Abstract
The leader region of the human immunodeficiency virus type 1 (HIV-1) genome has a highly folded structure, comprising at least two RNA stem-loops [the transactivation response (TAR) and poly(A) hairpins] near its 5′ end and four others (SL1 to SL4) downstream. Each of these stem-loops contributes to the function of the HIV-1 packaging signal, which efficiently targets genomic RNA into nascent virions. The central 140-base region of the leader, which includes the U5 and primer binding site (PBS) sequences, is also believed to adopt a complex structure, but the nature of this structure and its possible role in RNA packaging have not been extensively explored. Here we report a mutational analysis identifying at least three separate loci within the U5-PBS region which, when mutated, impair both HIV-1 packaging specificity and infectivity in a single-round proviral assay. In common with those of all previously described packaging signals in the leader, the function of one of these loci appeared to depend on secondary structure rather than on sequence alone. By contrast, the activity of the other two loci did not correlate with any predicted conformations. Moreover, unlike SL1 to SL4, the TAR, poly(A), and U5-PBS hairpins were not bound with high affinity by the nucleocapsid portion of the HIV-1 Gag protein in vitro, implying that they contribute to packaging through a mechanism distinct from that of SL1 to SL4. Our findings confirm the existence and importance of secondary structure around the PBS and demonstrate that functional packaging signals are distributed across the entire HIV-1 leader.
The process by which two strands of genomic RNA are specifically incorporated into an assembling retrovirus particle is known as packaging or encapsidation. This process is highly specific in that the genomic RNA accounts for a much greater proportion of RNA in virions than in the cytoplasm of infected cells, where virion assembly occurs. Most cellular and spliced viral RNAs, moreover, tend to be excluded from virions. The specificity of packaging is believed to depend largely on direct binding interactions between the viral polyprotein Gag—the main structural component of human immunodeficiency virus type 1 (HIV-1) capsids—and a cis-acting region known as the packaging signal (or ψ site) located in the 5′ leader region of the genomic RNA (reviewed in references 5 and 14). These interactions are mediated by the nucleocapsid (NC) domain of Gag, which includes a pair of zinc fingers flanked by several basic amino acid residues (1, 19, 21, 37, 40) and which binds specifically to HIV-1 ψ RNA sequences in vitro (3, 6, 8, 9, 16, 18, 20, 28, 38).
The HIV-1 ψ locus, in turn, has been shown to comprise several RNA stem-loop elements that each contribute individually to overall packaging efficiency (Fig. 1A) (10, 11, 17, 22, 29-31). Among these is a cluster of four stem-loops, designated SL1 to SL4, that span residues 243 to 352 of the genomic RNA and that have each been shown to serve as a specific, high-affinity binding site for Gag in vitro (3, 9, 12, 28). A strong correlation has been observed between the in vitro Gag-binding and in vivo packaging activities of these stem-loops. In addition, it has previously been shown that an artificially selected RNA aptamer that bound Gag with high affinity could functionally serve as a substitute for at least one of these native ψ elements (12). These and other findings support the view that the packaging activity of stem-loops SL1 to SL4 depends on their folded conformations and their ability to bind, perhaps cooperatively, to multiple copies of Gag. SL1 may also contribute to HIV-1 packaging by virtue of its unique capacity to initiate 5′ dimerization of the genome (13, 27, 33, 35).
FIG.1.
