Abstract
Recruitment of the 40S ribosome to the 5′ end of a eukaryotic mRNA requires assembly of translation initiation factors eIF4E, the cap-binding protein, together with eIF4A and eIF4G into a complex termed eIF4F. While the translational repressor 4E-BP1 regulates binding of eIF4E to eIF4G, the forces required to construct an eIF4F complex remain unidentified. Here, we establish that the herpes simplex virus-1 (HSV-1) ICP6 polypeptide associates with eIF4G to promote eIF4F complex assembly. Strikingly, release of eIF4E from the 4E-BP1 repressor is insufficient to drive complex formation, suggesting that ICP6 is an eIF4F-assembly chaperone. This is the first example of a translation initiation factor-associated protein that promotes active complex assembly and defines a new, controllable step in the initiation of translation. Homology of the N-terminal, eIF4G-binding segment of ICP6 with cellular chaperones suggest that factors capable of interacting with eIF4G and promoting eIF4F complex assembly may play important roles in a variety of processes where translation complexes need to be remodeled or assembled on populations of newly synthesized or derepressed mRNAs, including development, differentiation, and the response to a broad spectrum of environmental cues.
Keywords: Translational control, eIF4F assembly, eIF4E phosphorylation, HSV-1 replication
Among the many interactions between viruses and their hosts, those that recruit the cellular translational machinery to viral mRNAs play a decisive role in viral replication (for review, see Sarnow 2003; Schneider and Mohr 2003). As animal viruses do not encode the set of components required to initiate mRNA translation, they not only must commandeer cellular translation initiation factors to conscript ribosomes into service, but must also manipulate the regulatory circuitry that controls complex assembly. Failure to engage the core translational machinery and properly control its activity would not only severely curtail the production of virus-encoded polypeptides, but likely either impede progression of the viral life cycle or slow it down to such an extent that the infection is easily contained by innate host defenses, both of which would have a serious impact on the spread or survival of the virus within its host.
A critical step in the recruitment of the translational machinery to the 5′ end of most mRNAs is the recognition of the unique 7-methyl guanine structure by eIF4E, the cellular cap-binding protein. Through an association with eIF4G, a large scaffolding protein that coordinates interactions among numerous translation factors including eIF4A, an RNA unwinding enzyme, the eIF4F complex is formed, which, in turn, recruits the 40S ribosomal subunit bound to eIF3 (for review, see Gingras et al. 1999). Binding of eIF4G to eIF4E triggers a conformational change in both polypeptides that enhances the association of eIF4E with the cap and stabilizes the complex (Gross et al. 2003). In addition to mediating an association among the core eIF4F components, eIF4G also physically associates with the cellular kinase mnk along with PABP, the poly(A)-binding protein (Imataka et al. 1998; Pyronnet et al. 1999; Waskiewicz et al. 1999; Scheper et al. 2001). The interaction between PABP and eIF4G stimulates translation and is thought to form a topological “closed loop” between the 5′ and 3′ ends of the mRNA, possibly serving as a checkpoint that ensures each mRNA is a suitable substrate for translation initiation (Wells et al. 1998; Kahvejian et al. 2001). Mnk is the cellular eIF4E kinase, and is poised to integrate proliferative environmental signals from MAP kinases and stress-induced signals mediated by p38 (Waskiewicz et al. 1997). While the exact mechanism remains elusive, eIF4E phosphorylation can regulate translation (for review, see Scheper and Proud 2002; Mohr 2006).
The ability of eIF4E to functionally associate with eIF4G in an eIF4F complex appears to be a major point at which translation is regulated. This is thought to be achieved primarily through the actions of a family of small, eIF4E-binding proteins that negatively regulate translation, the 4E-BPs, and is regulated by protein phosphorylation (for review, see Gingras et al. 2001). In its hypophosphorylated state, 4E-BP1 binds eIF4E and occludes the binding site for eIF4G, preventing the incorporation of eIF4E into the eIF4F complex. However, upon phosphorylation by mTOR, eIF4E is released from the 4E-BP1 repressor and is subsequently available to associate with eIF4G (Haghighat et al. 1995; Mader et al. 1995; Marcotrigiano et al. 1999). The effectiveness of this model requires that the abundance of 4E-BP1 is sufficiently high to produce meaningful changes in the availability of eIF4E, and that eIF4E, once released from the 4E-BP1 repressor, is efficiently incorporated into the eIF4F complex. Significantly, the existence of factors that are required to actively promote the assembly of eIF4F complexes has not been explored.
Recently, we and others demonstrated that herpes simplex virus-1 and human cytomegalovirus, members of the medically important herpesvirus family, are able to stimulate the assembly of eIF4F complexes in primary human cells (Kudchodkar et al. 2004; Walsh and Mohr 2004; Walsh et al. 2005). In part, this involves the phosphorylation and inactivation of the translational repressor 4E-BP1. In addition, interfering with eIF4E phosphorylation by inhibiting the activity of the eIF4G-associated kinase mnk reduced viral replication between 100- and 300-fold (Walsh and Mohr 2004; Walsh et al. 2005). In HSV-1-infected cells, both of these activities require the master regulator encoded by the ICP0 gene (Walsh and Mohr 2004). One of the first polypeptides produced in HSV-1 infected cells (Elshiekh et al. 1991; Lium and Silverstein 1997), ICP0 is a multifunctional transactivator of viral gene expression with ubiquitin E3 ligase activity important for both vegetative viral replication and reactivation of latent infections (for review, see Everett 2000; Hagglund and Roizman 2004). However, while ICP0 is required for phosphorylation of both eIF4E along with 4E-BP1 and stimulates eIF4F complex assembly in infected cells, whether it achieves this by acting directly, indirectly, or through some combination of these two alternatives has remained obscure. Here, we establish that these translation control mechanisms require the products of ICP0-responsive genes, one of which, the ICP6 polypeptide, associates with eIF4G to promote eIF4F complex assembly. Remarkably, release of eIF4E from the 4E-BP1 repressor in the absence of the ICP6 polypeptide is insufficient to propel its incorporation into the eIF4F complex, suggesting that ICP6 is an eIF4F-assembly chaperone. This is the first example of a translation initiation factor-associated protein that promotes active eIF4F complex assembly and defines a new, controllable step in the initiation of translation.
