Abstract
Chloroplast genes of higher plants are transcribed by two types of RNA polymerase that are encoded by nuclear (NEP (nuclear-encoded plastid RNA polymerase)) or plastid (PEP (plastid-encoded plastid RNA polymerase)) genomes. NEP is largely responsible for the transcription of housekeeping genes during early chloroplast development. Subsequent light-dependent chloroplast maturation is accompanied by repression of NEP activity and activation of PEP. Here, we show that the plastid-encoded transfer RNA for glutamate, the expression of which is dependent on PEP, directly binds to and inhibits the transcriptional activity of NEP in vitro. The plastid tRNAGlu thus seems to mediate the switch in RNA polymerase usage from NEP to PEP during chloroplast development.
Keywords: chloroplast biogenesis, glutamyl-tRNA, NEP, PEP, SIG2
Introduction
Plastids of higher plants are cellular organelles that have their own genome and machinery for gene expression. Plastid DNA contains ∼120 genes, which encode four ribosomal RNAs, 30 transfer RNAs and ∼80 proteins that are required for transcription, translation and plastid functions such as photosynthesis (Sugita & Sugiura, 1996). Two types of RNA polymerase, termed NEP (nuclear-encoded plastid RNA polymerase) and PEP (plastid-encoded plastid RNA polymerase), are found in plastids of higher plants (Hess & Börner, 1999). NEP is a T3–T7 bacteriophage-type, singlesubunit enzyme (Hedtke et al, 1997) that predominantly mediates transcription of housekeeping genes such as those for components of the gene-expression machinery (Hajdukiewicz et al, 1997), whereas PEP is responsible for transcription of genes related to photosynthetic functions (De Santis-MacIossek et al, 1999). PEP consists of core subunits that are encoded by the genes rpoA, rpoB, rpoC1 and rpoC2 in the plastid genome and which assemble with one of several promoter specificity (sigma) factors encoded by nuclear genes. Nuclear genes encoding chloroplast sigma factors were first identified in a red alga (Liu & Troxler, 1996; Tanaka et al, 1996) and were subsequently detected in several higher plants (Allison, 2000). So far, six genes for putative PEP sigma factors (SIG1–SIG6) have been identified in Arabidopsis thaliana (Isono et al, 1997; Tanaka et al, 1997; Fujiwara et al, 2000).
We previously isolated a T-DNA insertion line of SIG2 (sig2-1) in A. thaliana, the phenotype of which is characterized by pale green leaves, presumably as a result of impaired chloroplast development (Shirano et al, 2000). The abundance of a specific class of chloroplast tRNAs, including those encoded by trnV, trnM, trnE and trnD, is reduced in this mutant (Fig 1A; Kanamaru et al, 2001), whereas the amounts of messenger RNAs of various photosynthesis-related genes are unaffected (Kanamaru et al, 2001). In contrast, the mRNAs of several NEP-dependent genes accumulate during the late stage of chloroplast development in the sig2-1 mutant. In wild-type plants, mRNAs of the NEP-dependent genes, including rpoB and accD, decreased during late chloroplast development; however, they remained markedly increased 72 h after illumination in the sig2-1 mutant (Fig 1A,B; Kanamaru et al, 2001). Similar observations have been described for a virescent mutant of rice (Kusumi et al, 1997), a barley mutant (albostrians) deficient in plastid ribosomes (Hess et al, 1993) and tobacco mutants that are deficient in individual core subunits of PEP (Hajdukiewicz et al, 1997; De Santis-MacIossek et al, 1999). However, the molecular mechanism of the decrease of NEP-dependent transcripts during chloroplast development remains to be determined. In this work, we performed an in vitro transcription assay to identify a key factor involved in this regulation. Surprisingly, we found that one of the plastid-encoded tRNAs, tRNAGlu, mediates the switch in RNA polymerase usage from NEP to PEP during chloroplast development.
Figure 1.
