Abstract
The mechanisms that control energy homeostasis and tissue growth during development are closely linked through the insulin signal transduction pathway. Changes in the level of insulin and other hormones reflect the nutritional status of the animal to control circulating sugar levels and fat metabolism. Systemic defects in insulin responsiveness can lead to elevated circulating glucose levels and fat accumulation. Here we present evidence that the microRNA miR-278 plays a role in the control of energy homeostasis in Drosophila. miR-278 mutants have elevated insulin production and are correspondingly lean. Despite the elevated insulin levels, miR-278 mutants have elevated circulating sugar, mobilized from adipose-tissue glycogen stores. We provide evidence that miR-278 mutants are insulin resistant and that miR-278 acts through regulation of the expanded transcript.
Keywords: miRNA, lipid, tissue growth
The insulin signal transduction pathway has well-characterized roles in the control of energy metabolism and tissue growth during animal development (for review, see Hafen 2004). Through a relay of phosphorylation events, activation of the Insulin Receptor leads to activation of PI3K, Akt, and finally TOR kinases. The TOR pathway is also independently regulated by cellular glucose and amino acid levels, thereby integrating information about the nutritional status of the cell with that of the organism (for review, see Avruch et al. 2005). This signaling cassette has two main output branches. One is transcriptional, via Akt-mediated phosphorylation, which inactivates FOXO-family transcription factors by promoting their cytoplasmic retention (Puig et al. 2003; Wolfrum et al. 2003). The second output controls cellular protein synthesis via TOR, which acts on 4E-BP and S6K, both important regulators of translation (for review, see Hay and Sonenberg 2004). The FOXO and TOR output branches of the insulin pathway have each been implicated in control of fat metabolism in insects and mammals (Wolfrum et al. 2004; Arquier et al. 2005; Reiling et al. 2005; Teleman et al. 2005b; Tettweiler et al. 2005).
microRNAs (miRNAs) are small noncoding RNAs that serve as post-transcriptional repressors of gene expression. miRNAs are estimated to comprise 1%–5% of animal genes (Bartel 2004; Bentwich et al. 2005; Berezikov et al. 2005), making them an abundant class of regulators. Target prediction methods have provided a means to infer possible functions for individual miRNAs, but to date, definitive biological functions have been assigned to only a few miRNAs by genetic analysis (Lee et al. 1993; Reinhart et al. 2000; Brennecke et al. 2003; Johnston and Hobert 2003; Xu et al. 2003; Sokol and Ambros 2005) or by other methods (Poy et al. 2004; Giraldez et al. 2005; Krutzfeldt et al. 2005). Here we present evidence that the Drosophila miRNA miR-278 contributes to control of energy balance principally by regulating insulin responsiveness.
Results and Discussion
The miR-278 locus was identified in a gain-of-function screen for genes affecting tissue growth during Drosophila development. Expression of P(EP)26043 in the posterior compartment of the wing imaginal disc caused robust overgrowth (Fig. 1A,B). P(EP)26043 was inserted in a region of ∼50 kb that did not contain annotated protein-coding genes in the correct orientation to allow their expression under Gal4 control. Although the nearest correctly oriented gene, miR-278, was located ∼45 kb from the EP element (Fig. 1A), Northern blot analysis showed that miR-278 was overexpressed by the EP element under Gal4 control (Fig. 1E).
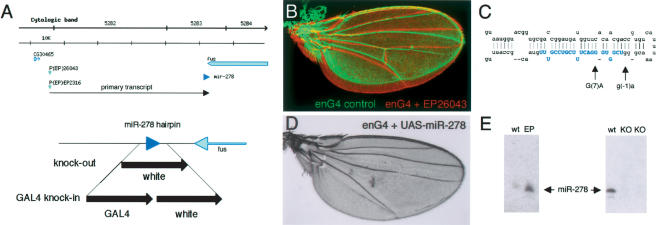
Figure 1.