Results
Genetic separation of discrete viral functions downstream of ICP0 required for phosphorylation of eIF4E and 4E-BP1
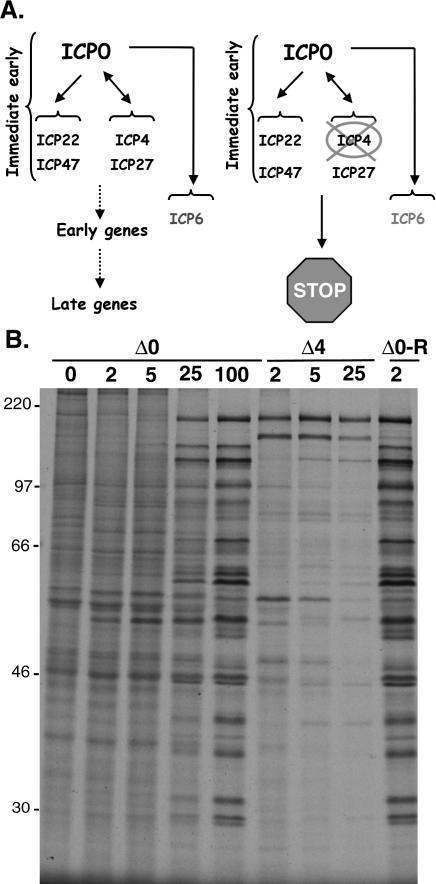
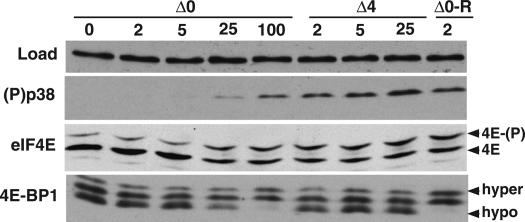
To evaluate how ICP0 promotes eIF4E and 4E-BP1 phosphorylation and to genetically define the functions involved, we made use of ICP0 and ICP4 mutant viral strains. The ICP0 gene specifies a master regulator of gene expression that is required for reactivation of latent infections in animal models and growth at low input doses of virus in cultured cells (for review, see Everett 2000; Hagglund and Roizman 2004). At lower input doses of virus, the replication of ICP0 mutant viruses is significantly impaired because in the absence of the ICP0 master regulatory protein (Fig. 1A), other critical regulators required for productive replication are not produced (Stow and Stow 1986; Sacks and Schaffer 1987). Importantly, the viral life cycle can proceed without ICP0 at higher input doses of virus. This is evident in Figure 1B, where viral protein production in growth-arrested, primary human fibroblasts is examined. At the lower multiplicities of infection (MOI) of 2 and 5, little change in the overall pattern of cellular polypeptide synthesis is observed in cells infected with the ICP0 mutant virus. However, at the higher MOIs of 25 and 100, the pattern of proteins synthesized changes dramatically, as the translation of host mRNAs is radically curtailed, while viral mRNAs are efficiently translated in the absence of the ICP0 gene product. The impaired translation of viral proteins observed at low MOI, is indeed, a result of the ICP0 mutation because a recombinant in which the mutant ICP0 allele has been repaired to its wild-type state efficiently directs high-level viral protein synthesis and impairs host protein synthesis at an MOI of 2 in a manner indistinguishable from wild-type virus (Fig. 1B; data not shown). Under conditions of low MOI, 4E-BP1 remains mostly hypophosphorylated in cells infected with an ICP0 mutant virus as evidenced by the more abundant immunoreactive bands that migrate most rapidly in high percentage SDS–polyacrylamide gels (Fig. 2). Equivalent low viral doses in which the ICP0 allele has been repaired to a wild-type state induce both p38 activation and 4E-BP1 phosphorylation (Fig. 2, Δ0-R lane). However, at high input doses of an ICP0 mutant virus, the requirement for ICP0 is obviated, productive replication ensues, and 4E-BP1 becomes hyperphosphorylated in the absence of the ICP0 protein (Fig. 2). Relative to a loading control antigen, ICP0 mutants are likewise unable to activate the cellular kinase p38 at low MOI, but do so efficiently at high MOI. Indeed, cells infected with an ICP0 mutant virus at lower MOIs are unable to stimulate eIF4E phosphorylation above basal levels present in uninfected cells. However, as the MOI of the ICP0 mutant increases, eIF4E phosphorylation ensues as evidenced by the accumulation of a slower migrating species of eIF4E upon isoelectric focusing (Fig. 2). Thus, at high MOI, phosphorylation of eIF4E and 4E-BP1 can proceed without ICP0, suggesting that ICP0 might act by regulating the expression of another viral gene expressed later in the life cycle.
Figure 1.
Comparison of proteins synthesized in cells infected with an ICP0 mutant versus an ICP4 mutant. (A) Regulation of HSV-1 gene expression by ICP0 and ICP4. (Left) ICP0 is a master regulatory protein required for proper immediate-early (IE) gene expression. Together, ICP0, ICP4, ICP27, and ICP22 establish the cascade pattern of viral gene expression necessary to complete the productive growth cycle. IE proteins activate early genes and early gene products promote viral DNA synthesis, which, in turn, activates the final class of late genes. (Right) In the absence of ICP4, the productive growth cycle arrests at the immediate-early (IE) stage. ICP0, together with ICP22, ICP47, ICP27, and ICP6, accumulate. ICP6 represents a special class of early genes regulated directly by ICP0. (B) Primary human fibroblasts (NHDF cells) were growth-arrested by serum starvation and either mock-infected (0 pfu [plaque-forming units]) or infected with the indicated virus (ΔICP0, ΔICP4, or Δ0-R) at an MOI of 2, 5, 25, or 100 pfu per cell. At 8 h post-infection, cells were radio-labeled for 1 h with 35S-methionine and cysteine. Total protein was subsequently isolated and fractionated by SDS-PAGE. The mobility of molecular weight standards (in kilodaltons) is shown to the left of the panel.
Figure 2.
p38 activation and eIF4E phosphorylation, but not 4E-BP1 phosphorylation, occur in cells infected with an ICP4 mutant. Primary human fibroblasts (NHDF cells) were growth-arrested by serum starvation and either mock-infected (0 pfu) or infected with the indicated virus (ΔICP0, ΔICP4, or Δ0-R) at an MOI of 2, 5, 25, or 100 pfu per cell. After 9 h, total protein was isolated, fractionated by SDS-PAGE or IEF (for eIF4E phosphorylation), and analyzed by immunoblotting with the indicated antibodies. Anti-PABP served as the control (LOAD) to ensure that equivalent amounts of protein were present in each lane. Phosphorylated [4E-(P)] and unphosphorylated (4E) forms of eIF4E resolved by IEF are indicated to the right of the panel, as are hyperphosphorylated (hyper) and hypophosphorylated (hypo) forms of 4E-BP1 resolved by SDS-PAGE on high-percentage gels.
The ICP4 gene encodes another critical HSV-1 regulatory protein, in addition to ICP0. The ICP4 gene product is a site-specific DNA-binding protein required to activate transcription of the viral early genes, many of which are important for viral DNA replication (for review, see Roizman and Knipe 2001). In the absence of the ICP4 gene product, a limited subset of viral polypeptides is produced in growth-arrested, primary human cells (Fig. 1B) before the viral life cycle arrests in the immediateearly stage, regardless of the MOI, imposing an absolute block on the expression of later genes and preventing the completion of the viral life cycle (Fig. 1A; DeLuca et al. 1985; DeLuca and Schaffer 1988). Unlike ICP0 mutants, which can replicate at high MOI, ICP4 is essential for viral replication in cultured cells. Significantly, whereas ICP4 mutant viruses are able to efficiently activate p38 and promote eIF4E phosphorylation in infected cells, they do not induce the hyperphosphorylation of 4E-BP1 (Fig. 2). This suggests that the limited subset of viral proteins produced in cells infected with an ICP4 mutant virus is responsible for stimulating p38 activation and that these cells arrest prior to expressing a distinct, independent function that promotes 4E-BP1 phosphorylation. Furthermore, it genetically separates the activity responsible for stimulating eIF4E phosphorylation from that which promotes 4E-BP1 phosphorylation. Thus, it establishes that HSV-1 encodes at least two discrete effectors capable of regulating the cellular eIF4F complex.
Identification of ICP6 as a 140-kDa protein associated with eIF4F in HSV-1-infected cells
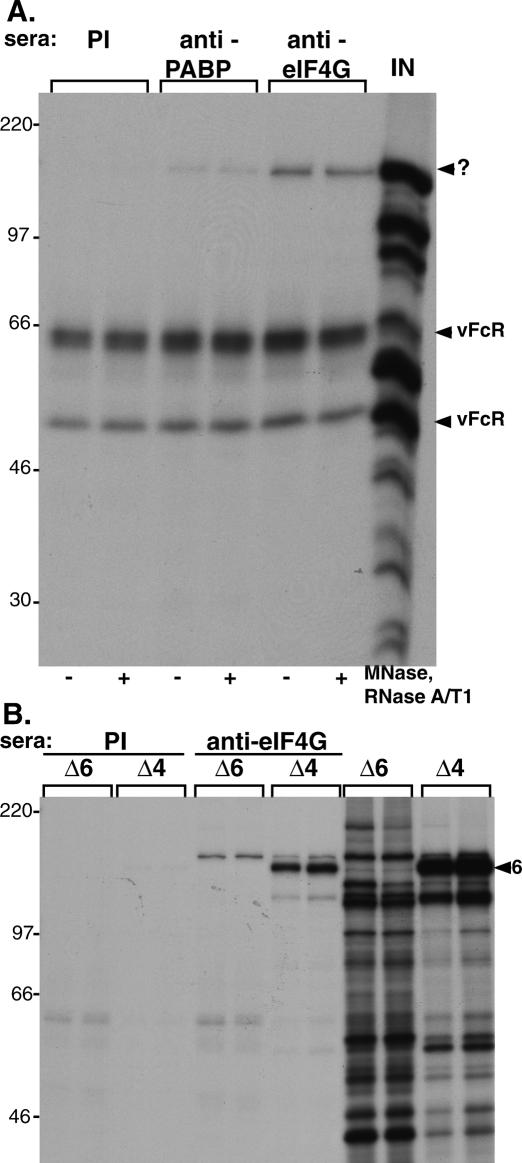
Given that the eIF4E kinase mnk is associated with eIF4G in the eIF4F complex, we tested the hypothesis that the factor responsible for stimulating eIF4E phosphorylation and eIF4F complex assembly might be associated with this very same structure. Radio-labeled extracts were prepared from growth-arrested, primary human cells infected with wild-type virus and subsequently immunoprecipitated with antisera directed against eIF4G, PABP, or preimmune sera. Following fractionation of the isolated immune complexes by SDS-PAGE, a 140-kDa polypeptide was detected in anti-eIF4G and anti-PABP immune complexes, but not in complexes isolated with preimmune sera (Fig. 3A). Given that host protein synthesis is profoundly suppressed in HSV-1-infected cells at the time of labeling, it is likely that this radio-labeled protein represents either a virus-encoded factor or a host component that is highly induced upon infection and escapes the virus-imposed host shut-off. The very same suppression of host protein synthesis in HSV-1-infected cells explains why eIF4G, eIF4E, and eIF4A, the core eIF4F components, are not radio-labeled under these conditions and are therefore not detected in the immune complex by this procedure. Furthermore, the association of the 140-kDa protein with eIF4G appears not to involve the independent binding of both components to the same RNA molecule, as the abundance of the 140-kDa protein in immune complexes was not significantly reduced by treatment of the extracts with a cocktail of nucleases under conditions known to eliminate >99% of RNA (Poppers et al. 2003).