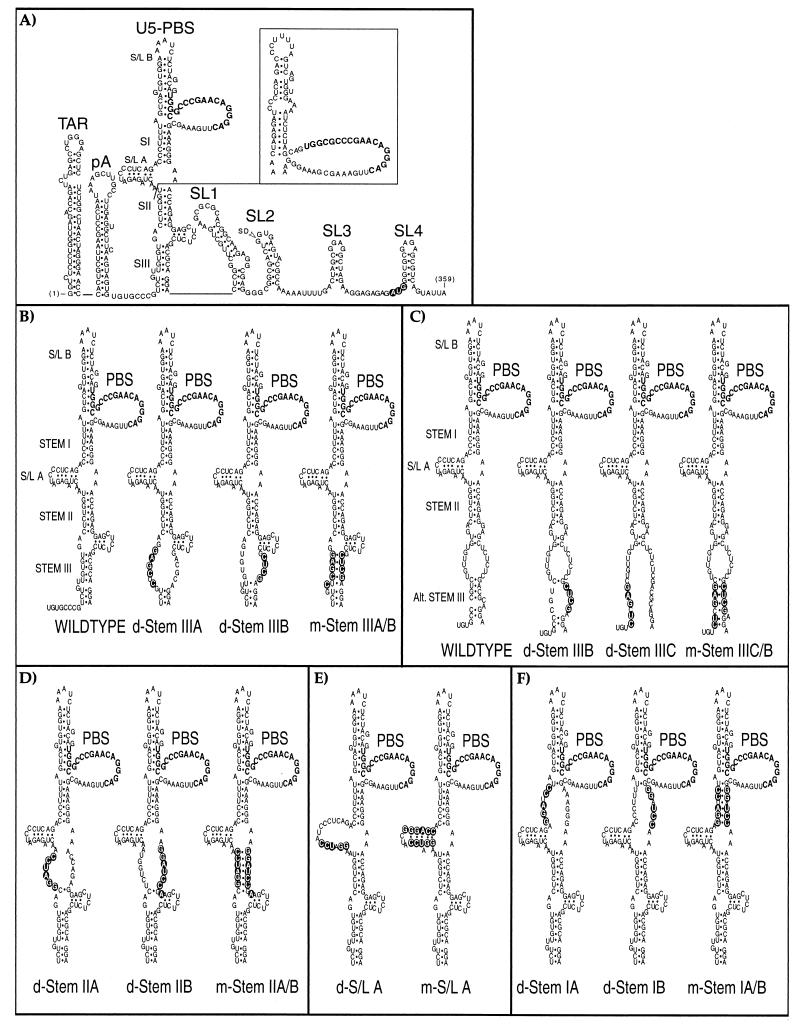
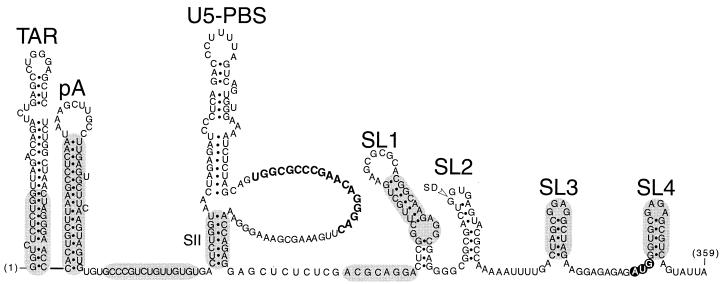
Mutant HIV-1 RNAs characterized in this study. (A) The 5′ leader region of wild-type HIV-1 RNA from strain HXB2, with the gag initiation codon shown with open lettering and the major 5′ splice donor (SD) indicated. The conformations shown for the TAR, poly(A) (pA), and SL1 to SL4 stem-loops were extensively validated by earlier genetic, functional, and structural data. The U5-PBS region, which is the focus of this study, is shown in two proposed conformations that are compatible with the results of published accessibility mapping studies (2, 15). The three major postulated stems (SI, SII, and SIII) are identified as described previously (2); two associated stem-loop elements are designated S/L A and S/L B in this study, and the 18 nucleotides (nt) of the PBS are shown in boldface type. An alternative conformation (15) for the distal sequences is shown as an insert. (B to F) The sequences and putative structural consequences of the 13 mutations tested in this study. Mutations predicted to disrupt base pairing are designated with the prefix “d-” whereas compensatory mutations designed to preserve potential base pairing have “m-” as the prefix. In most cases, compensatory mutations were constructed by combining a pair of disruptive mutations. The mutations shown in panels B and C were designed to test two alternative conformations of stem III. BspEI was used to linearize the plasmid DNA before the riboprobes used in the RNase protection assays were synthesized. An additional nucleotide outside stem II was altered in d-Stem IIB and m-Stem IIA/B to avoid creating a second BspEI restriction enzyme site; for the same reason, one nucleotide outside the stem of S/L A was altered in the m-S/L A mutant.
