobe complementary to sequences in gag (therefore detecting exclusively unspliced RNA), as well as with the aforementioned 32P-labeled eGFP riboprobe (Fig. 1B). In addition, each virion RNA was analyzed in three different amounts (1×, 3×, and 9×). The wild type and the two mutant plasmids transcribed similar amounts of cellular mRNA (Fig. 1B, lanes 3 to 5). Mutant viral RNAs CT5Δpsi1 (lanes 10 to 12) and CT5Δpsi2 (lanes 13 to 15) were packaged at 76 and 82%, respectively, of the amount of the wild-type CT5efs RNA (lanes 7 to 9). These data suggested that a 70% deletion between the MSD and the gag ATG codon minimally attenuates encapsidation of genomic RNA. Figure 1B also shows that the eGFP control mRNA was detected equivalently in each of the cell lysates. This important control verifies that the pelleted virion RNAs are not contaminated with cellular RNAs.
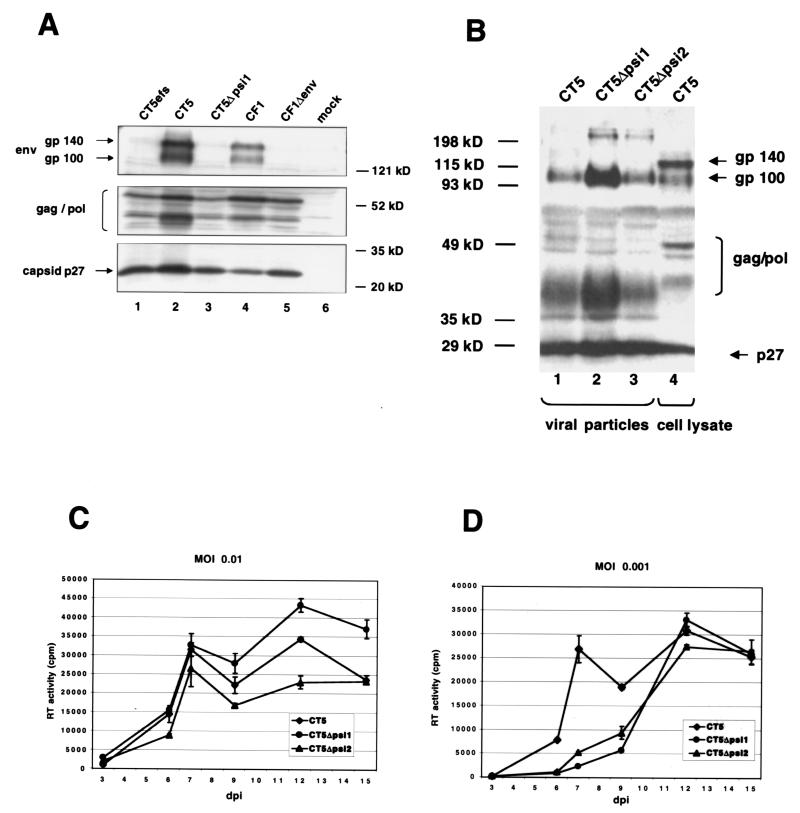
To determine whether the deletions in CT5Δpsi1 and CT5Δpsi2, which are in close proximity to the MSD, had an effect on FIV protein expression (and, by inference, the elaborate, Rev-regulated lentiviral splicing program), Western blotting was performed with cell lysates (Fig. 2A) and with lysates from sucrose-purified viral particles (Fig. 2B). Both the wild type (Fig. 2A, lanes 1 and 2, and B, lane 1) and the mutant (Fig. 2A, lane 3, and B, lanes 2 and 3) showed the same qualitative pattern of viral protein synthesis and processing in 293T cells. Moreover, RT levels in transfected 293T cell supernatants were equivalent to wild type for each plasmid (data not shown). Note that uncleaved Env glycoprotein (gp140) is prominent in infected-cell lysates (Fig. 2B, lane 4) but only the fully processed SU (gp100) is detectable in viral particles (lanes 1 to 3), demonstrating further that the sucrose purification methods we used result in particles that are not contaminated with cell debris.
FIG. 2.
Deletions in CT5Δpsi1 and CT5Δpsi2 have no effect on FIV protein expression and viral replication. (A) Western blot with cell lysates from 293T cells transfected with the indicated plasmids. (B) Western blot with lysates from viral particles CT5, CT5Δpsi1, and CT5Δpsi2 (lanes 1 to 3), harvested from CrFK.CXCR4 cells at 12 days postinfection, and a cell lysate from CT5-transfected 293T cells (lane 4). Both membranes were probed with Petaluma serum, a FIV-infected-cat serum. The secondary antibody was a peroxidase-conjugated goat anti-feline immunoglobulin G. (C and D) CrFK.CXCR4 cells were infected with CT5, CT5Δpsi1, or CT5Δpsi2 viral preparations at an MOI of 0.01 (C) or 0.001 (D). At different days postinfection (dpi), supernatant was collected and virus growth was measured by an RT assay. Error bars indicate standard deviations.
The minimal effects of the MSD-gag region deletions on packaging and lack of effects on viral expression led us to examine the replication of the two mutant viruses. In the viruses used in this experiment the env gene was restored, allowing for virus spread. CrFK.CXCR4 cells (32) were infected with CT5, CT5Δpsi1, or CT5Δpsi2 viral preparations at a multiplicity of infection (MOI) of 0.01 (Fig. 2C) or 0.001 (Fig. 2D), respectively. At different days postinfection, supernatant was collected and virus growth was measured by an RT assay. As illustrated in Fig. 2C and D, both mutants CT5Δpsi1 and CT5Δpsi2 showed productive replication comparable to that of the wild-type CT5 virus. At the lowest MOI, a modest delay in peak viral production, but not in the magnitude of the peak, was discernible (Fig. 2D). In addition, equivalent syncytium formation by all three viruses was observed (data not shown).