The miR-278 locus. (A) Schematic representation of the miR-278 locus. (Top) Overview of the genomic region (bar, 10 kb). P(EP)26043 and P(EP)EP2316 produced the tissue overgrowth phenotype shown in B. Both are ∼45 kb from the miR-278 sequence (blue arrowhead). (Bottom) Detail of the miR-278 region. miR-278 is located 300 bp from the 3′ end of the fus transcript. “knock-out” indicates the region replaced with the mini-white gene by homologous recombination; “Gal4 knock-in,” indicates the same region replaced by Gal4 and mini-white. (B) Adult wings from an engrailedGal4 control fly (green) and a fly expressing miR-278 in the posterior compartment under engrailedGal4 control from EP26043 (red). The wings are aligned in the anterior to show overgrowth of the posterior compartment expressing miR-278. (C) The predicted miR-278 hairpin, showing mutations identified as EMS-induced revertants of the overexpression phenotype. Blue shading indicates mature miR-278. (D) Adult wing expressing UAS-miR-278 in the posterior compartment under engrailedGal4 control. Note the more severe overgrowth and loss of the posterior cross-vein caused by UAS-miR-278. This resembles the phenotype of expanded mutant wings (not shown). (E) Northern blots showing miR-278 RNA. (Left) Overexpression in tubulinGal4 EP26043 flies compared with wild-type controls. (Right) Loss of miR-278 RNA in homozygous knock-out mutant flies (two independent isolates) compared with wild type. Thirty micrograms of total RNA per lane.
Two independent lines of evidence confirmed that miR-278 was responsible for the tissue overgrowth phenotype. First, we screened 60,000 chromosomes for EMS-induced mutations that suppressed the phenotype caused by GMR-GAL4 driven overexpression of P(EP)26043. One suppressor was a deletion removing miR-278 and three adjacent genes. A second suppressor altered position 7 of the mature miRNA (G7A) (Fig. 1C). A third suppressor affected the sequence of the hairpin at position –1, adjacent to the mature miRNA. Both point mutations only partially rescued the miR-278 overexpression phenotype and are therefore considered to be hypomorphic. Second, a UAS transgene expressing a 500-base pair (bp) genomic fragment containing the miR-278 hairpin also produced a strong tissue overgrowth phenotype (Fig. 1D).
Mutants lacking miR-278 were produced by homologous recombination, replacing 160 bp, including the miR-278 hairpin with the mini-white gene (Fig. 1A). miR-278 was absent in flies homozygous for two independent targeting events (Fig. 1E). A second null allele was produced by introducing Gal4 and mini-white in place of miR-278 (Fig. 1A). Both alleles were homozygous viable and produced flies of normal body size (Fig. 2A). miR-278 is expressed at low levels during early development, becoming detectable in L2 larvae and increasing to peak levels in pupae and adult flies (Fig. 2B). Examination of tissues dissected from mature third instar larvae showed no detectable expression in imaginal discs, but miR-278 was expressed in brain, gut, salivary gland, and fat body, the adipose tissue of the fly (Fig. 2B). Furthermore, expression of GFP under control of a 7-kb miR-278 promoter Gal4 fusion transgene showed predominantly fat body expression (Fig. 2C).
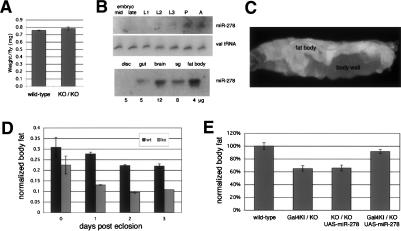
Figure 2.
miR-278 mutants are lean. (A) Average body weight of miR-278 mutant and control flies. Batches of 40 2-d-old males were assayed; experiment was done in quadruplicate. (B) Northern blot showing expression of miR-278 at different developmental stages. Loading control indicates the same filter probed to detect valine tRNA. (Bottom) Northern blot showing miR-278 expression in tissues dissected from third instar larvae. The amount of total RNA in each sample is indicted below. (C) Promoter GAL4 fusion driving GFP shows predominant fat body expression and weak body wall expression. (D) Total body triglycerides and total body protein were measured after controlled growth and aging. Plotted values are triglyceride levels normalized to protein levels. Batches of eight males were assayed; the experiment was done in triplicate. (E) Total body triglycerides normalized to total body protein for 8-d-old flies of the indicated genotypes. Batches of eight males were assayed; the experiment was done in triplicate. The low body fat level of the mutant was rescued by expression of UAS-miR-278 under control of miR-278-Gal4, which expresses Gal4 under control of the endogenous miR-278 regulatory elements.