Figure 3.
The 140-kDa protein associated with eIF4G in HSV-1-infected cells is the product of the ICP6 gene. (A) Coimmunoprecipitation of a 140-kDa protein with eIF4G and PABP in HSV-1-infected cells. Growth-arrested NHDFs were infected with HSV-1 (MOI = 10) and at 12 h post-infection, the cultures were radio-labeled with 35S amino acids for 1 h. Soluble extracts were prepared (lane labeled IN) and immunoprecipitated with the indicated antisera. Extracts were treated where indicated with a cocktail of micrococcal nuclease (MNase), RNaseA, and RNase T1 prior to immunoprecipitation. The mobility of the 140-kDa band (?) and the HSV-1-encoded Fc receptors (vFcR) are indicated to the right of the panel. (B) The 140-kDa protein does not associate with eIF4G in cells infected with an ICP6-null mutant. Duplicate cultures of growth-arrested NHDFs were infected (MOI = 10) with either an HSV-1 ICP6 deletion mutant (Δ6) or an ICP4 deletion mutant (Δ4). Extracts prepared from labeled cultures were immunoprecipitated with either anti-eIF4G or preimmune (PI) sera. Samples of input material prior to immunoprecipitation are included on the right (lanes marked Δ6, Δ4). The arrowhead marked 6 to the right of the panel points to the ICP6 polypeptide.
Proteomic analysis of infected cells suggested that the 140-kDa protein might be the product of the ICP6 gene, the large subunit of the HSV-1 ribonucleotide reductase. Significantly, the ICP6 gene is known to specifically require ICP0 for its expression (Fig. 1A; DeLuca et al. 1985; Sacks and Schaffer 1987; DeLuca and Schaffer 1988; Goldstein and Weller 1988a), and our earlier work had established a role for ICP0 in promoting eIF4F complex assembly (Walsh and Mohr 2004). In addition, as an ICP0-responsive gene, the ICP6 protein is produced in cells infected with an ICP4 mutant virus (Fig. 1A). To test if the 140-kDa protein associated with eIF4G was, indeed, encoded by the ICP6 gene, radio-labeled extracts prepared from cells infected with either an ICP6 mutant virus or an ICP4 mutant virus were immunoprecipitated with anti-eIF4G or preimmune sera. Whereas the 140-kDa protein is clearly seen in samples from duplicate cultures infected with an ICP4 mutant, it is absent from samples prepared from cells infected with an ICP6-null mutant (Fig. 3B). This suggests that the 140-kDa protein associated with eIF4G in virus-infected cells is, in fact, the product of the viral ICP6 gene. The autoradiograph in Figure 3B was intentionally overexposed to emphasize the complete absence of the ICP6 protein in anti-eIF4G immune complexes isolated from cells infected with an ICP6 mutant virus. As a result of this overexposure, slower-migrating background bands are visible in anti-eIF4G immune complexes isolated from cells infected with either an ICP4 or an ICP6 mutant virus.
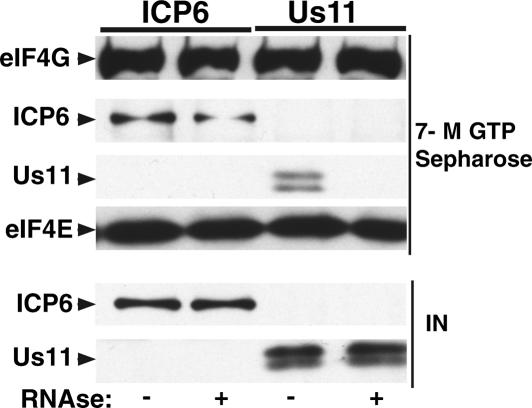
To determine if ICP6 alone was sufficient to associate with eIF4G in an eIF4F complex, purified bacterially expressed ICP6 protein was added to 293 cell-free extracts, and eIF4F complexes were isolated by batch adsorption to 7-methyl GTP Sepharose. As a control for nonspecific protein–protein together with protein–RNA interactions, Us11, a purified viral RNA-binding protein, was added to separate reactions (Khoo et al. 2002). Whereas both the ICP6 and Us11 proteins cofractionated with the eIF4F complex prior to the addition of RNases, only the ICP6 protein was capable of stable incorporation into an eIF4F complex in the presence of an RNase cocktail (Fig. 4). This demonstrates that ICP6 can bind to eIF4G in an eIF4F complex without any other viral factors and that this association is resistant to RNase treatment, strongly suggesting that it represents an interaction between protein components.
Figure 4.
Association of ICP6 with eIF4G in vitro does not require additional viral components. Four-hundred nanograms of purified His-tagged ICP6 protein or purified His-tagged Us11 protein were incubated on ice for 1 h with 200 μL of untreated or RNAse-treated 293 cell extract (10 mg/mL). eIF4F complexes were isolated by adsorption to 7-methyl GTP Sepharose, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. The signals from unfractionated cell extracts containing added purified proteins are shown below (IN). ICP6 and Us11 were detected with an anti-polyhistidine monoclonal antibody.
The N terminus of the multifunctional ICP6 protein associates with eIF4G
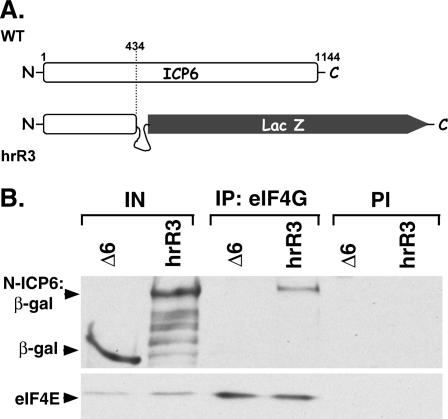
Many viral proteins are multifunctional, able to participate in a staggering array of seemingly disparate biological processes through the action of discrete domains juxtaposed onto a single polypeptide chain. While the 140-kDa ICP6 gene product is a subunit of the viral ribonucleotide reductase holoenzyme, this activity has been mapped to the C-terminal domain of the protein (Goldstein and Weller 1988a). A function of the large N-terminal extension, however, has not been elucidated. To discern if the N-terminal segment of the ICP6 protein could mediate the observed association with eIF4G, cells were first transfected with a β-galactosidase expression plasmid or the parental expression plasmid lacking the β-galactosidase gene. Subsequently, cells transfected with the plasmid expressing β-galactosidase were infected with an ICP6-null mutant (Δ6), whereas those transfected with the vector alone were infected with an ICP6 mutant that only produced the N-terminal 434 residues of ICP6 as a β-galactosidase fusion protein (hrR3) (Fig. 5A). Extracts were prepared from each of the cultures and immunoprecipitated with anti-eIF4G sera. Whereas both β-galactosidase and the ICP6:β-galactosidase fusion protein are present at similar levels in the cell-free extracts, only the ICP6:β-galactosidase fusion protein is found in eIF4G immune complexes (Fig. 5B). Since β-galactosidase was not observed to interact with eIF4G, this establishes that the interaction between the ICP6:β-galactosidase fusion protein was, indeed, mediated through the ICP6 protein sequences. Equal quantities of eIF4E, a known eIF4G-associated protein, in the immune complexes isolated from infected cells serve as a control for the effectiveness of the coimmunoprecipitation procedure. Thus, the N-terminal 434 residues of ICP6 are sufficient to mediate a specific interaction with eIF4G, establishing that at least some of the determinants important for associating with eIF4G reside in the ICP6 N terminus.