The region comprising SL1 to SL4 alone, however, does not appear to be sufficient to target RNA into HIV-1 virions (7). Instead, the minimal region thus far shown to confer autonomous packaging activity spans the first 350 to 400 contiguous bases of the genome, and thus includes not only SL1 to SL4 but also 242 bases upstream (22, 25, 31, 36). The functionally relevant features upstream of SL1 and their precise contributions to packaging have not been characterized fully. We and others have shown that mutations that disrupt the proximal stems of two structures in the R region—the transactivation response (TAR) and poly(A) stem-loops, respectively—can selectively interfere with genomic packaging (10, 17, 22). The activity of these packaging loci within the TAR and poly(A) stems, like those of SL1 to SL4, depends on their base-paired structures, but it is not yet known whether they act as binding sites for Gag or might instead subserve another, as-yet-undetermined function. Moreover, deletion analyses have suggested that unspecified sequences located between the poly(A) and SL1 hairpins also contribute to packaging (10, 31). This 138-base region, which includes the primer binding site (PBS), is predicted to fold into a complex, folded structure (Fig. 1A) (2, 4, 15). At least two alternative conformations of the U5-PBS region have been proposed (2, 15), however, and there has been little direct evidence to support the existence or biological significance of either conformation in vivo or to indicate which portions of the U5-PBS region may contribute to packaging.
We created a series of 13 disruptive and compensatory mutations within the U5-PBS region in order to map features that contribute to the specificity of RNA packaging in a single-round replication assay. The design of the mutations was based primarily on a model proposed by Beerens et al. (2) (Fig. 1A), which depicts the proximal U5-PBS region as forming three major stems (designated stems I, II, and III) and a short internal stem-loop (S/L A). Targeting those proposed stems individually, we mutated clusters of residues on the 5′ or 3′ strand of each, producing mutants (designated d-Stem mutants) that were predicted to disrupt stem formation; complementary pairs of these mutations were then combined in an attempt to restore each predicted stem through compensatory mutation (creating mutants designated m-Stem). We avoided targeting sequences in and around the PBS itself, including an adjacent, predicted stem-loop (S/L B), as such mutations might be expected to cause defects in tRNA binding and reverse transcription that would confound our analysis.
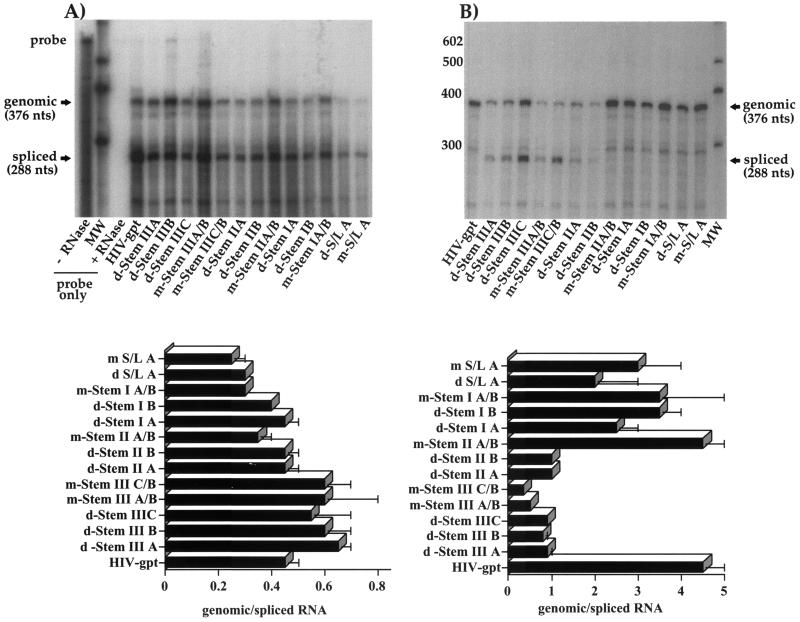
The 13 mutations (Fig. 1B to F) were introduced into the ψ region of HIV-gpt, a cloned, full-length HIV-1 proviral vector in which a portion of the env gene has been replaced by a selectable drug resistance marker (26). When HIV-gpt is cotransfected into cells together with a vector that expresses the amphotropic murine leukemia virus Env protein, viral particles are produced that can infect target cells and transduce the drug resistance trait but that cannot undergo further rounds of replication. Under selective conditions, each infected cell can give rise to a drug-resistant colony, whose frequency provides a measure of the infectivity of the virus preparation. When transiently cotransfected into human 293T cells, each of our mutant constructs directed the synthesis and release of approximately wild-type levels of the viral p24 core antigen to the culture supernatant (data not shown); therefore, these mutations did not appear to interfere with transcription of the viral genomic RNA or with the subsequent expression of proviral genes. We then isolated cytoplasmic RNA from the transfected 293T cells and probed them for HIV-1 RNA using a previously described quantitative RNase protection assay (RPA) that distinguishes between genomic and spliced viral transcripts (10-12, 34). As shown in Fig. 2A, we found that although the total abundance of viral RNA varied somewhat among mutants, the steady-state ratio of genomic to spliced RNAs was fairly constant, never differing by as much as twofold from that of wild-type HIV-gpt. Thus, none of the mutations greatly affected the pattern of viral RNA expression in transfected cells.