In view of the connection between control of tissue growth and energy metabolism, and the prominent expression of miR-278 in fat body, we asked whether miR-278 mutants might have metabolic defects. Drosophila stores fat as triglycerides, mainly in the fat body. Homozygous mutant animals lacking miR-278 were lean, measured as the normalized ratio of total body triglycerides to total protein. The magnitude of the difference increased during the first days of adult life (Fig. 2D). A comparable lean phenotype was found in flies trans-heterozygous for the knock-out allele and the Gal4 knock-in (Fig. 2E). Use of the Gal4 knock-in allele allowed us to test whether this phenotype was attributable to loss of miR-278. This proved to be the case, as expression of UAS-miR-278 under miR-278 Gal4 control suppressed the lean phenotype of the mutant flies (Fig. 2E).
Fat storage and mobilization are controlled by circulating hormones, including insulin-like peptides (ILPs) and adipokinetic hormone (AKH). AKH induces mobilization of glycogen and fat reserves (Lee and Park 2004). To investigate whether elevated AKH could explain the leanness of miR-278 mutants, we tested transcript levels by quantitative RT–PCR (Q-PCR). AKH transcript levels were not elevated (Fig. 3A), but the ILPs expressed in brain neurosecretory cells (Ikeya et al. 2002) were upregulated in heads from miR-278 mutants (Fig. 3B). Elevated ILP expression should lead to systemic activation of the insulin signaling pathway. Previous reports have indicated that systemic reduction of insulin production or signaling produces obesity in flies (Böhni et al. 1999; Rulifson et al. 2002 Hwangbo et al. 2004). To investigate whether the elevated insulin production in miR-278 mutants is responsible for their lean phenotype, we measured fat levels in flies in which we simultaneously reduced insulin signaling by removing one copy of the catalytic subunit of PI3K (dp110) (Fig. 3C). This reduction of insulin signaling capacity was sufficient to largely suppress the leanness of the miR-278 mutants. Flies with a stronger reduction of insulin signaling produced by removing chico, the adaptor subunit for PI3K, show reduced viability in addition to obesity (Böhni et al. 1999). Elevated insulin levels in the miR-278 mutant proved to be sufficient to counteract the reduced viability of chico mutants (Fig. 3D), but not to overcome their obesity (data not shown). These findings suggest that the miR-278 mutant phenotypes can, to a large extent, be attributed to elevated insulin signaling due to overexpression of the ILP genes.
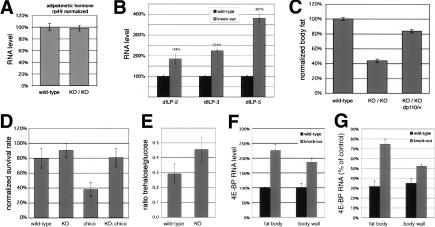
Figure 3.
Elevated insulin and circulating sugar in miR-278 mutants. (A,B) Q-PCR on RNA from adult flies (A) or adult heads (B) normalized to rp49 RNA. Animals were grown under controlled conditions. AKH RNA was unaltered, whereas the Drosophila insulin-like peptides ILP2, ILP3, and ILP5 were upregulated. (C) Total body triglycerides normalized to total body protein for flies of the indicated genotypes. (D) Survival rates. Fifty first instar larvae were seeded per tube and eclosing adult flies counted. Samples indicate the average of 10 tubes. The reduced survival of the chico mutant was significantly rescued by reduction of miR-278 (t-test = 1.2 × 10–7). Error bars indicate standard deviation. (E) Ratio of glucose and trehalose levels in adult males grown under controlled conditions (Teleman et al. 2005b). Trehalase was omitted for the glucose measurement. (F) Expression level of 4E-BP in control and mutant tissues by Q-PCR. (G) Insulin-induced changes in gene expression in wild-type and mutant larvae by Q-PCR. Values show 4E-BP levels 1 h post-insulin injection as percentage of mock injection.