To determine whether the replication of these mutant viruses resulted in selection for revertants, RNA was prepared from the virions isolated from the day 15 supernatants of the experiments shown in Fig. 2D. After DNase treatment, RT-PCR was performed with primers that amplify from the first nucleotide of the R repeat to nt 713 (the amplicon extends 394 nt upstream and 302 nt downstream of the MSD-gag interval). Control PCRs without RT showed that reverse transcription was required for PCR product formation (data not shown). Five amplicons were cloned and sequenced for each virus. The original mutant sequences were found to be present in all clones for both Δpsi1 and Δpsi2. Compensatory (second-site) mutations were not found elsewhere within the amplified region. Therefore, reversion of the mutant MSD-gag intervals did not occur.
The 5′ LTR and 154 nt of the leader are required for FIV encapsidation.
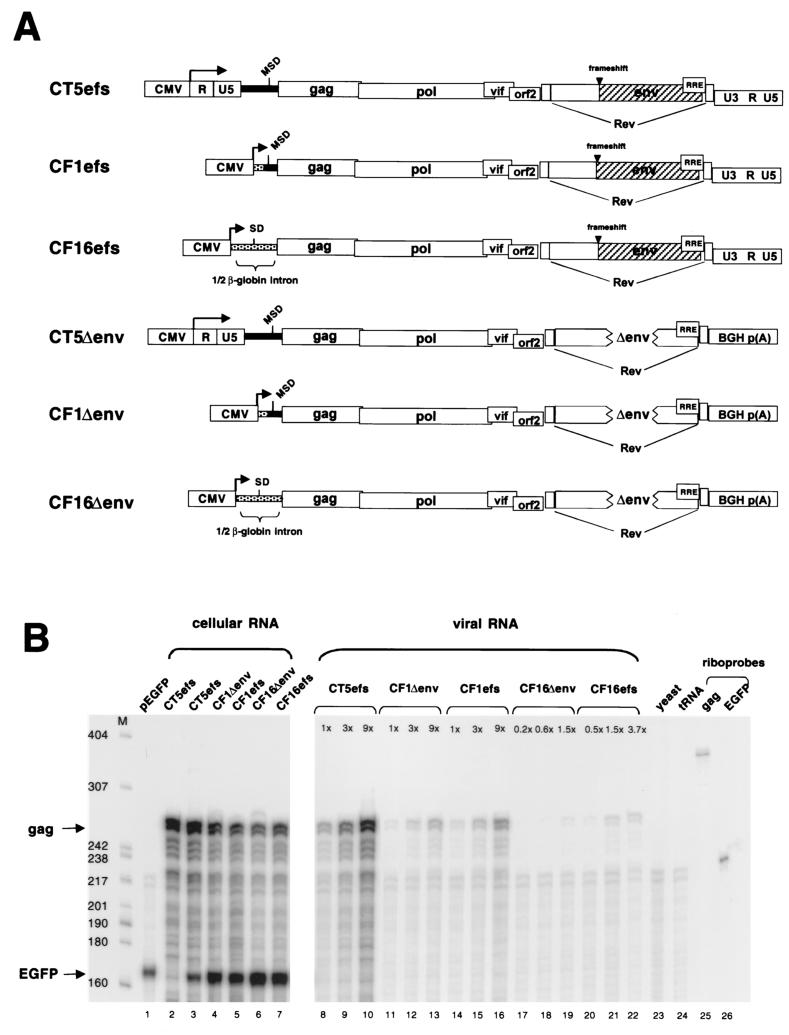
To address whether sequences upstream of the MSD contribute to packaging, mutants CF1efs and CF16efs were constructed (Fig. 3A). In CF1efs, which is an SU-frameshifted variant of first-generation FIV vector packaging plasmid precursor CF1 (32, 33), the left-hand U3, R, and U5 elements and 154 nt of the 5′ leader are replaced by the CMV promoter, leaving 119 nt of FIV leader upstream of gag. In CF16efs, all viral sequences between the CMV promoter and the ATG gag codon are deleted; a β-globin intron is immediately upstream of gag. Encapsidation efficiencies of those two mutant RNAs were determined (Fig. 3B). Compared to encapsidation of wild-type RNA (CT5efs, lanes 8 to 10), packaging of CF1efs RNA was severely reduced (18% of wild-type level) (lanes 14 to 16) and that of CF16efs was even further impaired (7% of wild-type level) (lanes 20 to 22). These results implicated regions upstream of the MSD in encapsidation.
FIG. 3.
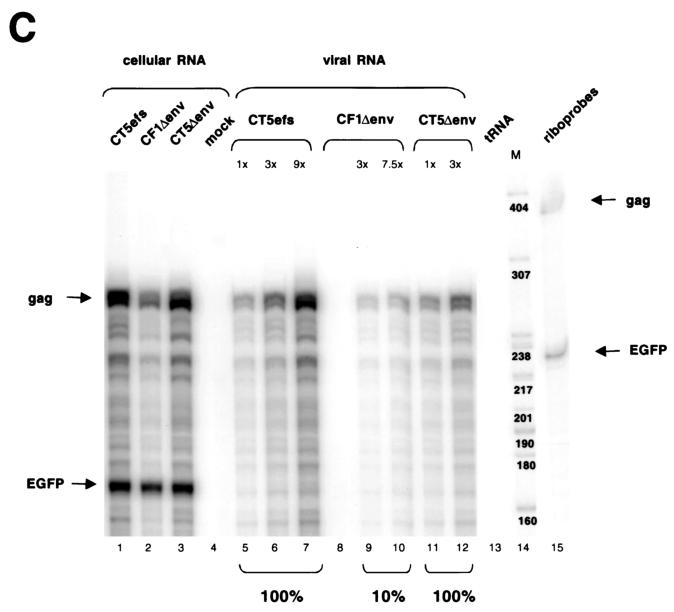
The 5′ LTR and 154 nt of the leader are required for FIV encapsidation. (A) Schematic representation of plasmids analyzed. CT5efs, CF1efs, and CF16efs have a frameshifting mutation in the env gene. In CT5Δenv, CF1Δenv, and CF16Δenv, 875 nt is deleted from the env gene and the 3′ LTR is replaced by the bovine growth hormone (BGH) polyadenylation signal. The FIV leader is represented by a black bar; non-FIV sequences are stippled. In CF1efs and CF1Δenv, the 5′ LTR and 154 nt of the leader are replaced by the CMV promoter. In CF16efs and CF16Δenv, all viral sequences between the CMV promoter and the ATG gag codon are replaced by β-globin intron sequences. (B) RPA of a noncompetitive encapsidation experiment with CT5efs, CF1Δenv, CF1efs, CF16Δenv, and CF16efs transfected with pEGFP-N1 into 293T cells. Antisense riboprobes pSPT-CT5PB and pSPT-EGFP were used to detect viral gag mRNA and eGFP mRNA, respectively. (C) Noncompetitive encapsidation assay with CT5efs, CF1Δenv, and CT5Δenv. The same riboprobes as for panel B were used.