FIG. 2.
Quantitative RNase protection assays. Cytoplasmic (A) or virion-derived (B) RNAs were annealed to an excess of radiolabeled mutant-specific riboprobe and treated with single-strand-specific RNases, and the resulting protected fragments were separated on denaturing polyacrylamide gels. For each construct, the largest protected fragment (376 nt) corresponds to genomic RNA sequences whereas the second major fragment (288 nt) corresponds to spliced sequences. All riboprobes were also mixed with 2 μg of Escherichia coli tRNA and subjected to the assay with (+) or without (−) RNase treatment. Panel A includes an aliquot (1/20) of the wild-type probe without RNase treatment. MW, molecular weight markers (indicated in nucleotides). Each of the top panels shows an autoradiogram from one representative experiment; bottom panels depict composite data and standard errors from two independent RNase protection assays. Standard errors too small to depict graphically are not shown. Assays were performed exactly as previously described (10-12, 34) and were quantitated by phosphorimaging.
This same RPA was then used to quantify viral RNAs within the virions released from these cells. We harvested cell supernatants at 48 h posttransfection, partially purified virions by pelleting through 20% sucrose cushions, extracted RNA from equal numbers of pelleted virions (as assayed by p24 antigen content) carrying the various ψ mutations, and then measured the ratio of genomic to spliced viral transcripts in each mutant population, which we and others have found provides a reliable measure of packaging efficiency. As shown in Fig. 2B, we found that each of the five mutations targeting stem I or stem-loop A had only modest effects on virion RNA content, which provided no conclusive evidence as to whether either proposed structure actually forms. By contrast, both mutations designed to disrupt stem II (d-Stem IIA and d-Stem IIB) caused marked defects in packaging which were fully corrected when these two complementary mutations were combined to restore the proposed structure (m-Stem IIA/B). These findings confirm that stem II folds as predicted and that its structure, but not its sequence, contributes to RNA packaging.
Mutations targeting the putative stem III region (d-Stem IIIA, IIIB, and IIIC) also caused packaging defects of a severity comparable to that seen with disruption of stem II (Fig. 2B). Each of these mutations significantly reduced the ratio of genomic to spliced RNAs, and two of the three (d-Stem IIIA and IIIB) also reduced the absolute quantity of genomic RNA found in virions. At this location, however, compensatory mutations failed to reverse the defect, regardless of whether they were designed to restore the predicted stem III fold (m-Stem IIIA/B) or an alternative, less thermostable conformation (m-Stem IIIC/B) (Fig. 1B and C, respectively). Indeed, both of the putative compensatory mutations were more severely defective than their disruptive counterparts, perhaps reflecting the greater number of mutated bases contained by these mutants. Sequences in the putative stem III thus contribute significantly to packaging function, but their effects do not correlate with or support the existence of the proposed secondary structure in this region.