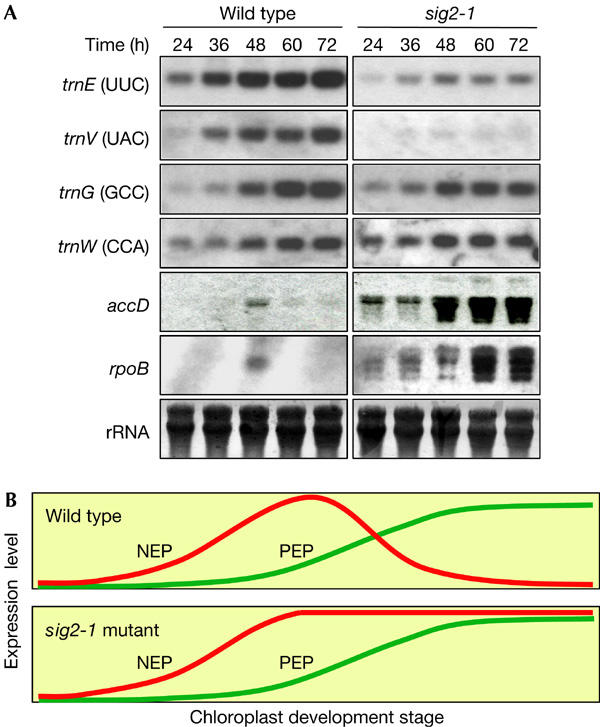
Time course of expression of plastid tRNA and NEP-dependent genes at the early developmental stage of chloroplasts in the sig2-1 mutant of Arabidopsis thaliana. (A) Northern blot analysis with probes specific for the transcripts of SIG2-dependent (trnE, trnV), or SIG2-independent (trnG (GCC), trnW) plastid tRNA genes, or of NEP-dependent genes (accD, rpoB). The beginning of illumination for seed germination after release from the cold treatment is taken as zero. (B) Hypothetical scheme for expression profiles of NEP- or PEP-dependent genes during plastid development. Accumulation of NEP-dependent transcripts is transiently observed during the early stage of chloroplast development. Transcription of PEP subsequently increases in association with the light-dependent chloroplast development; at this time, the expression of NEP-dependent genes decreases in wild-type plants but not in the sig2-1 mutant. The expression levels are not to scale.
Results and Discussion
To characterize the molecular mechanism responsible for the repression of NEP activity during the late stage of chloroplast development, we performed in vitro plastid transcription assays. For the detection of NEP-dependent transcription, we used A. thaliana cultured cells derived from root calli (Umeda et al, 1998), the plastids of which seem to consist mostly of undifferentiated proplastids. Preparation of proplastid extracts and in vitro transcription assays were modified from methods established for tobacco (Kapoor & Sugiura, 1999). Primer extension analysis showed that an in vitro transcription reaction performed with a proplastid extract and template DNA, which contain the promoter region of the NEP-dependent gene accD, generated transcripts from the accD promoter. In Arabidopsis, the 5′ end of the NEP-dependent accD transcript has not yet been determined. Here, we mapped the 5′ end of the accD transcript at the nucleotide position −89 relative to the ATG translation initiation site (Fig 2A,B). To confirm that this 5′ end of transcript was derived from a promoterspecific, in vitro-transcribed product, we performed primer extension analyses using three different primers, including that located on the plasmid part of the template DNA, all of which detected the accD transcripts with expected sizes (Fig 2C,D); these transcripts are not detected in the absence of extract, nucleoside 5′-triphosphates or template DNA on in vitro transcription reaction (Fig 2D, lanes 2–4). This transcription from the accD promoter was not affected by the addition of the recombinant SIG2 protein (Fig 2E) whereas it was inhibited by the addition of in vitro-synthesized tRNAGlu (Fig 2F)—one of the SIG2-dependent plastid tRNA species. In contrast, the addition of other SIG2-dependent (tRNAVal) or SIG2-independent (tRNAGly, tRNATrp) plastid tRNAs did not affect the transcriptional activity of NEP (Fig 2F). These results suggest that tRNAGlu specifically represses the activity of NEP during the late phase of chloroplast development. This finding is consistent with a recent in vivo study showing that NEP activity is upregulated during chloroplast development to a lesser extent in tobacco transgenic lines that overexpress RPOT3 (the catalytic subunit of NEP) than in PEP-deficient plants (Liere et al, 2004), because the overexpressed RPOT3 would still be inhibited by tRNAGlu.
Figure 2.
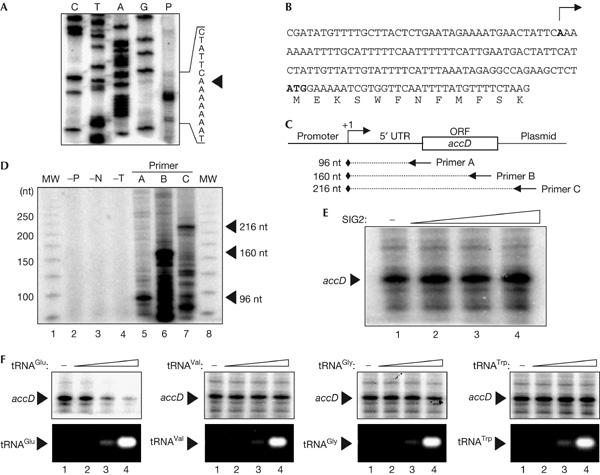
Primer extension analysis of NEP-dependent transcription in vitro in the presence of plastid tRNAs. (A) In vitro transcription was performed with a supercoiled DNA template containing the promoter region of accD and the 5′ end of the transcript was mapped by primer extension analysis (lane P). The cDNA sequences (C, T, A and G) obtained with the same primers are also shown. (B) Nucleotide sequence of the regions around the transcription initiation sites (arrow) of accD. (C) Partial structure of template DNA containing the accD promoter region. The primer C locates on the plasmid part of the template DNA. UTR, untranslated region; ORF, open reading frame. (D) Primer extension analysis was performed using primers with different locations (primers A, B and C, lanes 5–7). The result of in vitro transcription without protein extract (−P), nucleoside 5′-triphosphates (−N) or template DNA (−T) is also shown (lanes 2–4). MW shows the 25-basepair ladder marker of sizes indicated on the left (lanes 1,8). nt, nucleotides. (E) In vitro transcription was performed in the presence of SIG2 at 0, 1, 10 or 100 nM (lanes 1–4, respectively). (F) In vitro transcription was performed in the presence of in vitro-synthesized tRNAGlu, tRNAVal, tRNAGly or tRNATrp at 0, 40, 400 or 4000 nM (lanes 1–4, respectively).