To examine whether the 3′ U3 and R elements contribute to encapsidation, the bovine growth hormone polyadenylation signal was used to replace the 3′ LTR in plasmids having variably intact 5′ cis regions, generating mutants CT5Δenv, CF1Δenv, and CF16Δenv (Fig. 3A). In addition, 875 nt is deleted from the env gene, eliminating envelope glycoprotein expression (Fig. 2A, lane 5) (CF1Δenv is a first-generation FIV vector packaging plasmid [33]). The packaging efficiencies of CF1Δenv RNA (Fig. 3B, lanes 11 to 13) were comparable to those of CF1efs RNA (lanes 14 to 16), and those of CF16Δenv (lanes 17 to 19) were similar to those of CF16efs (lanes 20 to 22). These data suggest that the 3′ cis-acting (LTR) sequences, as well as the 875 nt in env, do not contain packaging determinants. This was corroborated by the finding that encapsidation efficiencies of CT5efs and CT5Δenv RNAs are identical (Fig. 3C, compare lanes 5 and 6 with lanes 11 and 12).
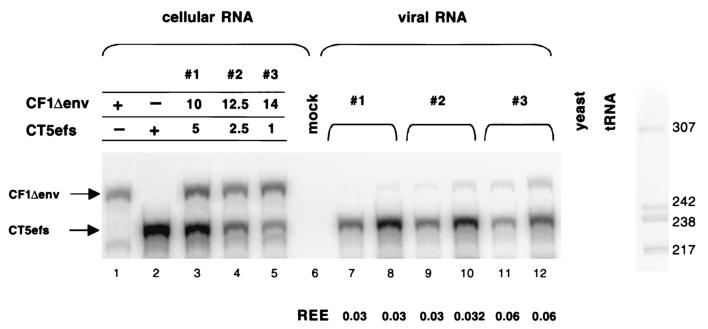
The noncompetitive assay described above demonstrated that transcripts from packaging plasmid CF1Δenv were minimally encapsidated into particles. We further analyzed encapsidation of CF1Δenv RNA in a directly competitive assay, which measures the relative encapsidation efficiencies (REEs) of coexpressed wild-type and mutant RNAs. The benefits of a directly competitive assay are that the same Gag-Pol precursor pools and nascent particles are available to the intracellular viral RNAs and that analysis of both RNAs within the same cellular and virion samples obviates problems with intersample variations accrued incrementally during transfection, RNA recovery, and RPAs (26). CF1Δenv was cotransfected into 293T cells with the wild-type CT5efs plasmid at different ratios (Fig. 4, lanes 3 to 5). Cellular and virion RNAs were subjected to an RPA with a riboprobe capable of detecting both wild-type and mutant RNAs. The REE of the mutant was determined by calculating the ratio of mutant RNA to wild-type RNA in the virion, relative to the ratio of the two RNAs in the cytoplasm (28). The REE of CF1Δenv RNA was between 3.0 and 6.0% of that of the wild-type CT5efs RNA (lanes 7 to 12). This result confirms that obtained in the noncompetitive assay (Fig. 3B, lanes 11 to 13, and C, lanes 9 and 10), where CF1Δenv packaging was reduced to 10% of that of the wild type. It is important to note that the REE is independent of the amount of DNA transfected and therefore independent of the amount of RNA transcribed in the cell (Fig. 4) (28). These data show that the 5′ LTR (R and U5) and the 154 nt of the leader contain an important packaging determinant. The 3′ LTR, in contrast, does not contribute significantly to encapsidation of FIV RNA.
FIG. 4.
RPA of a competitive encapsidation experiment. CF1Δenv (10, 12.5, or 14 μg) and CT5efs (5, 2.5, or 1 μg) were cotransfected into 293T cells. Antisense riboprobe pSPT-CF1MM was used to detect 5 μg of cellular RNA (lanes 1 to 5) and 5 μl (lanes 7, 9, and 11) or 15 μl (lanes 8, 10, and 12) of viral RNA. Protected RNA species are 281 nt for CF1Δenv and 233 nt for CT5efs. The REE was determined by calculating the ratio of mutant RNA to wild-type RNA in the virion, relative to the ratio of the two RNAs in the cytoplasm (28).
Encapsidation determinants are located in R and U5.