Why are insulin levels elevated in the miR-278 mutant? Under normal circumstances, insulin levels increase in response to elevated circulating sugar after feeding, leading to renormalization of circulating sugar levels. Interestingly, miR-278 mutants showed elevated circulating sugar in the form of trehalose (Fig. 3E), which is sufficient to explain their elevated insulin production. However, despite high insulin levels, the mutant flies remain hyperglycemic. Since trehalose is generated by mobilization of glycogen reserves in the fat body, this suggested that the fat body might be insulin resistant.
To test this, we measured insulin-dependent transcriptional response in miR-278 mutants. Impaired insulin signaling leads to nuclear translocation of the transcription factor Foxo and activation of its target 4E-BP (Puig et al. 2003; Teleman et al. 2005b). 4E-BP levels were higher in mutant fat body, indicating reduced insulin sensitivity, despite higher insulin production in the mutant (Fig. 3F). Insulin injection into wild-type larvae strongly reduced 4E-BP levels in the fat body but caused only modest reduction in the miR-278 mutant fat body (Fig. 3G). Peripheral insulin-sensitive tissues, where miR-278 is expressed at lower levels, were less strongly affected in the mutant (larval body wall muscle and epidermis) (Fig. 2C). A second Foxo target, InR, behaved similarly. Thus we observe a greater degree of insulin resistance in the fat body than in peripheral tissues. Insulin resistance in fat body has previously been shown to cause leanness in Drosophila (Teleman et al. 2005b). This resembles the situation in mice, in which selective impairment of insulin signaling in adipose tissue causes leanness, whereas systemic insulin resistance causes obesity (for review, see Kitamura et al. 2003). Thus, the differential impairment of insulin responsiveness that we observe in adipose vs peripheral tissues in the miR-278 mutant may be sufficient to explain the lean phenotype.
miRNAs are small noncoding RNAs that repress gene expression by recruiting effector complexes (miRNPs) to miRNA complementary sites on mRNAs. miRNP recruitment depends on sequence complementarity to the 5′ end of the miRNA, which provides much of the information needed for target recognition (Brennecke et al. 2005; Lewis et al. 2005). We therefore looked for possible miR-278 target genes with sequences complementary to the 7-nucleotide (nt) seed region of miR-278 in their 3′ untranslated regions (UTRs). miR-278 is unusual in having only 91 seed matches: The average miRNA generated 479 such matches (Supplementary Table 1). This raises the possibility that miR-278 has relatively few biologically important targets. miR-278 seed matches were distributed in the 3′UTRs of 63 genes, three of which had two sites (multiple transcript isoforms account for the difference between 91 total sites and 66 sites in 63 unique genes) (see Supplementary Table 2). Most of these genes were detectably expressed in adipose tissue (57/63). Among these expanded was noteworthy because of its known role in growth regulation and because its RNA level was most elevated in miR-278 mutants (Supplementary Table 2).
expanded loss-of-function mutants cause tissue overgrowth (Boedigheimer et al. 1997; Blaumueller and Mlodzik 2000), consistent with the expected effects of target repression by miR-278. The expanded 3′UTR contains two potential miR-278 sites (Fig. 4A). Their ability to confer regulation by miR-278 was tested using a luciferase reporter linked to the expanded 3′UTR, expressed in S2 cells with or without miR-278. As a positive control, we used a reporter with two perfect RNA interference (RNAi)-type miR-278 sites in its 3′UTR. miR-278 expression reduced luciferase activity from this reporter to 10% and from the expanded 3′UTR reporter to 60% of control levels (Fig. 4B). To confirm that miR-278 can regulate expanded at the post-transcriptional level, we made use of sevenless-Gal4 to direct expression the endogenous expanded gene in the developing eye. This causes a small externally roughened eye. Coexpression of miR-278 with expanded suppressed this defect to a considerable extent (Fig. 4C). miR-278 overexpression was able to reduce expression of the endogenous Expanded protein in the wing imaginal disc (Fig. 4D,E), as well as Expanded overexpressed from a UAS transgene, confirming that this reflects post-transcriptional regulation (Supplementary Fig. 1). Reduced Expanded expression provides an explanation for the tissue overgrowth caused by miR-278 overexpression. Indeed, coexpression of Expanded from a transgene suppressed the overgrowth caused by miR-278 overexpression (Fig. 4F).