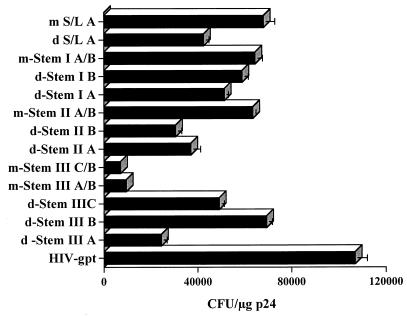
Viral supernatants were then used to infect human HOS cell cultures, and the infectivity of each mutant was assessed (Fig. 3) by enumeration of drug-resistant colonies as previously described (10-12, 34). None of the mutants proved as infectious as wild-type HIV-gpt, perhaps reflecting inefficient primer binding or other effects on PBS function (see below). Relative infectivities within most groups of mutants, however, tended to mirror their packaging activities. In particular, infectivity was reduced by each of the two disruptive mutations in stem II (d-Stem IIA and IIB) but was partially restored when these two complementary mutations were combined (m-Stem IIA/B), confirming the existence and function of this stem. Interestingly, mutations in S/L A suggested a similarly beneficial effect of compensatory mutation which might be accounted for by their parallel but statistically nonsignificant effect on packaging (Fig. 2B). All mutations in stem I produced only modest effects, as did two of the three disruptive mutations in stem III. As in the case of packaging, the most severe infectivity defects were observed with complementary mutants m-Stem IIIA/B and IIIB/C, which had been intended to preserve putative stem III structures.
FIG. 3.
The infectivities of the various mutants were assayed in HOS cells and expressed as the number of Escherichia coli guanine phosphoribosyltransferase-positive (gpt+) CFU per microgram of viral p24 antigen. Assays of viral stocks from at least two transfection and infection assays yielded similar results. Standard errors too small to depict graphically are not shown. Colony formation assays were performed exactly as previously described (10-12, 34).
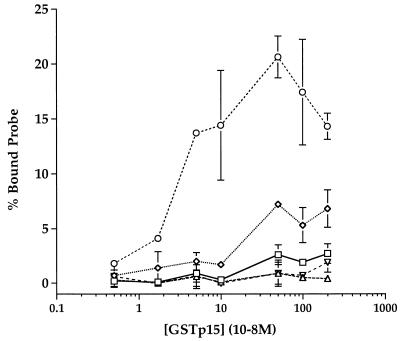
The foregoing results indicated that point mutations at certain discrete sites in the U5-PBS region could impair HIV-1 RNA packaging and infectivity to a degree similar to that described previously for mutations in the TAR, poly(A), and SL1 to SL4 stem-loops (10, 11, 34). Because the packaging activity of SL1 to SL4 is correlated with their ability to bind HIV-1 Gag or NC protein in vitro, we investigated whether the same was true of the upstream packaging elements. For this purpose, we synthesized short RNAs corresponding to various portions of the wild-type HIV-1 leader and measured their binding affinities for a bacterially expressed NC fusion protein by using a nitrocellulose filter-binding assay. It has been reported previously that, in the presence of excess nonspecific RNA, this NC derivative can bind a transcript encompassing SL1 to SL4 with an apparent dissociation constant (Kd) of 10 to 100 nM in this assay and that it can bind any one of these four stem-loops individually with only four- to eightfold-lower affinity but that it does not bind appreciably to the antisense forms of the same RNAs (9). For the present study (Fig. 4), we confirmed high-affinity binding to a 114-base polyvalent construct of SL1 to SL4 but found only weak affinity (Kd > 1 μM) for a comparably sized (139-base) U5-PBS transcript and detected no appreciable binding to the antisense U5-PBS sequence or to shorter RNAs containing the TAR or poly(A) stem-loop alone. Thus, in stark contrast to SL1 to SL4, the elements upstream of SL1 that influence HIV-1 RNA packaging show little or no affinity for HIV-1 NC protein in vitro.
FIG. 4.
Filter-binding assay of glutathione S-transferase (GST)-p15 protein binding to radiolabeled HIV-1 leader-derived RNAs. The GST-p15 fusion protein was expressed and purified as described previously (9). Five radiolabeled RNAs were in vitro transcribed with T7 RNA polymerase from PCR-generated DNA products containing the T7 promoter positioned directly upstream from the desired nucleotide in the HIV-1 leader region. The TAR transcript (▵) extended from nucleotide positions +1 to 57 (57 nt), the poly(A) transcript (▿) extended from positions 58 to 104 (47 nt), the U5-PBS transcript (◊) extended from positions 104 to 242 (139 nt), and the SL1-to-SL4 transcript (○) extended from positions 241 to 354 (114 nt). An antisense RNA transcript which was the exact complement of the U5-PBS sequence, and hence extended from nt 242 to 104 (139 nt), was also synthesized (anti-U5-PBS) (□). Binding reactions were performed as described previously (9). Briefly, assays were performed in duplicate with a 30-μl reaction mixture containing the indicated amounts of GST-p15, 10 μg of E. coli tRNA, 5 U of RNase inhibitor, and 50,000 cpm (about 5 ng) of 32P-labeled transcript in 30 mM HEPES, 50 mM KCl, 10 μM ZnCl2, and 2 mM dithiothreitol, pH 7.5. After 15 min at 25°C, reaction mixtures were placed on prewetted nitrocellulose disks (0.45-μm pore size) and filtered through with 3 ml of the above-mentioned buffer before the filters were dried and quantitated by liquid scintillation counting. All data were corrected for background binding of the probe to filters in the absence of protein. Each data point and error bar indicates the mean ± standard error for duplicate determinations in a single representative experiment. All binding assays were repeated at least three times with similar results.