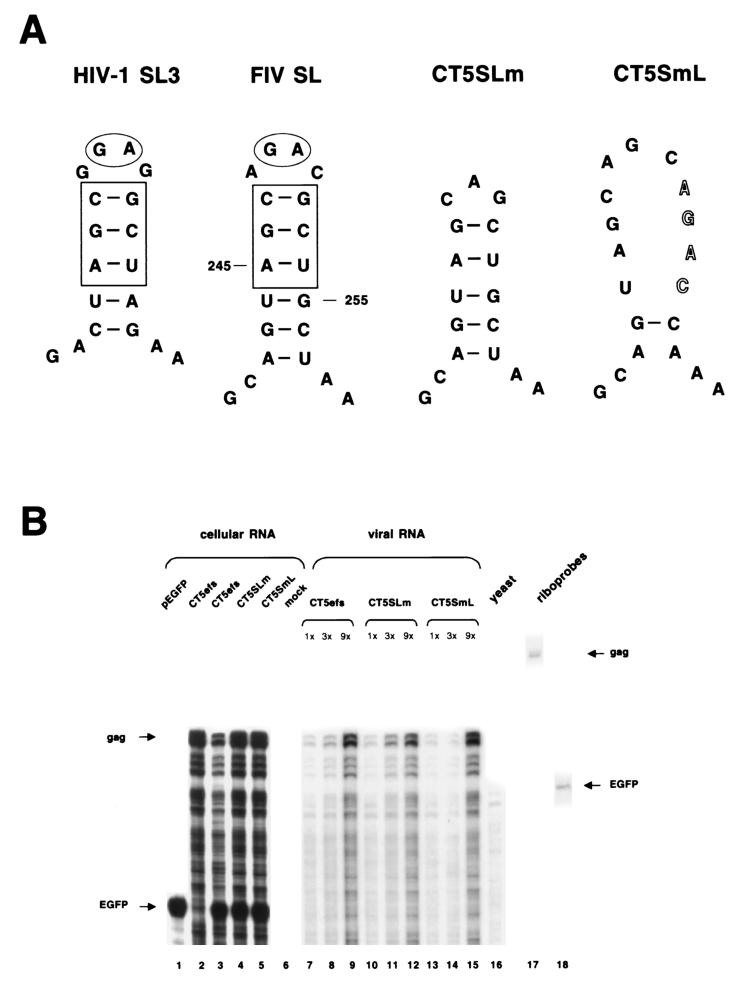
RNA secondary-structure analyses (42) were performed for the 154-nt leader sequence shown to contribute to FIV RNA encapsidation. A potential stem-loop structure was found from position 242 to 257 in the RNA, consisting of six base pairs in the stem and four bases in the loop (Fig. 5A). Remarkably, three base pairs in the stem and two of the four nucleotides in the loop are identical to stem-loop 3 in HIV-1 (Fig. 5A). SL3 has been shown to contribute to encapsidation and to bind HIV-1 nucleocapsid protein (6, 8, 23). We therefore tested the importance of this stem-loop in FIV RNA encapsidation, by introducing mutations in either the loop (CT5SLm) or the stem (CT5SmL). In the latter, four bases in the 3′ strand of the putative stem were mutated to prevent base pairing (Fig. 5A). Encapsidation efficiencies were determined (Fig. 5B). Compared to wild-type CT5efs (lanes 7 to 9), neither mutating the loop (lanes 10 to 12) nor disrupting the stem (lanes 13 to 16) reduced packaging of RNA significantly. The same result was obtained when the mutants were placed in the context of CT5Δpsi2 (data not shown).
FIG. 5.
A putative FIV stem-loop does not contribute to FIV mRNA encapsidation. (A) Schematic representation of HIV-1 SL3 and a putative stem-loop structure in the FIV leader (FIV SL). In CT5SLm the loop was mutated, and in CT5SmL the stem was disrupted. (B) RPA of a noncompetitive encapsidation experiment with CT5efs, CT5SLm, and CT5SmL transfected with pEGFP-N1 into 293T cells. Antisense riboprobes pSPT-CT5PB and pSPT-EGFP were used to detect viral gag mRNA and eGFP mRNA, respectively.
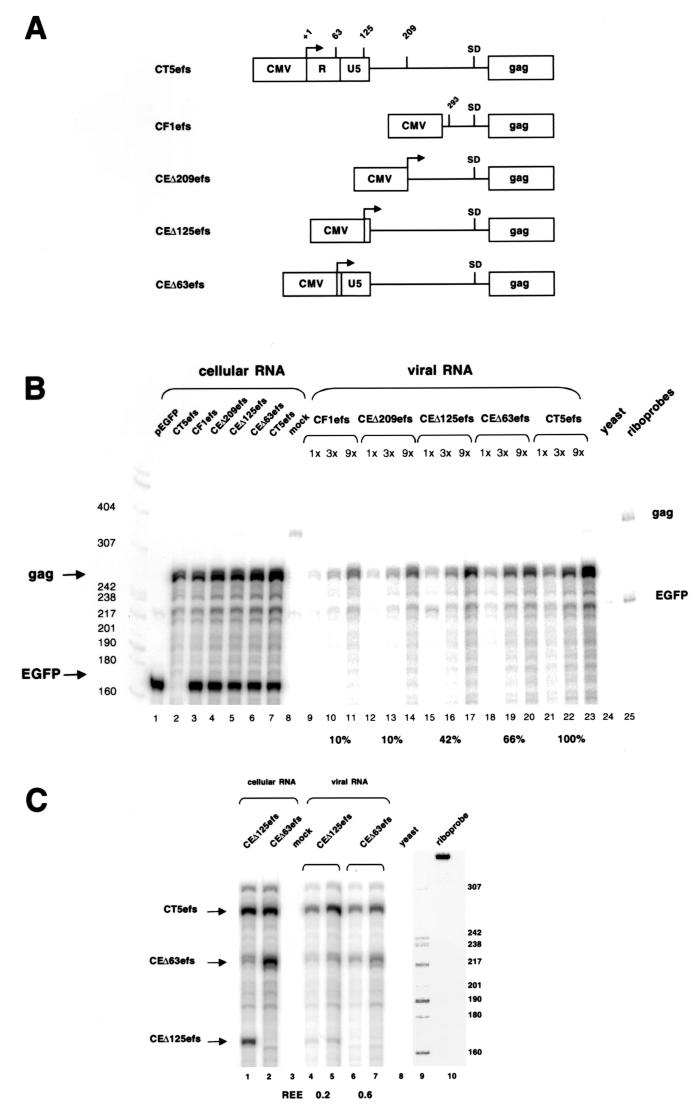
To determine systematically which sequences contribute most to encapsidation, the 5′ LTR leader was restored stepwise in CF1efs, generating plasmids CEΔ209efs, CEΔ125efs, and CEΔ63efs (the numbers refer to the amount of left-hand genomic mRNA sequence that is missing). In this series of mutants, the FIV mRNA cap site was preserved, as in CT5efs. Packaging efficiencies were analyzed in noncompetitive (Fig. 6B) and competitive (Fig. 6C) assays. Figure 6B shows that the encapsidation efficiencies increased as leader sequences were restored. Packaging of CEΔ209efs RNA (lanes 12 to 14) was indistinguishable from that of CF1efs (lanes 9 to 11), despite the presence in this mRNA of 84 additional upstream leader nucleotides. This result is consistent with the finding that the above-described stem-loop structure, situated in those 84 nt, does not contribute to packaging. CEΔ125efs RNA (lanes 15 to 17), which contains all of the non-LTR leader, was packaged at 42% relative to wild-type CT5efs RNA (lanes 21 to 23). Incremental addition of the U5 element (CEΔ63efs) resulted in a further increase in packaging to 66% (lanes 18 to 20). A similar result was obtained in the competitive assay (Fig. 6C), where CEΔ125efs or CEΔ63efs was cotransfected with CT5efs. In CEΔ125efs, which had all of the leader restored, the REE was 0.2 (lanes 4 and 5). Strikingly, when the U5 element was present in CEΔ63efs, the REE increased threefold, reaching 0.6 (lanes 6 and 7).