Figure 5.
The N terminus of the HSV-1 ICP6 protein associates with eIF4G. The N-terminal 434 residues of ICP6 are sufficient to allow a heterologous protein to associate with eIF4G. (A) Cells infected with the hrR3 mutant produce an ICP6 derivative in which the N-terminal 434 residues of ICP6 are fused to lacZ (Goldstein and Weller 1988a). The ICP6 protein encoded by wild-type (WT) and the hrR3 mutant are depicted for comparative purposes. (B) 293 cells were transfected with either a lacZ expression plasmid or the expression plasmid lacking an insert (vector alone). Twenty-four hours later, the cells transfected with the lacZ expression plasmid were infected (MOI = 10) with an ICP6 deletion mutant (Δ6), whereas the cells transfected with vector alone were infected with a virus that produces the N-terminal 434 residues of ICP6 fused to β-galactosidase (hrR3). At 10 h post-infection, cell extracts were prepared and immunoprecipitated with anti-eIF4G or preimmune (PI) sera. Isolated immune complexes were fractionated by SDS-PAGE and analyzed by immunoblotting with anti-β-galactosidase (top panel) or anti-eIF4E (bottom panel). Samples of input material prior to immunoprecipitation (IN) are also included on the left.
Impaired assembly of eIF4F complexes and eIF4E phosphorylation in cells infected with an ICP6 mutant virus
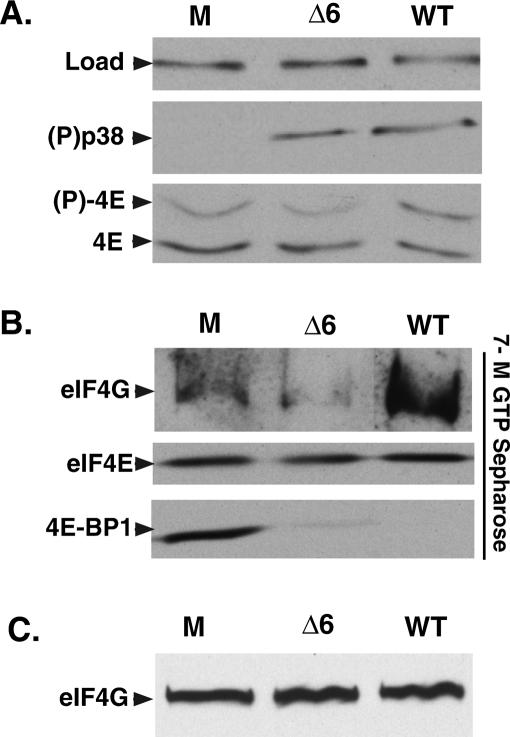
To address the functional consequences of the association between ICP6 and eIF4F, the phenotype of an ICP6 mutant virus in growth-arrested, primary human cells was examined in detail. Remarkably, even though p38 was activated to wild-type levels in cells infected with an ICP6-null mutant, the abundance of phosphorylated eIF4E relative to unphosphorylated eIF4E remained identical to that observed in mock-infected cells (Fig. 6A). This is surprising because activated p38 usually stimulates mnk to phosphorylate eIF4E, as is seen in cells infected with wild-type virus (Fig. 6A). Infection with wild-type virus also resulted in the release of 4E-BP1 and the subsequent recruitment of eIF4G into the eIF4F complex (Fig. 6B). However, in cells infected with an ICP6-null mutant, while the 4E-BP1 repressor is released from eIF4E, eIF4G does not appear to be recruited into the eIF4F complex (Fig. 6B). This suggests that ICP6 is required to recruit eIF4G and stimulate eIF4F complex assembly, as the total abundance of eIF4G in the lysates remains constant (Fig. 6C). It further demonstrates that the release of eIF4E from the 4E-BP1 translational repressor is not, in and of itself, sufficient to promote eIF4F complex assembly in HSV-1-infected cells; in addition, it provides evidence from an independent mutant that 4E-BP1 phosphorylation can be genetically separated from eIF4E phosphorylation and eIF4F complex assembly, corroborating and extending our observation that ICP4 mutants can stimulate eIF4E phosphorylation but are unable to promote 4E-BP1 phosphorylation (Fig. 2). Finally, these results also serve to link eIF4E phosphorylation to eIF4F complex assembly, a phenomenon first reported in uninfected cells (Tuazon et al. 1990; Pyronnet et al. 1999). ICP6 mutants fail to promote eIF4E phosphorylation despite the presence of activated p38 due to their failure to assemble eIF4F complexes. Since eIF4G, which is bound to the cellular eIF4E kinase mnk, is not recruited into the eIF4F complex, the absence of ICP6 prevents the correct apposition of the kinase with its substrate, eIF4E.
Figure 6.
ICP6 mutants are unable to promote assembly of eIF4F complexes in infected cells. p38 activation occurs without detectable eIF4F complex assembly and eIF4E phosphorylation in cells infected with an ICP6 mutant virus. Growth-arrested primary NHDF cells were either mock-infected (M) or infected with wild-type (WT) HSV-1 or an ICP6 deletion mutant (Δ6). (A) After 10 h, total protein was isolated, fractionated by SDS-PAGE or IEF (eIF4E phosphorylation), and analyzed by immunoblotting with the indicated antibodies. The loading control (Load) samples were probed with anti-PABP. (B) Soluble extracts were prepared and proteins bound to 7-methyl GTP Sepharose 4B were fractionated by SDS-PAGE, analyzed by immunoblotting, and visualized with the indicated antibodies. (C) A sample of each soluble extract representing the starting input material for the 7-methyl GTP Sepharose-binding experiment shown in B was fractionated by SDS-PAGE and analyzed by immunoblotting with anti-eIF4G antisera.
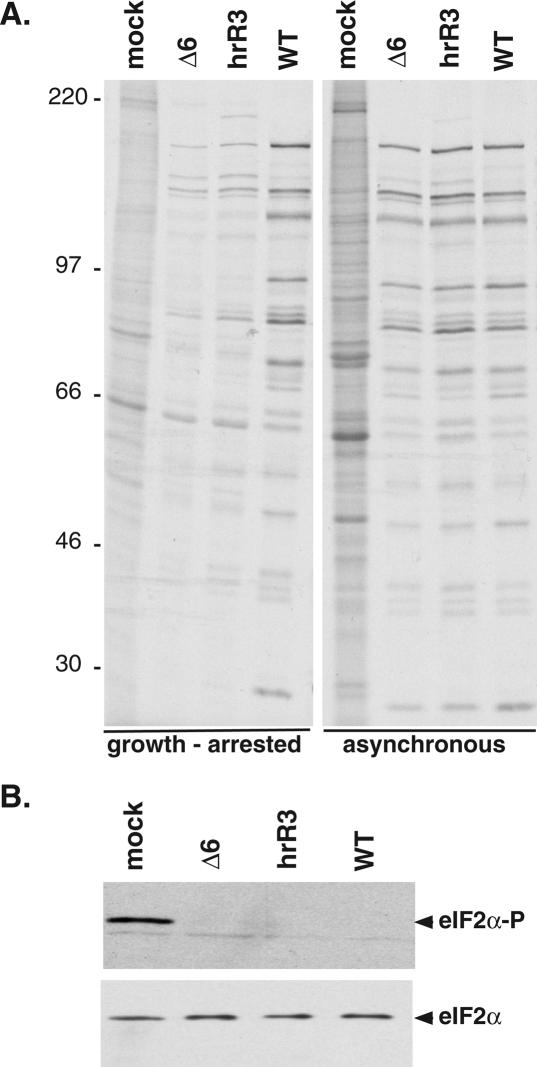
Both the inability to stimulate eIF4E phosphorylation and assemble eIF4F complexes may reflect the reduced rates of translation observed in growth-arrested primary human cells infected with either an ICP6-null mutant or a mutant that only produces the N-terminal 434 residues of the protein (hrR3) (Fig. 7A). Although this same segment of the polypeptide is sufficient to associate with eIF4G, it is not sufficient to promote eIF4F complex formation (data not shown) or direct wild-type levels of translation, suggesting that other determinants of the ICP6 protein are required to promote eIF4F complex assembly. Furthermore, while eIF4F complex assembly is impaired in growth-arrested cells infected with an ICP6 mutant virus, an increase in eIF2α phosphorylation was not observed (Fig. 7B) as multiple HSV-1-encoded functions act to prevent the accumulation of phosphorylated eIF2α (He et al. 1997; Mulvey et al. 2003). Thus, the reduction in translation observed in growth-arrested cells infected with an ICP6 mutant correlates with a defect involving eIF4F complex assembly together with eIF4E phosphorylation and does not appear to impinge on any of the characterized pathways that inhibit translation by inactivating the critical initiation factor eIF2 through phosphorylation of its α subunit (for review, see Schneider and Mohr 2003; Mohr 2006). Strikingly, an activity in cycling cells is able to complement the reduced translation rates exhibited by the ICP6 mutants in growth-arrested cells, raising the possibility that cellular functions capable of promoting eIF4F complex assembly can compensate for the loss of ICP6 under these conditions (Fig. 7A). The activity of such cellular eIF4F assembly factors is likely to be impaired in growth-arrested cells, consistent with their reduced abundance of assembled eIF4F complexes and lower global rates of translation (Stanners and Becker 1971; Thomas and Gordon 1979; Thomas et al. 1981; Thomas and Thomas 1986).