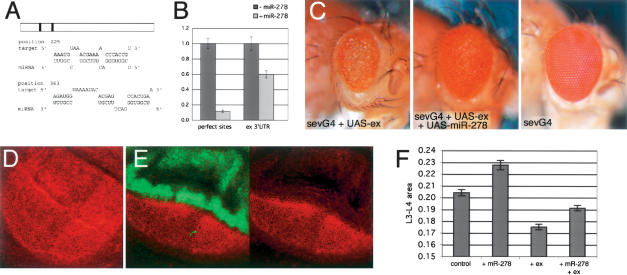
Figure 4.
miR-278 regulates expanded. (A) Potential miR-278 target sites in the expanded 3′UTR. (B) Normalized luciferase activity for a reporter transgene containing the expanded 3′UTR compared with a control reporter with (black bars) and without (gray bars) coexpression of miR-278. Error bars indicate standard deviations. (C) Eyes from flies overexpressing expanded under sevenless-Gal4 control (left) and overexpressing expanded and miR-278 together (middle), and a control eye (right) for comparison. (D) Expanded protein in a wild-type wing imaginal disc visualized by antibody labeling (red). (E) Down-regulation of Expanded protein in the dorsal compartment of a wing disc expressing miR-278 under apterous-Gal4 control (green). The dorsal compartment has an extra fold due to tissue hyperplasia. (F) Expanded expression suppresses miR-278 overgrowth. Intervein L3–L4 area as percentage of total wing size for dpp-GAL4 driving indicated transgenes.
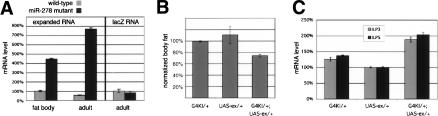
Three lines of evidence suggest that misregulation of expanded contributes to the lean phenotype of miR-278 mutants. expanded mRNA levels increased several fold in miR-278 mutant tissues (Fig. 5A). This effect is post-transcriptional, as expression of a lacZ-reporter inserted in the expanded gene did not increase in the absence of miR-278. Elevated expression of expanded under miR-278-Gal4 control in otherwise wild-type flies was sufficient to cause elevated expression of the brain-expressed ILP genes and to produce a lean phenotype (Fig. 5B,C). This indicates that increased expanded expression in tissues that express miR-278 is sufficient to cause the changes observed in the miR-278 mutant. It therefore follows that the elevated expanded RNA level observed in the mutant can explain the observed defects.
Figure 5.
expanded regulation and the miR-278 mutant phenotype (A) Expanded mRNA levels and expanded locus transcription levels, as assayed by lacZ expression from an insertion, in control and mutant tissues, measured by Q-PCR. (B) Total body triglycerides normalized to total body protein for flies of the indicated genotypes. Batches of eight males were assayed. The experiment was done in triplicate. (C) ILP3 and ILP5 mRNA levels measured by Q-PCR in adult flies of the indicated genotypes.
Expanded is a membrane-associated protein that acts together with the ERM-like protein Merlin, the fly ortholog of the NF2 tumor suppressor (McCartney et al. 2000). Merlin mediates regulatable association between membrane and cytoskeleton to organize membrane domains that regulate adhesion and signaling. While there are many possible modes of action for the Expanded–Merlin complex, it has been reported to modulate growth-factor receptor signaling. A number of molecular associations between Merlin and proteins involved in RTK signaling have been reported, as has a role in endocytosis, all of which may affect signaling (McClatchey and Giovannini 2005). Elevated Expanded levels may be responsible for the reduced insulin sensitivity observed in miR-278 mutants, perhaps identifying a novel cause of insulin resistance.