FIG. 6.
(A) Schematic representation of CT5efs, CF1efs, CEΔ209efs, CEΔ125efs, and CEΔ63efs (the numbers refer to the amount of left-hand genomic mRNA sequences missing). (B) RPA of a noncompetitive encapsidation experiment. The plasmids depicted in panel A were cotransfected with pEGFP-N1. Antisense riboprobes were the same as in Fig. 5B. (C) RPA of a competitive encapsidation experiment. Wild-type CT5efs was cotransfected with either CEΔ125efs or CEΔ63efs. Antisense riboprobe pSPT-CT5SR was used. Protected RNA species are 293 nt for CT5efs, 230 nt for CEΔ63efs, and 168 nt for CEΔ125efs.
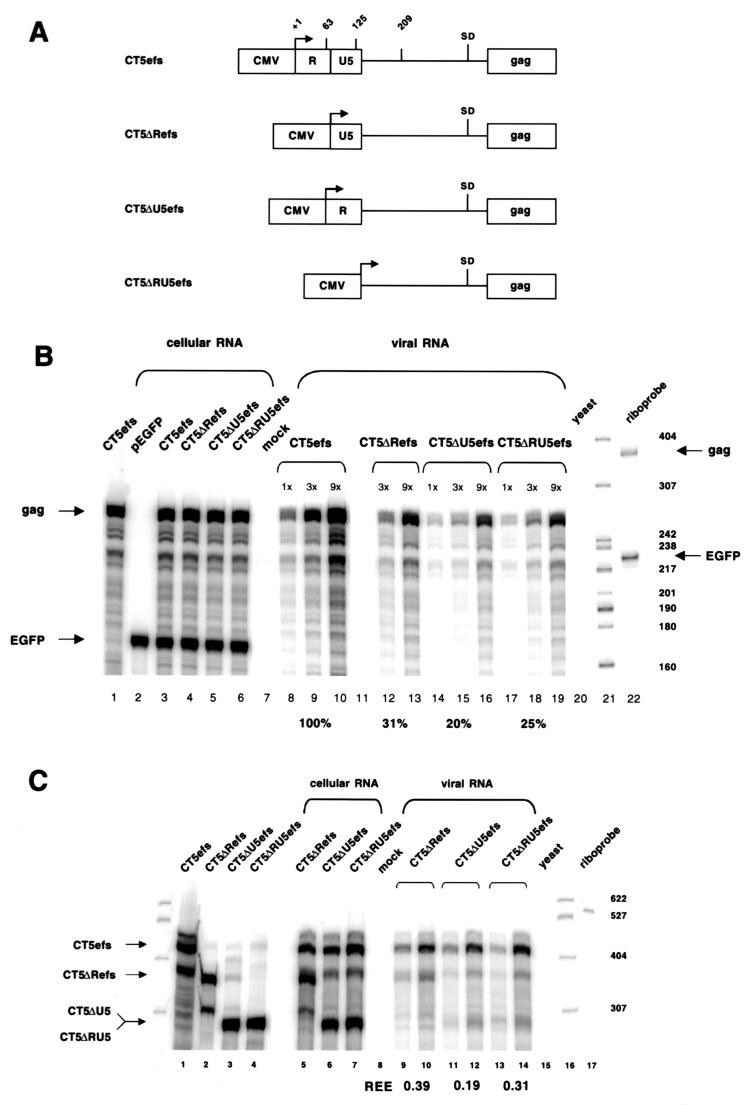
These results prompted us to precisely delete R, U5, or both elements, generating plasmids CT5ΔRefs, CT5ΔU5efs, and CT5ΔRU5efs, respectively (Fig. 7A ). Deletion of the R element (Fig. 7B, lanes 12 and 13) reduced packaging of its RNA to 31% of that of wild-type CT5efs (lanes 8 to 10). Deletion of the U5 element resulted in 20% packaging relative to that of the wild type (lanes 14 to 16), and deletion of both elements produced relative encapsidation of 25% (lanes 17 to 19). These results were corroborated by the competitive assay (Fig. 7C). The REEs were 0.39 for CT5ΔRefs (lanes 9 and 10), 0.19 for CT5ΔU5efs (lanes 11 and 12), and 0.31 for CT5ΔRU5efs (lanes 13 and 14). Note that to ensure proper transcriptional initiation, we preserved the first 14 nt of the R repeat at the start of the R repeat deletion constructs (in addition to preserving the length of the TATA box-to-transcriptional start interval). In lanes 2 and 5 of Fig. 7C, single discrete bands are seen with a probe that overlaps the 5′ end of the FIV mRNA, rather than smears that would reflect heterogeneous transcriptional initiation, and these bands are the precise size expected from initiation at the proper FIV cap site. These data strongly suggest that deletion of sequences upstream of the tRNA PBS, particularly of U5, are deleterious for FIV encapsidation.