Figure 7.
Reduction in translation rates in quiescent cells infected with ICP6 mutants. (A) Growth-arrested or asynchronous, growing cells were infected (MOI = 10) with the indicated viruses. At 9 h post-infection, cells were radio-labeled for 1 h with 35S-methionine and cysteine. Total protein was subsequently isolated and fractionated by SDS-PAGE. The mobility of molecular weight standards (in kilodaltons) is shown to the left of the panel. (B) eIF2α phosphorylation in cells infected with ICP6 mutant viruses. Growth-arrested cells were infected with the indicated viruses as in A. After 10 h, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with antibodies specific for either total eIF2α or phosphorylated eIF2α (eIF2α-P).
Purified ICP6 stimulates the binding of eIF4G to eIF4E in vitro
To gain insight into the underlying mechanism by which ICP6 promotes the assembly of eIF4F complexes in HSV-1-infected cells, we focused our attention on how ICP6 might impact on the association of eIF4E with eIF4G. Not only does the association between eIF4E and eIF4G represent a critical step in eIF4F assembly, it can be reconstituted in vitro with defined, purified components expressed in bacteria. Indeed, when binding reactions containing sufficient quantities of purified eIF4E and eIF4G derivatives are allowed to proceed to equilibrium, stable complexes suitable for structural analysis can be isolated (Gross et al. 2003). This strongly suggested that any attempts to visualize the effect of ICP6 on this association in a reconstituted system with purified proteins would most likely be apparent under suboptimal, nonequilibrium binding conditions.
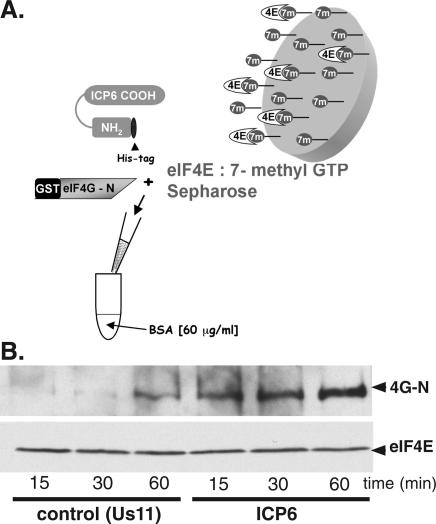
Bacterially expressed eIF4E was isolated by batch adsorption to 7-methyl GTP Sepharose beads, and a fixed quantity of the protein-laden beads was incubated with a purified eIF4G-N-terminal fragment in the presence of purified, bacterially expressed His-tagged ICP6 or an equimolar quantity of a His-tagged control protein (His-Us11) expressed and affinity-purified in an identical manner (Fig. 8A). At various times, the polypeptides bound to the 7-methyl GTP beads were collected, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera. Strikingly, greater quantities of eIF4G remained stably associated with the 7-methyl GTP beads in reactions programmed with ICP6 compared with those that received equivalent amounts of the Us11 control protein (Fig. 8B). The enhanced retention of eIF4G on the beads occurred at all the time points examined (Fig. 8B) and was dependent on prior incubation of the beads with eIF4E (data not shown). Furthermore, the kinetics of eIF4G binding to the eIF4E–7-methyl GTP beads were substantially faster in reactions containing ICP6 compared with reactions that contained a control protein. In particular, the quantity of eIF4G associated with eIF4E after 1 h incubation with a control protein was similar to the amount of eIF4G bound to the affinity resin after 15 min in the presence of ICP6. Thus, ICP6 appears to stimulate the assembly of eIF4F complexes by directly promoting the association of eIF4E with the N terminus of eIF4G. No other viral or cellular components appear to be required.
Figure 8.
Binding of the eIF4G N-terminal domain to eIF4E is stimulated by ICP6 in vitro using purified components. (A) Illustration of the reconstituted in vitro system developed to assess the impact of ICP6 on the eIF4E–eIF4G interaction. Bacterially expressed eIF4E (4E) bound to 7-methyl GTP Sepharose beads (eIF4E:7-methyl GTP Sepharose) was incubated with a purified eIF4G N-terminal fragment fused to GST (GST eIF4G-N) in the presence of purified, His-tagged full-length ICP6 or a His-tagged control protein (Us11) in a buffered solution containing bovine serum albumin (BSA). (B) Analysis of eIF4G retained by eIF4E bound to 7-methyl GTP Sepharose. At the indicated times, the beads were collected by centrifugation and washed. Bound proteins were fractionated by SDS-PAGE and analyzed by immunoblotting using antibodies directed against either eIF4E or GST.
Discussion
By promoting the assembly of eIF4F complexes together with phosphorylation of eIF4E and the inactivation of the translational repressor 4E-BP1, HSV-1 uses multiple strategies to stimulate the translational machinery in growth-arrested cells. Moreover, the function responsible for 4E-BP1 phosphorylation is genetically separable and therefore independent from the activity required to assemble eIF4F complexes and promote eIF4E phosphorylation. These latter functions appear to be linked, as they require activities intrinsic to the 140-kDa protein product of the viral ICP6 gene.
While the regulation of eIF4F activity has been proposed to be a key target for translational control, models accounting for the assembly of eIF4F complexes have largely focused on how 4E-BP1 negatively regulates the ability of eIF4E to engage eIF4G (Gingras et al. 2001). Notably, the release of eIF4E mediated by 4E-BP1 phosphorylation has been proposed to expose the binding site for eIF4G on eIF4E and subsequently allow these two components of eIF4F to associate (Haghighat et al. 1995; Mader et al. 1995; Marcotrigiano et al. 1999). Little consideration was given to the possibility that other trans-acting factors might be required to facilitate or chaperone this molecular association. Our genetic and biochemical analysis establishes, for the first time, that the inactivation of the 4E-BP1 translational repressor is not, in and of itself, sufficient to promote eIF4F complex assembly, as 4E-BP1 is efficiently jettisoned from eIF4E in cells infected with an ICP6 mutant virus, but the subsequent recruitment of eIF4G into an eIF4F complex is not observed. Surprisingly, this critical construction step requires the activity of a virus-encoded function, the product of the ICP6 gene.
Not only is the ICP6 polypeptide required to promote the assembly of eIF4F complexes, it also facilitates eIF4E phosphorylation. Despite the fact that the p38 kinase pathway, which is entirely responsible for eIF4E phosphorylation in HSV-1-infected cells (Walsh and Mohr 2004), is activated, phosphorylated eIF4E does not accumulate in cells infected with an HSV-1 ICP6 mutant and eIF4G is not recruited into an eIF4F complex. This is consistent with a model wherein eIF4E phosphorylation is contingent on the proper assembly of the eIF4F complex whose molecular architecture results in the correct apposition of the kinase mnk with its substrate, eIF4E. Similar findings limiting eIF4E phosphorylation to assembled complexes have been likewise found in other systems that do not involve viral infection (Tuazon et al. 1990; Pyronnet et al. 1999). Thus, the manner in which ICP6 promotes eIF4E phosphorylation appears to be similar to observations made in uninfected cells.