Our findings have focused on the role of expanded as a target of miR-278 action in vivo. While these results indicate that much of the miR-278 mutant phenotype can be attributed to failed regulation of a single target, we do not exclude the possibility that other target genes might also contribute. It is noteworthy that miR-278 appears to have relatively few potential target sites, and thus may act via fewer target genes than most “average” miRNAs, which are thought to regulate hundreds of genes (Brennecke et al. 2005; Grun et al. 2005; Krek et al. 2005; Lewis et al. 2005; Xie et al. 2005).
The role of miR-278 in keeping expanded mRNA at very low levels in adipose tissue, where the miRNA is expressed, is intriguing in light of the emerging view that target RNAs are often expressed at very low or undetectable levels in the miRNA-expressing tissues (Farh et al. 2005; Stark et al. 2005). In this case, because of the strong reduction in expanded transcript level, it can be argued that the miRNA contributes meaningfully to shaping the expression domain of its target RNA.
miRNAs are thought to control many processes in development and disease (for review, see Alvarez-Garcia and Miska 2005; see also Poy et al. 2004; Lu et al. 2005; Sokol and Ambros 2005; Zhao et al. 2005). It is intriguing that several miRNAs are though to regulate energy homeostasis. miR-14 has been shown to regulate fat metabolism by an unknown mechanism (Xu et al. 2003). Our finding that miR-278 regulates insulin production parallels a recent report that miR-375 controls insulin secretion (Poy et al. 2004). Inhibition of miR-375 activity in cultured pancreatic β-cells using anti-sense oligonucleotides increased insulin secretion. We have provided evidence that miR-278 acts by a different mechanism, at least in part through regulation of expanded to control insulin sensitivity in adipose and peripheral tissues.
Both in mammals and in flies, the total amount of body fat depends on the balance of insulin signaling in adipose tissue relative to other insulin responsive tissues (e.g., muscle). Mice in which the insulin receptor is specifically knocked-out in adipose tissue have a 50% decrease in fat pad mass. In contrast, mice in which the insulin receptor is specifically removed from muscle have increased fat stores (for review, see Kitamura et al. 2003). This is likely to reflect whether circulating glucose is consumed in muscle or in adipose tissue to be stored—and insulin signaling affects this balance. The same holds true in flies. Flies with impaired insulin signaling in adipose tissue are lean (Teleman et al. 2005b). In contrast, flies in which insulin signaling is decreased systemically—by ablating the neurosecretory cells which secrete ILPs; by removing the insulin receptor substrate, chico, throughout the whole animal; or by feeding flies rapamycin—are obese (Böhni et al. 1999; Broughton et al. 2005; Teleman et al. 2005a). This must be caused by reduced insulin signaling in tissues other than the adipose tissue. In this context, miR-278 helps tip the balance toward fat accumulation. miR-278 increases insulin sensitivity, and in the wild-type animal, it is most strongly expressed in adipose tissue.
Materials and methods
Reagents
EP(2)26043 was isolated using Gal4 directed overexpression of EPg elements as described (Teleman et al. 2005b). UAS-expanded was described (Blaumueller and Mlodzik 2000). EP(2)2316, Apterous-Gal4, engrailed-Gal4, and sevenless-Gal4, are described at the Berkeley Drosophila Genome Project (http://www.fruitfly.org). Antibody to Expanded protein was as described in McCartney et al. (2000).
Targeted homologous recombination
Ends-out homologous recombination was performed essentially as described (Gong and Golic 2003). Left and right arms of the targeting vector were prepared by PCR of 3.6-kb fragments from BAC DNA using oligos gcggccgcGCCACATTGCGCCTTCAGTTT + gcggccgcAAAATTGTCG CAGGTGAACGG for the upstream flank and ggatccCTGTCTTGC ACCTGCCGTCCA + ggatccAGTTGCGGCGCTTTAGTGTCT for the downstream flank. Flies carrying the targeting construct were crossed to flies with heat-inducible FLP recombinase and I-SceI endonuclease (70FLP and 70I-SceI, Bloomington Stock Center), and the resulting progeny were heat-shocked for 45 min at 38°C on day 3 of development. Mosaic-eyed females were crossed to If/CyO males, and targeting events were mapped to the second chromosome in the next generation. Absence of the miR-278 locus was tested first by genomic PCR and then by Northern blot for the mature product. Targeted mutants were tested for the presence of a function copy of the nearby fus gene by complementation.