FIG. 7.
Encapsidation determinants are located in R and U5. (A) Schematic representation of CT5efs, CT5ΔRefs, CT5ΔU5efs, and CT5ΔRU5efs. (B) RPA of a noncompetitive encapsidation experiment with CT5efs, CT5ΔRefs, CT5ΔU5efs, and CT5ΔRU5efs transfected with pEGFP-N1. Antisense riboprobes pSPT-CT5PB and pSPT-EGFP were used. (C) RPA of a competitive encapsidation experiment. Wild-type CT5efs was cotransfected with either CT5ΔRefs, CT5ΔU5efs, or CT5ΔRU5efs. Antisense riboprobe pSPT-CT5MM was used. Protected RNA species are 447 nt for CT5efs, 373 nt for CT5ΔRefs, and 307 nt for CT5ΔU5efs and CT5ΔRU5efs.
The proximal 311 nt of gag are necessary for efficient FIV RNA encapsidation: a mechanism to identify unspliced mRNA.
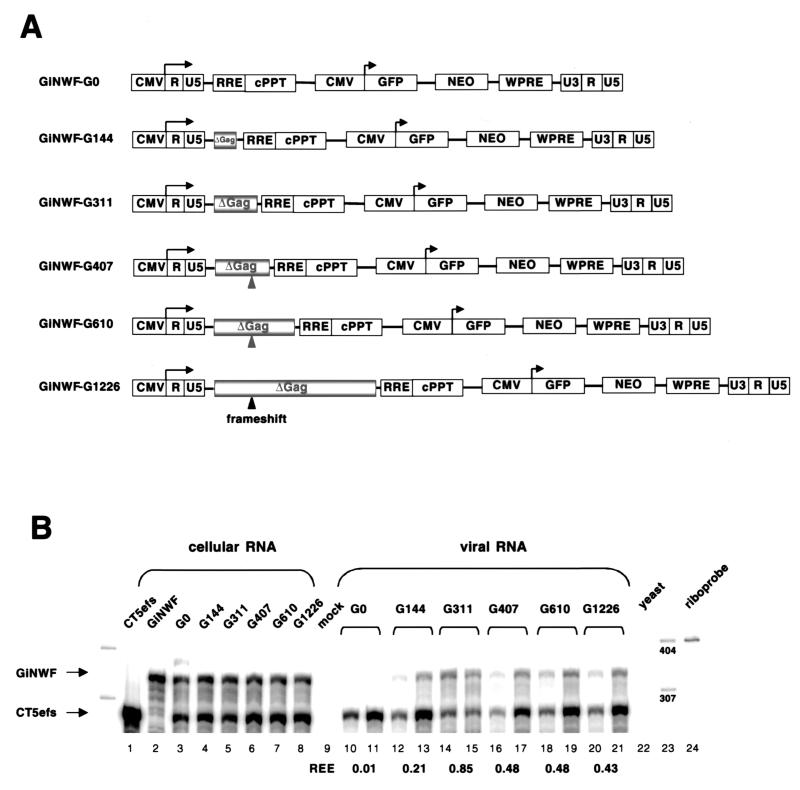
While the above-described results provided evidence that FIV encapsidation determinants reside upstream of the MSD, they left unanswered how the packaging apparatus distinguishes between spliced and unspliced mRNAs. Inclusion of intronic sequences within retroviral encapsidation determinants allows such discrimination. Indeed, in HIV-1 (12, 24) and MoMLV (5), packaging determinants extend into gag. We therefore investigated the contribution of FIV gag sequences to packaging with six subgenomic FIV transfer vectors containing variable portions (0, 144, 311, 407, 610, and 1,226 nt) of gag (Fig. 8A). The three transfer vectors with the longest gag fragments have a frameshift generated by a single-nucleotide insertion at position 298 of gag to prevent expression of potentially interfering Gag protein fragments. Encapsidation efficiencies of the vector RNAs were analyzed in a competitive assay with wild-type FIV (CT5efs), which was the source of all virion proteins. When no gag sequences were present in the vector RNA (GiNWF-G0), packaging was abolished (Fig. 8B, lanes 10 and 11). Inclusion of the first 144 nt (GiNWF-G144) increased the REE of vector RNA to 0.21 (lanes 12 and 13). With 311 nt of gag sequences present (GiNWF-G311), almost-wild-type levels were reached (REE, 0.85; lanes 14 and 15). However, addition of further gag sequences (GiNWF-G407, GiNWF-G610, and GiNWF-G1226) was suboptimal (lanes 16 to 21), resulting in REEs of 0.43 and 0.48, respectively. Consistent with these results, FIV vectors lacking gag sequences had a negligible ability to transduce marker genes (5-log-unit reduction in titer [data not shown]). These results were not an artifact of the particular FIV transfer vector context. In an experiment analogous to that of Adam and Miller (1), we placed the entire 5′ untranslated region and proximal 33 nt of FIV gag upstream of a heterologous test RNA (eGFP) lacking any other viral sequences; no detectable packaging was conferred (data not shown). Since the first 311 nt of gag is necessary (but not sufficient) for efficient packaging, the results identify a mechanism whereby FIV can encapsidate its genomic mRNA in preference to subgenomic mRNAs. As a further control for specificity, HIV-1 and HIV-2 packaging plasmids did not enable detectable cross-packaging of FIV vectors, and FIV packaging plasmids did not enable detectable transfer of HIV-1 or HIV-2 vectors (data not shown).