Our mutational analysis confirms that at least one base-paired stem structure (stem II) in the U5-PBS region forms as predicted and that its formation is biologically significant, promoting both RNA packaging and infectivity of HIV-1. The two halves of this stem are separated by 85 bases in the linear sequence of the leader, making it one of the longest-range structural interactions yet detected in the HIV-1 genome. In addition, we found that mutations immediately 5′ or 3′ of stem II can also impair packaging activity but that these two flanking regions appear to function independently rather than together as a helix (stem III) or other evident secondary structure.
Our findings extend those of Beerens et al. (2), who introduced mutations into the putative stem I, II, and III regions of an infectious HIV-1 clone and assessed the effects on replication kinetics. In accord with our results, those authors found that mutations targeting the postulated stem I had little functional effect and that disruptive mutations in the stem III region impaired replication whereas mutations intended to restore its structure exacerbated the defect. Their results also demonstrated a biological effect of mutations targeting stem II but did not confirm the existence of this stem in vivo. It is not clear why their compensatory stem II mutation failed to restore replication while ours corrected the packaging defect and partially restored infectivity, but this discrepancy may have resulted from differences in the specific mutant sequences used or between our single-round replication system and their spreading-virus assay. In the same study, Beerens et al. (2) also identified an 8-base sequence (GACUCUGG), which they termed the primer activation signal (PAS), that overlapped all but one base pair of stem II and appeared to promote the initiation of reverse transcription. It is interesting that our compensated mutant m-Stem II A/B, in which five bases of the PAS are altered, has a persistent 30 to 40% defect in infectivity despite nearly wild-type packaging, perhaps indicating a defect in PAS function. More recently, Huthoff and Berkhout (24) have proposed that the HIV-1 leader as a whole can interconvert between the folded structure depicted in Fig. 1A and an alternative conformation and that this refolding serves as a switch to promote either encapsidation or translation of the viral RNA. Because both proposed conformers share stem II in common, however, our results can neither support nor refute this regulatory model nor the existence of either large-scale conformation in vivo. Thus, although chemical and enzymatic accessibility data (2, 15) indicate that the U5-PBS region is highly structured in vitro, stem II and, arguably, stem-loop A are the only features of its structure that have so far been verified biologically.
Figure 5 presents a schematic model of the HIV-1 leader, indicating regions (shaded) where point mutations have been found, in this or previous studies (10, 11), to impair packaging. Eight such regions have been identified, six of which coincide with stem or stem-loop structures that have proven functional significance. The downstream sites, in SL1 to SL4, contribute to packaging by virtue of their ability to bind Gag, but this does not appear to be the case for any of the sites upstream, whose mechanisms of action remain unknown. One possibility is that these upstream sites influence the global folding of the leader, indirectly promoting access of Gag to its binding sites in SL1 to SL4. Another is that certain upstream sites might contribute, along with SL1, to dimerization of the genome and thereby perhaps double the effective valency of the RNA for Gag. In this regard, a secondary dimer contact has been observed upstream of SL1 in the HIV-1 genome (23), and two deletion mutations in the U5-PBS region—involving the 3′ sides of stems I and II or that of stem III—were each recently reported to reduce RNA dimer yield in HIV-1 virions (39), though the extent to which any packaging defect might have contributed to this phenotype is unknown. Finally, it is also possible that one or more of these upstream sites represent binding sites for other, as-yet-unspecified viral or host proteins that serve as cofactors for efficient RNA packaging (32, 41).
FIG. 5.