RNA analysis
Northern blots for miRNAs were prepared as in Brennecke et al. (2003). Q-PCR was performed as in Teleman et al. (2005a).
Triglyceride and trehalose measurements were as described in Teleman et al. (2005b).
Reporter gene assays
Luciferase 3′UTR reporter and miRNA plasmids were expressed under control of the tubulin promoter. miR-278 was expressed from a plasmid containing a 500-bp genomic region, including the hairpin. S2 cells were transfected in six-well plates with 0.1 μg of the firefly luciferase reporter plasmid, 0.1 μg of a Renilla luciferase expressing plasmid (transfection control), and 1 μg of the miRNA expression plasmid or empty vector. Transfections were performed in triplicate. Dual-luciferase assays (Promega) were performed 2.5 d after transfection according to the manufacturer's protocol.
Q-PCR
PCRs were performed using the ABI7000 or ABI7500 machines and the ABI SYBR green system. All measurements were normalized to rp49. To assay transcription levels of the expanded locus, mRNA levels of lacZ were measured for the exe1 allele. This allele is an imprecise excision of a pLacW insert at the transcription start site of expanded, leaving behind the lacZ (Boedigheimer et al. 1993).
Insulin injections
Prewandering third instar larvae were injected with either PBS + 0.1% phenyl red (“mock”), or the same buffer + 200 nM bovine insulin (“insulin”) into their hemolymph. Larvae were then returned to normal food for 1 h before tissue dissection and RNA extraction.
Acknowledgments
We thank Ann-Mari Voie and Eva Loeser for technical assistance.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.374406.
References
- Alvarez-Garcia I. and Miska, E.A. 2005. MicroRNA functions in animal development and human disease. Development 132: 4653–4662. [DOI] [PubMed] [Google Scholar]
- Arquier N., Bourouis, M., Colombani, J., and Leopold, P. 2005. Drosophila Lk6 kinase controls phosphorylation of eukaryotic translation initiation factor 4E and promotes normal growth and development. Curr. Biol. 15: 19–23. [DOI] [PubMed] [Google Scholar]
- Avruch J., Lin, Y., Long, X., Murthy, S., and Ortiz-Vega, S. 2005. Recent advances in the regulation of the TOR pathway by insulin and nutrients. Curr. Opin. Clin. Nutr. Metab. Care 8: 67–72. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37: 766–770. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H., and Cuppen, E. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21–24. [DOI] [PubMed] [Google Scholar]
- Blaumueller C.M. and Mlodzi, M.K. 2000. The Drosophila tumor suppressor expanded regulates growth, apoptosis, and patterning during development. Mech. Dev. 92: 251–262. [DOI] [PubMed] [Google Scholar]
- Boedigheimer M., Bryant, P., and Laughon, A. 1993. Expanded, a negative regulator of cell proliferation in Drosophila, shows homology to the NF2 tumor suppressor. Mech. Dev. 44: 83–84. [DOI] [PubMed] [Google Scholar]
- Boedigheimer M.J., Nguyen, K.P., and Bryant, P.J. 1997. expanded functions in the apical domain to regulate the growth rate of imaginal discs. Dev. Genet. 20: 103–110. [DOI] [PubMed] [Google Scholar]
- Böhni R., Riesgo-Escovar, J., Oldham, S., Brogiolo, W., Stocker, H., Andruss, B.F., Beckingham, K., and Hafen, E. 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 97: 865–875. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the pro-apoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark, A., Russell, R.B., and Cohen, S.M. 2005. Principles of microRNA-target recognition. PLoS Biol. 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S.J., Piper, M.D., Ikeya, T., Bass, T.M., Jacobson, J., Driege, Y., Martinez, P., Hafen, E., Withers, D.J., Leevers, S.J., et al. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. 102: 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., and Bartel, D.P. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310: 1817–1821. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli, R.M., Glasner, M.E., Enright, A.J., Thomson, J.M., Baskerville, S., Hammond, S.M., Bartel, D.P., and Schier, A.F. 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308: 833–838. [DOI] [PubMed] [Google Scholar]