FIG. 8.
The proximal 311 nt of gag is necessary for efficient FIV RNA encapsidation. (A) Schematic representation of GiNWF-G clones, subgenomic FIV transfer vectors containing increasing amounts of gag sequences. (B) RPA of a competitive encapsidation experiment. Wild-type CT5efs was cotransfected with GiNWF-G0, -G144, -G311, -G407, -G610, or -G1226. Antisense riboprobe pSPT-FLAP was used. Protected RNA species are 358 nt for all GiNWF-G clones and 296 nt for CT5efs.
DISCUSSION
These experiments identify major determinants of FIV genomic RNA encapsidation. Encapsidation determinants are present within a large span of the 5′ end of the genome, requiring regions located upstream of the tRNA PBS, and within the proximal gag gene. Since intron (gag) sequences are required, the results disclose a mechanism whereby FIV can encapsidate its genomic mRNA in preference to subgenomic mRNAs.
Theoretically, any first intron sequences (from the MSD to the first splice acceptor just upstream of vif), could function to allow the encapsidation apparatus to selectively identify full-length FIV mRNA, because this region is absent from all subgenomic mRNAs. It was therefore plausible that the gag region that we identified in the subgenomic FIV vector experiments (Fig. 8) could function as a stand-alone encapsidation determinant. However, it is clear from the lack of encapsidation of CF1Δenv, CF16Δenv, and CF1efs transcripts that this is not the case. Each was effectively excluded from encapsidation in competitive and noncompetitive experiments. However, discriminating between spliced and unspliced FIV mRNAs is likely to be only one of the constraints on the packaging signal recognition and related interactions with nucleocapsid or other proteins derived from the Gag-Pol precursor; i.e., there may be other selection pressures for the apparent dispersal of encapsidation elements within the 5′ end of the RNA. For example, some recent evidence suggests that recombinant FIV nucleocapsid protein may bind to FIV RNAs at or upstream of the PBS (within U5) and may initiate dimerization and facilitate minus-strand initiation at this location (30). That study did not examine encapsidation. If FIV differs from HIV, murine retroviruses, and avian sarcoma-leukosis virus (ASLV) in having a main locus for NC interaction in the vicinity of the tRNA PBS, this would be consistent with our observations that deletions upstream of the PBS, particularly of U5, are deleterious for FIV encapsidation. Whether retroviral dimerization precedes or is causally related to packaging remains controversial (37).
A 70% deletion between the MSD and gag did not substantially attenuate FIV encapsidation. Consistent with the encapsidation data, this virus (CT5Δpsi2) was also replication competent and reached nearly wild-type levels of RT production in tissue culture. The deletion in CT5Δpsi2 extends to within 2 nt of the GU dinucleotide involved in MSD intron lariat formation but did not appear to affect the intricate lentiviral splicing program, since Western blot patterns of FIV proteins in cell lysates and levels of RT production were indistinguishable from those of the wild type, and robust productive replication of this virus was observed. The region is highly conserved in FIV genomes. Although proper splicing may be inferred from the unperturbed appearance of Rev-dependent structural proteins, splicing was not examined directly in our study, and contributions of the region to optimal splicing or encapsidation remain possible. HIV-1 encapsidation and replication are blocked by deletions between the MSD and the gag ATG codon (2, 20); HIV-2 encapsidation is reduced by deleting half of this region (29), and larger deletions highly attenuate encapsidation and block replication (31). In this regard, it is notable that the MSD-gag interval is shorter in FIV than in HIV-1 and HIV-2 (20 versus 44 and 75 nt, respectively). In contrast, the principal identified packaging determinant in the ASLV group of avian retroviruses is located upstream of the MSD (3, 17). Nevertheless, full-length ASLV RNA is highly favored over subgenomic mRNAs for encapsidation, a paradox that remains unexplained (4).
The results of the noncompetitive and competitive RPAs for FIV encapsidation were consistent, although the competitive assays in some cases showed slightly greater attenuation of mutants for packaging (for example, compare packaging efficiencies for CF1Δenv in Fig. 3 and 4). Thus, it is clear that for FIV, encapsidation under noncompetitive conditions is substantially reduced. Parallel assays for HIV-1 have produced somewhat contrasting results, since in the absence of a wild-type mRNA (noncompetitive assay), encapsidation of mutant RNAs lacking important packaging signals can be substantial (26). The consistent results obtained with both assays for FIV notwithstanding, we view competitive assays for encapsidation as inherently more convincing, because by definition the same Gag-Pol precursor pools and assembling particles are available to the competing intracellular viral RNAs. In addition, the competitive assay reduces problems with intersample variation, since both RNAs are processed and analyzed within the same sample. Random degradation and other artifacts that may accrue incrementally during transfection, preparation of RNAs, and RNase protection assays are thus less likely to confound comparisons of one mRNA with another.
The FIV encapsidation determinants identified here are favorably situated for constructing optimal replication-defective lentiviral vector systems. Definition of the minimum sequences that are necessary and sufficient for encapsidation is required for this purpose, because these systems depend on efficient packaging of transfer vector mRNAs by viral proteins supplied in trans. Simultaneously, preventing encapsidation of viral protein-encoding mRNAs, as well as minimizing and preferably eliminating homologous overlap between these and transfer vector components, is important to prevent formation of replication-competent recombinants. The documented exclusion from encapsidation of FIV packaging construct transcripts suggests that minimizing FIV replication-competent recombinant potential is feasible. Since the critical overlap in gag in plasmids such as CF16Δenv is short (311 nt), one simple way to eliminate significant 5′-end homology between transfer vector and packaging constructs would be to change codons from lentiviral usage to optimal human usage at these nucleotides.