While translation of viral mRNAs is impaired in growth-arrested cells infected with an ICP6-null mutant, actively dividing cells support efficient translation of HSV-1 mRNAs in the absence of the ICP6 protein. The presence of factors in cycling cells that promote eIF4F complex assembly in a manner analogous to ICP6 could, in principle, allow for viral mRNA translation. Alternatively, the greater concentration of limiting translation factors normally resident in dividing cells may be sufficient to allow for unassisted assembly of eIF4F complexes. Factors capable of promoting eIF4F complex assembly might have significant impact when the intracellular abundance of initiation factors is reduced beyond a given threshold level. In this regard, it is intriguing that HCMV, a related herpesvirus, radically increases the abundance of core eIF4F components in resting cells (Walsh et al. 2005). Further work is necessary to ascertain whether the increase in eIF4F components seen in HCMV-infected cells obviates the need for a discrete eIF4F-assembly factor similar to the ICP6 polypeptide.
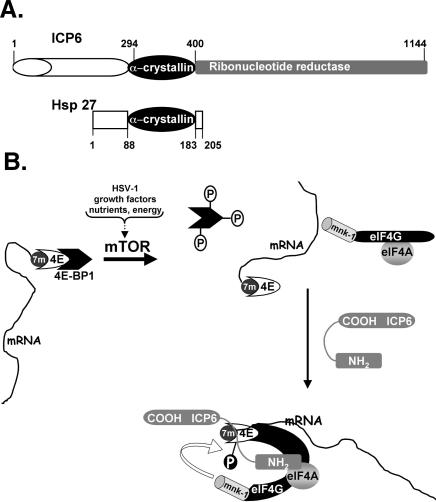
Like many viral regulatory proteins, ICP6 appears to be multifunctional, highlighting the plasticity through which evolution melds together what appear to be unrelated functional domains into a single polypeptide chain. While the C terminus of the ICP6 polypeptide has long been known to comprise one subunit of the viral ribonucleotide reductase holoenzyme, the function of the 434-residue N terminus has remained a mystery (Goldstein and Weller 1988a). Recently, the ICP6 N terminus has been shown to contain an α-crystallin domain homologous to small heat-shock proteins (Fig. 9A; Chabaud et al. 2003). The α-crystallin domain is important for hsp oligomerization and cryoprotective activity (for review, see Barral et al. 2004). Significantly, whereas the purified viral ribonucleotide reductase R1 subunit displays chaperone activity comparable to hsp27 in a thermal denaturation assay with citrate synthase, the cellular R1 from mammals does not contain an α-crystallin domain nor does it exhibit hsp27-like chaperone activity in vitro (Chabaud et al. 2003). In addition, the cellular chaperones hsp25 and hsp27 have been reported to associate with eIF4G (Cuesta et al. 2000; Cowan and Morley 2004). Based on our studies in infected cells, we suggest that one target of the ICP6 chaperone-like activity may, in fact, be the components of the eIF4F complex, as ICP6 is required for the virus-induced assembly of eIF4F complexes in growth-arrested primary human cells. Indeed, we have established with purified components in vitro that ICP6 enhances the association of the eIF4G N-terminal domain with eIF4E bound to 7-methyl GTP Sepharose and accelerates the rate of complex formation between these two critical eIF4F components. Prior analysis of a yeast eIF4G fragment associated with cap-bound eIF4E revealed major structural transitions in both subunits, whereupon a segment of eIF4G forms a right-handed helical ring that wraps around the eIF4E N terminus forming a molecular bracelet that allosterically enhances the binding of eIF4E to the 7-methyl GTP cap (Gross et al. 2003). Perhaps ICP6 promotes this transition by accelerating the rate of complex formation or promoting other structural transitions in eIF4G and/or eIF4E that facilitate the stable association of these components. Both scenarios are consistent with the hypothesis that ICP6 functions as a chaperone in this reaction, promoting the assembly of eIF4F complexes. Not only does an assembly factor such as ICP6 ensure that the architecture of the eIF4F complex is correct, it also allows for the proper apposition of the cellular kinase mnk with its substrate, eIF4E (Fig. 9B). Such a model accounts for both the eIF4F complex assembly defect as well as the impaired phosphorylation of eIF4E observed in cells infected with an ICP6 mutant virus.
Figure 9.
Model illustrating how ICP6 promotes eIF4F complex assembly and eIF4E phosphorylation. (A) ICP6 has an α-crystallin domain with homology to the cellular chaperone, hsp27. ICP6 is a multifunctional polypeptide with a C-terminal domain (residues 400–1144, shaded in gray) that specifies one subunit of the virus-encoded ribonucleotide reductase holoenzyme (Goldstein and Weller 1988a,b). The α-crystallin homology domain (residues 294–400) present in ICP6 (Chabaud et al. 2003) and hsp27 (residues 88–183) is shown as a black oval. (B) The cellular cap-binding protein is depicted bound to the 7-methyl GTP cap (7m) at the mRNA 5′ end. eIF4E, in turn, is bound to the translational repressor 4E-BP1 and is unable to assemble into an eIF4F complex with the other translation initiation factors, eIF4G and eIF4A. Activation of the kinase mTOR in response to a variety of cues such as HSV-1 infection, growth factor signaling, alterations to the nutrient pool, or changes in cellular energy reserves results in phosphorylation of the translational repressor protein 4E-BP1 and the release of eIF4E from 4E-BP1. However, the release of eIF4E from the repressor 4E-BP1 is not sufficient for eIF4F assembly. In growth-arrested cells infected with HSV-1, the assembly of eIF4E together with eIF4G and eIF4A into an eIF4F complex requires the assistance of ICP6. Through its N-terminal domain, we propose that ICP6 recognizes eIF4F components and potentially acts as a chaperone to foster the assembly of an eIF4F complex in HSV-1-infected cells. The molecular architecture of this complex allows for the correct apposition of the cellular kinase mnk with its substrate, eIF4E, and its assembly facilitates eIF4E phosphorylation.
The requirement of viral factors to actively assemble eIF4F complexes may represent attractive new targets in the continuing quest for safe, effective antiviral agents. Moreover, it is likely that factors that promote eIF4F complex assembly and/or disassembly will play prominent roles in a variety of biological processes that require remodeling of existing eIF4F complexes on an ensemble of newly synthesized or derepressed mRNA transcripts. This would not, of course, be limited to viral infection, but may also play critical roles in development, cellular differentiation, and the response to environmental stress (Cuesta et al. 2000; Doerwald et al. 2003; Cowan and Morley 2004).
Materials and methods
Cells, viruses, and transfection
293 (ATCC) and primary human foreskin fibroblasts NHDF (Clonetics) were cultured, growth-arrested by serum starvation, and infected as described previously (Walsh and Mohr 2004). HSV-1 (wild-type strains Patton and KOS) was prepared in Vero cells. The ICP0-null mutant 7134 (Cai and Schaffer 1989) and the repaired 7134R virus were obtained from Dr. P. Schaffer (Harvard Medical School, Boston, MA). The n12 ICP4 mutant (DeLuca and Schaffer 1988) was obtained from Dr. N. DeLuca (University of Pittsburgh School of Medicine, Pittsburgh, PA). The ICP6 deletion virus ΔICP6 (Goldstein and Weller 1988b) and the N-terminal fusion mutant hrR3 (Goldstein and Weller 1988a) were obtained from Dr. S. Weller (University of Connecticut, Farmington, CT). Transfection of 293 cells was preformed using Liopfectamine (Invitrogen, catalog #11668-019) according to the manufacturer's instructions. The β-galactosidase expression plasmid pCMV-LacZ was a gift from Lai-Yee Wong and Dr. Angus Wilson (New York University School of Medicine, New York).
Antibodies and chemicals
Anti-eIF4G polyclonal antiserum was generated by immunizing rabbits (Cocalico Biologicals) with purified C-terminal eIF4GI polypeptide. The plasmid expressing the eIF4GI C terminus was a kind gift of Dr. R. Schneider (New York University School of Medicine, New York), and polyclonal anti-PABP antiserum was a kind gift of Dr. S. Morley (University of Sussex, Brighton, United Kingdom). Antibodies directed against the following antigens were purchased from the indicated commercial suppliers: eIF4E (BD Transduction Laboratories, catalog #E27620). 4EBP1 and phospho-specific p38 MAPK (Cell Signal Technologies, catalog #9452 and #9216), β-galactosidase antibody (Cortec Biochem, catalog #CR7001RP2), and poly-histidine (Sigma-Aldrich, catalog #H1029).