Working model of genetically verified RNA secondary structures and cis-acting packaging signals (shaded) within the first 359 nt of the HIV-1 genome. The shaded ovals represent those regions, identified here and in earlier studies (10-12), that have been shown genetically to be necessary for optimally efficient genomic RNA encapsidation.
Acknowledgments
We thank A. Mujeeb and S. Georgantis for the purified NC protein and Zeneida Mosquera for technical help.
This research was supported by NIH grants AI37036 and AI40317.
REFERENCES
- 1.Aldovini, A., and R. A. Young. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beerens, N., F. Groot, and B. Berkhout. 2001. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 276:31247-31256. [DOI] [PubMed] [Google Scholar]
- 3.Berglund, J. A., B. Charpentier, and M. Rosbash. 1997. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 25:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz, R. D., and S. P. Goff. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202:233-246. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz, R. D., M. L. Hammarskjold, C. Helga-Maria, D. Rekosh, and S. P. Goff. 1995. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology 212:718-723. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz, R. D., J. Luban, and S. P. Goff. 1993. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 67:7190-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clever, J., C. Sassetti, and T. G. Parslow. 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 69:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever, J. L., D. A. Eckstein, and T. G. Parslow. 1999. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J. Virol. 73:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever, J. L., R. A. Taplitz, M. A. Lochrie, B. Polisky, and T. G. Parslow. 2000. A heterologous, high-affinity RNA ligand for human immunodeficiency virus Gag protein has RNA packaging activity. J. Virol. 74:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 15.Damgaard, C. K., H. Dyhr-Mikkelsen, and J. Kjems. 1998. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 26:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das, A. T., B. Klaver, B. I. Klasens, J. L. van Wamel, and B. Berkhout. 1997. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71:2346-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 19.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 67:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geigenmuller, U., and M. L. Linial. 1996. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J. Virol. 70:667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helga-Maria, C., M. L. Hammarskjold, and D. Rekosh. 1999. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4127-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoglund, S., A. Ohagen, J. Goncalves, A. T. Panganiban, and D. Gabuzda. 1997. Ultrastructure of HIV-1 genomic RNA. Virology 233:271-279. [DOI] [PubMed] [Google Scholar]
- 24.Huthoff, H., and B. Berkhout. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaye, J. F., J. H. Richardson, and A. M. Lever. 1995. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J. Virol. 69:6588-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landau, N. R., K. A. Page, and D. R. Littman. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 28.Lochrie, M. A., S. Waugh, D. G. Pratt, Jr., J. Clever, T. G. Parslow, and B. Polisky. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 Gag polyprotein. Nucleic Acids Res. 25:2902-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride, M. S., and A. T. Panganiban. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70:2963-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride, M. S., M. D. Schwartz, and A. T. Panganiban. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouland, A. J., J. Mercier, M. Luo, L. Bernier, L. DesGroseillers, and E. A. Cohen. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 74:5441-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muriaux, D., P. M. Girard, M. B. Bonnet, and J. Paoletti. 1995. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J. Biol. Chem. 270:8209-8216. [DOI] [PubMed] [Google Scholar]
- 34.Ohi, Y., and J. L. Clever. 2000. Sequences in the 5′ and 3′ R elements of human immunodeficiency virus type 1 critical for efficient reverse transcription. J. Virol. 74:8324-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paillart, J. C., R. Marquet, E. Skripkin, B. Ehresmann, and C. Ehresmann. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269:27486-27493. [PubMed] [Google Scholar]
- 36.Parolin, C., T. Dorfman, G. Palu, H. Gottlinger, and J. Sodroski. 1994. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J. Virol. 68:3888-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon, D. T. K., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakaguchi, K., N. Zambrano, E. T. Baldwin, B. A. Shapiro, J. W. Erickson, J. G. Omichinski, G. M. Clore, A. M. Gronenborn, and E. Appella. 1993. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc. Natl. Acad. Sci. USA 90:5219-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, N., L. Jette, M. A. Wainberg, and M. Laughrea. 2001. Role of stem B, loop B, and nucleotides next to the primer binding site and the kissing-loop domain in human immunodeficiency virus type 1 replication and genomic-RNA dimerization. J. Virol. 75:10543-10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y., and E. Barklis. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 69:5716-5722. (Erratum, 71: 5712, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]