The observations of a graded increase in encapsidation for transfer vectors until 311 nt of gag was present, followed by a decrease of encapsidation with additional increments of gag (Fig. 8), provides an approach for improvements in transfer vector efficacy (24). We do not know why transfer vectors having more than 311 nt of gag are packaged less well. A possible explanation is secondary structure that arises with the artificially juxtaposed RRE. In addition, the first FIV transfer vectors (33) contained all of the Gag ORF (with a frameshift early in the reading frame). Corresponding to the lower packaging efficiency, these vectors resulted in significantly lower titers than those obtained with second-generation transfer vectors having the shorter gag fragment (24). Moreover, the competitive RPAs (Fig. 8) suggest that further optimization may be possible, since current transfer vector mRNA encapsidation is still not fully equivalent to that of the wild type.
Acknowledgments
We thank Tom Hope for pBluescriptIISK+WPREB11 and R. Cattaneo, M. Federspiel, V. von Messling, and T. Whitwam for critical reviews of the manuscript.
The support of NIH grant AI47536 (to E.M.P.) is gratefully acknowledged.
REFERENCES
- 1.Adam, M. A., and A. D. Miller. 1988. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 62:3802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini, A., and R. A. Young. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, J. D., and M. L. Linial. 2000. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 74:456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks, J. D., A. Yeo, K. Green, F. Cepeda, and M. L. Linial. 1998. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 72:6190-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, M. A., T. D. Palmer, R. E. Gelinas, and A. D. Miller. 1987. Evidence that the packaging signal of Moloney murine leukemia virus extends into the Gag region. J. Virol. 61:1639-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berglund, J. A., B. Charpentier, and M. Rosbash. 1997. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 25:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz, R. D., and S. P. Goff. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202:233-246. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz, R. D., M. L. Hammarskjold, C. Helga-Maria, D. Rekosh, and S. P. Goff. 1995. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology 212:718-723. [DOI] [PubMed] [Google Scholar]
- 10.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donello, J. E., J. E. Loeb, and T. J. Hope. 1998. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 72:5085-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elder, J. H., and T. R. Phillips. 1993. Molecular properties of feline immunodeficiency virus (FIV). Infect. Agents Dis. 2:361-374. [PubMed] [Google Scholar]
- 13.Harrison, G. P., and A. M. Lever. 1992. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 66:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison, G. P., G. Miele, E. Hunter, and A. M. Lever. 1998. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 72:5886-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helga-Maria, C., M. L. Hammarskjold, and D. Rekosh. 1999. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4127-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jewell, N. A., and L. M. Mansky. 2000. In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 81:1889-1899. [DOI] [PubMed] [Google Scholar]
- 17.Katz, R. A., R. W. Terry, and A. M. Skalka. 1986. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J. Virol. 59:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaye, J. F., and A. M. Lever. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaye, J. F., J. H. Richardson, and A. M. Lever. 1995. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J. Virol. 69:6588-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lever, A., H. Gottlinger, W. Haseltine, and J. Sodroski. 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 63:4085-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lever, A. M., J. H. Richardson, and G. P. Harrison. 1991. Retroviral RNA packaging. Biochem. Soc. Trans. 19:963-966. [DOI] [PubMed] [Google Scholar]
- 22.Linial, M. L., and A. D. Miller. 1990. Retroviral RNA packaging: sequence requirements and implications. Curr. Top. Microbiol. Immunol. 157:125-152. [DOI] [PubMed] [Google Scholar]
- 23.Lochrie, M. A., S. Waugh, D. G. Pratt, Jr., J. Clever, T. G. Parslow, and B. Polisky. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 25:2902-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewen, N., M. Fautsch, M. Peretz, C. Bahler, J. D. Cameron, D. H. Johnson, and E. M. Poeschla. 2001. Genetic modification of human trabecular meshwork with lentiviral vectors. Hum. Gene Ther. 12:2109-2119. [DOI] [PubMed] [Google Scholar]
- 25.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride, M. S., and A. T. Panganiban. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70:2963-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride, M. S., and A. T. Panganiban. 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 71:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride, M. S., M. D. Schwartz, and A. T. Panganiban. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCann, E. M., and A. M. Lever. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 71:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moscardini, M., M. Pistello, M. Bendinelli, D. Ficheux, J. T. Miller, C. Gabus, S. F. Le Grice, W. K. Surewicz, and J. L. Darlix. 2002. Functional interactions of nucleocapsid protein of feline immunodeficiency virus and cellular prion protein with the viral RNA. J. Mol. Biol. 318:149-159. [DOI] [PubMed] [Google Scholar]
- 31.Poeschla, E., J. Gilbert, X. Li, S. Huang, A. Ho, and F. Wong-Staal. 1998. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J. Virol. 72:6527-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poeschla, E., and D. Looney. 1998. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J. Virol. 72:6858-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poeschla, E., F. Wong-Staal, and D. Looney. 1998. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 34.Sakalian, M., and E. Hunter. 1998. Molecular events in the assembly of retrovirus particles. Adv. Exp. Med. Biol. 440:329-339. [DOI] [PubMed] [Google Scholar]
- 35.Sakalian, M., J. W. Wills, and V. M. Vogt. 1994. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J. Virol. 68:5969-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shields, A., W. N. Witte, E. Rothenberg, and D. Baltimore. 1978. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell 14:601-609. [DOI] [PubMed] [Google Scholar]
- 37.Swanstrom, R., and J. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Huges, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 38.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vicenzi, E., D. S. Dimitrov, A. Engelman, T. S. Migone, D. F. Purcell, J. Leonard, G. Englund, and M. A. Martin. 1994. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J. Virol. 68:7879-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitwam, T., M. Peretz, and E. M. Poeschla. 2001. Identification of a central DNA flap in feline immunodeficiency virus. J. Virol. 75:9407-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, J., and H. M. Temin. 1993. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science 259:234-238. [DOI] [PubMed] [Google Scholar]
- 42.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]