- Gong W.J. and Golic, K.G. 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. 100: 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Wang, Y.L., Langenberger, D., Gunsalus, K.C., and Rajewsky, N. 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen E. 2004. Interplay between growth factor and nutrient signaling: Lessons from Drosophila TOR. Curr. Top. Microbiol. Immunol. 279: 153–167. [DOI] [PubMed] [Google Scholar]
- Hay N. and Sonenberg, N. 2004. Upstream and downstream of mTOR. Genes & Dev. 18: 1926–1945. [DOI] [PubMed] [Google Scholar]
- Hwangbo D.S., Gersham, B., Tu, M.P., Palmer, M., and Tatar, M. 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429: 562–566. [DOI] [PubMed] [Google Scholar]
- Ikeya T., Galic, M., Belawat, P., Nairz, K., and Hafen, E. 2002. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12: 1293. [DOI] [PubMed] [Google Scholar]
- Johnston R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Kitamura T., Kahn, C.R., and Accili, D. 2003. Insulin receptor knockout mice. Annu. Rev. Physiol. 65: 313–332. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun, D., Poy, M.N., Wolf, R., Rosenberg, L., Epstein, E.J., MacMenamin, P., da Piedade, I., Gunsalus, K.C., Stoffel, M., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37: 495–500. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky, N., Braich, R., Rajeev, K.G., Tuschl, T., Manoharan, M., and Stoffel, M. 2005. Silencing of microRNAs in vivo with `antagomirs.' Nature 438 685–689. [DOI] [PubMed] [Google Scholar]
- Lee G. and Park, J.H. 2004. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz, G., Miska, E.A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B.L., Mak, R.H., Ferrando, A.A., et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838. [DOI] [PubMed] [Google Scholar]
- McCartney B.M., Kulikauskas, R.M., LaJeunesse, D.R., and Fehon, R.G. 2000. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development 127: 1315–1324. [DOI] [PubMed] [Google Scholar]
- McClatchey A.I. and Giovannini, M. 2005. Membrane organization and tumorigenesis: The NF2 tumor suppressor, Merlin. Genes & Dev. 19: 2265–2277. [DOI] [PubMed] [Google Scholar]
- Poy M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230. [DOI] [PubMed] [Google Scholar]
- Puig O., Marr, M.T., Ruhf, M.L., and Tjian, R. 2003. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes & Dev. 17: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling J.H., Doepfner, K.T., Hafen, E., and Stocker, H. 2005. Diet-dependent effects of the Drosophila Mnk1/Mnk2 homolog Lk6 on growth via eIF4E. Curr Biol 15: 24–30. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Rulifson E.J., Kim, S.K., and Nusse, R. 2002. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 296: 1118–1120. [DOI] [PubMed] [Google Scholar]
- Sokol N.S. and Ambros, V. 2005. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 19: 2343–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke, J., Bushati, N., Russell, R., and Cohen, S. 2005. Animal microRNAs cofer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Chen, Y.W., and Cohen, S.M. 2005a. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes & Dev. 19: 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2005b. Drosophila melted modulates FOXO and TOR activity. Dev. Cell 9: 271–281. [DOI] [PubMed] [Google Scholar]
- Tettweiler G., Miron, M., Jenkins, M., Sonenberg, N., and Lasko, P.F. 2005. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes & Dev. 19: 1840–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C., Besser, D., Luca, E., and Stoffel, M. 2003. Insulin regulates the activity of forkhead transcription factor Hnf-3β/Foxa-2 by Akt-mediated phosphorylation and nuclear/cytosolic localization. Proc. Natl. Acad. Sci. 100: 11624–11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C., Asilmaz, E., Luca, E., Friedman, J.M., and Stoffel, M. 2004. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature 432: 1027–1032. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu, J., Kulbokas, E.J., Golub, T.R, Mootha, V., Lindblad-Toh, K., Lander, E.S., and Kellis, M. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Samal, E., and Srivastava, D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220. [DOI] [PubMed] [Google Scholar]