35S-methionine labeling, gel electrophoresis, and immunoblotting
NHDF cells (6 × 105) were incubated for 1 h in 1 mL of methionine-free DMEM containing 77 μCi of a 35S-methionine/cysteine mixture (Amersham, catalog #NEG072). Total cellular protein was subsequently solubilized in 250 μL of sample buffer (62.5 mM Tris-HCl at pH 6.8, 2% SDS, 10% glycerol, 0.7 M β-mercaptoethanol), boiled for 3 min, and fractionated by SDS-PAGE. Labeled proteins were visualized by exposing the fixed, dried gel to X-ray film. Alternatively, where indicated, proteins were transferred to nitrocellulose following SDS-PAGE. Immunoblots were processed, incubated with primary antibody, and developed using the ECL reagent according to the manufacturer's instructions (Amersham). Vertical Slab Isoelectric Focusing (IEF) was performed as described (Morley and Pain 1995; Walsh et al. 2003; Walsh and Mohr 2004).
7-Methyl GTP Sepharose chromatography and coimmunoprecipitation of labeled HSV-1 proteins
Mock-infected or infected cultures (1.5 × 106 cells per 60-mm dish) were labeled as described above, except 2 mL of methionine-free medium containing 500 μCi of 35S-methionine was used. Cells were lysed in NLB and clarified as described previously (Walsh and Mohr 2004). Where indicated, RNase treatment was performed by adjusting the reactions to 1 mM CaCl2, 50 μg/mL RNase A, 300 units/mL RNase T1 (Fermentas), and 100 units/mL units micrococcal nuclease (Fermentas) and rocking at room temperature for 20 min before returning treated samples to ice. Supernatants were precleared with 20 μL of rabbit preimmune serum, which was collected by two consecutive incubations with 100 μL of settled bed volume (sbv) Pansorbin (Calbiochem, catalog #507858). Following transfer of the precleared extract to fresh microfuge tubes containing 2 μL of the appropriate preimmune or polyclonal antiserum, the reactions were incubated for 1 h at 4°C. Protein A Sepharose (12 μL sbv) that had previously been incubated in buffer containing 3% fraction V BSA to block nonspecific protein binding was subsequently added. The reactions were placed on a rocking platform and incubated for an additional 1 h at 4°C. Lastly, the beads were collected by brief centrifugation and washed three times in 0.5 mL of NLB. Immune complexes were fractionated by SDS-PAGE. 7-Methyl GTP Sepharose chromatography was described previously (Walsh and Mohr 2004).
Cloning and purification of His-tagged ICP6
To add a poly-histidine tag to the ICP6 N terminus, ICP6 coding sequences contained in the NotI–EcoRV segment of the genomic EcoRI A fragment were isolated and subcloned into pBSΔXho that had been previously digested with XbaI, end-filled with Klenow, then digested with NotI. The ICP6 gene was amplified from this plasmid using the Taq expand amplification kit (Roche) along with a primer adding sequences encoding a His tag at the 5′ end (5′-ATCTCGAGAATTCCATGGCCAGCC GCCCAGCCGCATCC-3′) and a primer introducing a SpeI site at the 3′ end (5′-GTACCGACTAGTTCACAGCGCGCAGCT CATGCAGAC-3′). Spe1/Not1 digestion was used to clone the amplified sequence into the commercial vector pFASTBAC (Invitrogen). pFASTBAC-His-ICP6 was then digested with PvuII and religated to generate pFASTBAC-His-ICP6-PvuII, which lacks 341 nucleotides in the middle region of the PCR-amplified ICP6. The BstEII–PmlI-digested fragment of the original NotI–EcoRV fragment of HSV-1 DNA containing ICP6 was then used to replace the same region in the PCR-amplified pFASTBAC-His-ICP6-PvuII. This replaced the vast majority of PCR-amplified ICP6, eliminating the possibility that mutations may have been introduced into this segment during the PCR reaction. Prior removal of the PvuII fragment from PCR-amplified ICP6 distinguished the amplified ICP6 product from the nonamplified cloned fragment. DNA sequencing confirmed the fidelity of the remaining regions of PCR-amplified ICP6. Spe1/Not1 digestion was then used to transfer the cloned ICP6 gene from pFASTBAC into an IPTG-inducible construct pET15b (Novogen) to generate pET-His-ICP6. Overnight cultures from colonies of BL21 cells expressing pET-His-ICP6 were used to inoculate 1-L cultures grown at room temperature. Once the cultures reached OD600 = 0.6, IPTG (0.2 mM) was used to induce protein expression for 1 h. Bacteria were recovered by centrifugation and His-tagged proteins were purified from sonicated cell extracts over Ni-Nta Agarose and dialyzed as described previously for His-Us11 (Peters et al. 2002).
Cell extracts and in vitro binding assays
Confluent dishes of 293 cells were washed in PBS and lysed in extraction buffer (50 mM HEPES-KOH at pH 7.4, 2 mM EDTA, 20 mM NaF, 75 mM KCl, 2 mM Na3VO4, 25 mM β-glycerophosphate, 1.5 mM MgCl2, 10% glycerol [v/v], 1× complete mini-EDTA free protease inhibitor tablet [Roche]) by a single round of freeze–thaw. When indicated, RNase treatment was performed as described above. Extracts were clarified by centrifugation at 10,000g for 10 min at 4°C. For binding assays, 400 ng of purified his-tagged protein was added to 200 μL of cell extract (10 mg/mL) for 1 h on ice. eIF4F complexes were then recovered from Sepharose 4B-precleared extracts by adsorption to 7-methyl GTP Sepharose as described above.
Assembly of eIF4E–eIF4G complexes in vitro
BL21(DE3)LysS bacteria (OD600 = 0.6) were induced to express mammalian eIF4E with 0.5 mM IPTG for 1 h at room temperature. Bacteria were harvested by centrifugation, resuspended in PBS containing protease inhibitors (1 EDTA-free complete protease inhibitor tablet from Roche per 15 mL of PBS), snap-frozen in a dry-ice–ETOH bath, and stored at –80°C. The suspension was thawed on ice and sonicated at the 30% cycle setting using a Branson probe 102C (5 pulses of 10 sec on, 10 sec off). Lysates were clarified by centrifugation at 20,000 rpm (Sorvall SS34 rotor) for 30 min, and aliquots were snap-frozen in a dry-ice–ETOH bath and stored at –80°C until needed. Soluble, cap-binding eIF4E in the lysate preparation was empirically quantified by comparing the amount of eIF4E bound to 7-methyl GTP Sepharose in different serial dilutions to known quantities of BSA standards following staining of SDS–polyacrylamide gels with Coomassie blue R250. To prepare eIF4E-bound 7-methyl GTP Sepharose for in vitro assembly reactions, 7-methyl GTP Sepharose beads (Amersham Pharmacia) equilibrated in binding buffer (50 mM HEPES-KOH at pH 7.4, 1 mM EDTA, 20 mM NaF, 75 mM KCl, 0.66 mM Na3VO4, 25 mM β-glycerophosphate, 1.5 mM MgCl2, 5% glycerol [v/v], 1× complete mini-EDTA free protease inhibitor tablet [Roche]) were mixed with eIF4E containing extract to achieve a concentration of 3.75 μg of eIF4E per milliliter of packed 7-methyl GTP Sepharose beads. After 3 h of rocking at 4°C, fraction V BSA was added to a final concentration of 3% to saturate the nonspecific binding sites, and the incubation was continued overnight. The next day, the beads were washed three times with binding buffer + 0.05% NP-40 and resuspended as a 20% slurry (v/v) in binding buffer + 0.05% NP-40 + 60 μg/mL fraction V BSA. Binding reactions were assembled on ice and contained 75 ng of eIF4E immobilized on 20 μL of packed 7-methyl GTP Sepharose beads, and either 30 nM purified, His-tagged ICP6 or His-tagged Us11 protein. Assembly was initiated after the addition of 25 ng of purified eIF4GN (residues 1–626) fused to GST. Reactions were incubated on ice and periodically manually agitated at regular intervals to maintain the beads in suspension. At various times, the beads were collected by centrifugation, washed three times in 0.5 mL of Binding Buffer + 0.05% NP-40 and subsequently boiled for 3 min in 40 μL of sample buffer (62.5 mM Tris-HCl at pH 6.8, 2% SDS, 10% glycerol, 0.7 M β-mercaptoethanol). Bound proteins were fractionated by SDS-PAGE and analyzed by immunoblotting with antisera directed against eIF4E or GST (Santa Cruz Biotechnology, catalog #SC138).
Acknowledgments
We thank Sandra Weller, Neal DeLuca, Priscilla Schaffer, Bob Schneider, and Simon Morley for generously providing reagents, and Matt Mulvey for invaluable advice on cloning strategies for ICP6. This work was supported by a grant from the National Institutes of Health and the Irma T. Hirschl Charitable Trust to I.M. D.W. was supported in part by NIH grant 2 P30 AI027742-16.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1375006.
References
- Barral J.M., Broadley, S.A., Schaffar, G., and Hartl, F.U. 2004. Roles of molecular chaperones in protein misfolding diseases. Sem. Cell Dev. Biol. 15: 17–29. [DOI] [PubMed] [Google Scholar]
- Cai W.Z. and Schaffer, P.A. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63: 4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud S., Lambert, H., Sassville, A.M.-J., Lavoie, H., Guibault, C., Massie, B., Landry, J., and Langelier, Y. 2003. The R1 subunit of herpes simplex virus ribonucleotide reductase has chaperone-like activity similar to Hsp27. FEBS Lett. 545: 213–218. [DOI] [PubMed] [Google Scholar]
- Cowan J.L. and Morley, S.J. 2004. The proteasome inhibitor MG132 promotes the reprogramming of translation in C2C12 myoblasts and facilitates the association of hsp25 with the eIF4F complex. Eur. J. Biochem. 271: 3596–3611. [DOI] [PubMed] [Google Scholar]
- Cuesta R., Laroia, G., and Schneider, R.J. 2000. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes & Dev. 14: 1460–1470. [PMC free article] [PubMed] [Google Scholar]
- DeLuca N. and Schaffer, P. 1988. Physical and functional domains of the herpes simplex virus. J. Virol. 62: 732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca N., McCarthy, A.M., and Schaffer, P. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerwald L., Onnekink, C., van Genesen, S.T., de Jong, W.W., and Lubsen, N.H. 2003. Translational thermotolerance provided by small heat shock proteins is limited to cap-dependent initiation and inhibited by 2-aminopurine. J. Biol. Chem. 278: 49743–49750. [DOI] [PubMed] [Google Scholar]
- Elshiekh N.A., Harris-Hamilton, E., and Bachenheimer, S.L. 1991. Differential dependence of herpes simplex virus immediate-early gene expression on de novo-infected cell protein synthesis. J. Virol. 62: 6430–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R.D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 22: 761–770. [DOI] [PubMed] [Google Scholar]
- Gingras A.-C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 15: 807–826. [DOI] [PubMed] [Google Scholar]
- Goldstein D.J. and Weller, S.K. 1988a. Herpes simplex virus type-1 ribonucleotide reductase activity is dispensible for virus growth and DNA synthesis: Isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 62: 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1988b. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of ribonucleotide reductase: Characterization of an ICP6 deletion mutant. Virology 166: 41–51. [DOI] [PubMed] [Google Scholar]
- Gross J.D., Moerke, N.J., von der Haar, T., Lugovskoy, A.A., Sachs, A.B., McCarthy, J.E., and Wagner, G. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750. [DOI] [PubMed] [Google Scholar]
- Hagglund R. and Roizman, B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78: 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A., Mader, S., Pause, A., and Sonenberg, N. 1995. Repression of cap-dependent translation by 4E-binding protein 1: Competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14: 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Gross, M., and Roizman, B. 1997. The γ(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1 α to dephosphorylate the α subunit of eukaryotic initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. 94: 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Gradi, A., and Sonenberg, N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17: 7480–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A., Roy, G., and Sonenberg, N. 2001. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 66: 293–300. [DOI] [PubMed] [Google Scholar]
- Khoo D., Perez, C., and Mohr, I. 2002. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J. Virol. 76: 11971–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar S.B., Yu, Y., Maguire, T.G., and Alwine, J.C. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78: 11030–11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lium E. and Silverstein, S. 1997. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J. Virol. 71: 8602–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S., Lee, H., Pause, A., and Sonenberg, N. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4G and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15: 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras, A.C., Sonenberg, N., and Burley, S.K. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 3: 707–716. [DOI] [PubMed] [Google Scholar]
- Mohr I. 2006. Phosphorylation and dephosphorylation events that regulate viral mRNA translation. Virus Res. (in press). [DOI] [PubMed]
- Morley S.J. and Pain, V.M. 1995. Translational regulation during activation of porcine peripheral blood lymphocytes: Association and phosphorylation of the α and γ sub-units of the initiation factor complex eIF4F. Biochem. J. 312: 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M., Poppers, J., Sternberg, D., and Mohr, I. 2003. Regulation of eIF2 α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J. Virol. 77: 10917–10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G.A., Khoo, D., Mohr, I., and Sen, G.C. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76: 11054–11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppers J., Mulvey, M., Perez, C., Khoo, D., and Mohr, I. 2003. Identification of a lytic-cycle Epstein-Barr virus gene product that can regulate PKR activation. J. Virol. 77: 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S., Imataka, H., Gingras, A.-C., Fukunaga, R., Hunter, T., and Sonenberg, N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. and Knipe, D. 2001. Herpes simplex viruses and their replication. In Field's virology, Vol. 2, 4th ed. (eds. D.M. Knipe and P.M. Howley), pp. 2239–2459. Lippincott, Williams and Wilkins, Philadelphia. [Google Scholar]
- Sacks W.R. and Schaffer, P.A. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61: 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P. 2003. Viral internal ribosome entry site elements: Novel ribosome–RNA complexes and roles in viral pathogenesis. J. Virol. 77: 2801–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper G.C. and Proud, C.G. 2002. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 269: 5350–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper G.C., Morrice, N.A., Kleijn, M., and Proud, C.G. 2001. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 21: 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.J. and Mohr, I. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28: 130–136. [DOI] [PubMed] [Google Scholar]
- Stanners C.P. and Becker, H. 1971. Control of macromolecular synthesis in proliferating and resting Syrian hamster cells in monolayer culture. I. Ribosome function. J. Cell. Physiol. 77: 31–42. [DOI] [PubMed] [Google Scholar]
- Stow N.D. and Stow, E.C. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67: 2571–2585. [DOI] [PubMed] [Google Scholar]
- Thomas G. and Gordon, J. 1979. Regulation of protein synthesis during the shift of quiescent animal cells into the proliferative state. Cell Biol. Int. Rep. 3: 307–320. [DOI] [PubMed] [Google Scholar]
- Thomas G. and Thomas, G. 1986. Translational control of mRNA expression during the early mitogenic response in Swiss mouse 3T3 cells: Identification of specific proteins. J. Cell. Biol. 103: 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Thomas, G., and Luther, H. 1981. Transcriptional and translational control of cytoplasmic proteins after serum stimulation of quiescent Swiss 3T3 cells. Proc. Natl. Acad. Sci. 78: 5712–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuazon P.T., Morley, S.J., Dever, T.E., Merrick, W.C., Rhoads, R.E., and Traugh, J.A. 1990. Association of initiation factor eIF-4E in a cap binding protein complex (eIF-4F) is critical for and enhances phosphorylation by protein kinase C. J. Biol. Chem. 265: 10617–10621. [PubMed] [Google Scholar]
- Walsh D. and Mohr, I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes & Dev. 18: 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D., Meleady, P., Power, B., Morley, S.J., and Clynes, M. 2003. Increased levels of the translation initiation factor eIF4E in differentiating epithelial lung tumor cell lines. Differentiation 71: 126–134. [DOI] [PubMed] [Google Scholar]
- Walsh D., Perez, C., Notary, J., and Mohr, I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79: 8057–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A.J., Flynn, A., Proud, C.G., and Cooper, J.A. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases mnk1 and mnk2. EMBO J. 16: 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A.J., Johnson, J.C., Penn, B., Mahalingam, M., Kimball, S.R., and Cooper, J.A. 1999. Phosphorylation of the cap-binding protein eukaryotic translation factor 4E by protein kinase mnk1 in vivo. Mol. Cell. Biol. 19: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Hillner, P.E., Vale, R.D., and Sachs, A.B. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]