To verify a role for tRNAGlu in repression of NEP activity, the electrophoretic mobility-shift assays (EMSAs) of tRNA–protein binding were performed on the basis of a previously described method of detecting binding of the yeast protein kinase GCN2 to tRNA (Dong et al, 2000). The recombinant RPOT3 protein with the His6 tag was purified by affinity chromatography (Fig 3A). A shifted band corresponding to a tRNA–protein complex was specifically detected on incubation of recombinant A. thaliana RPOT3 with 32P-labelled tRNAGlu (Fig 3B). tRNAVal, tRNAGly and tRNATrp did not interact efficiently with the recombinant RPOT3 protein in this binding assay (Fig 3B). The shifted band of the tRNAGlu–RPOT3 complex was not detected by the addition of recombinant His6–SIG2 or BSA (Fig 3C), but supershifted in the presence of the His-tag antibody (Fig 3D). Furthermore, the binding of RPOT3 to the 32P-labelled tRNAGlu was markedly reduced in the presence of excess unlabelled tRNAGlu, but was only slightly affected at higher concentrations of excess tRNAVal, tRNAGly or tRNATrp (Fig 3E). These findings show that tRNAGlu binds specifically to RPOT3, and that this interaction might be responsible for repression of NEP activity during chloroplast development.
Figure 3.
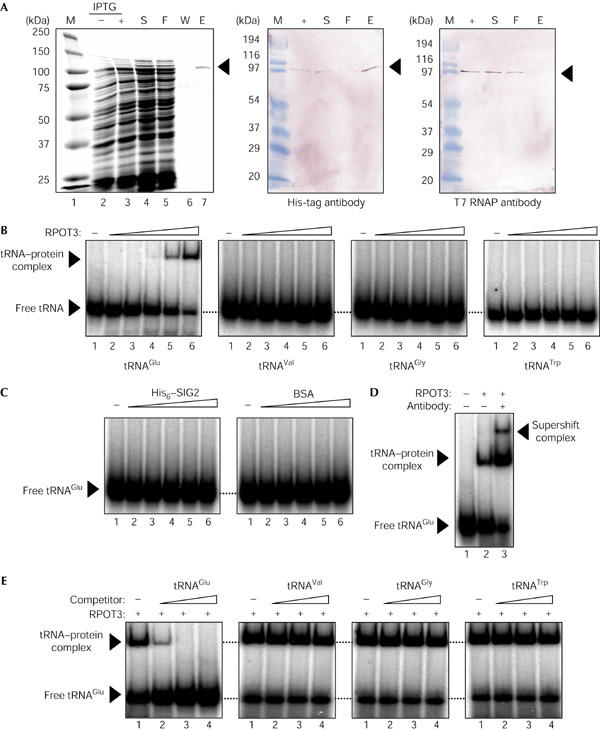
Specific interaction of plastid tRNAGlu with recombinant RPOT3. (A) Coomassie brilliant blue staining of purified RPOT3 (left panel). The recombinant RPOT3 protein was expressed by the addition of IPTG (− and +, lanes 2,3), and the soluble fraction (S, lane 4) was passed through a nickel column (F, lane 5). Binding protein was thoroughly washed by a buffer containing 40 mM imidazole (W, lane 6), followed by elution with the same buffer containing 50 mM imidazole (E, lane 7). M shows a protein marker of the sizes indicated on the left (lanes 1). Four samples (+, S, F and E) were also detected on western blotting with an antibody against His tag (middle panel) or against T7 RNA polymerase (RNAP, right panel). (B) The electrophoretic mobilityshift assays (EMSAs) were performed after incubation of 20 nM 32P-labelled tRNAGlu, tRNAVal, tRNAGly or tRNATrp, as indicated, with 0, 10, 20, 40, 60 or 80 nM recombinant RPOT3 (lanes 1–6, respectively). (C) EMSAs were performed after incubation of 20 nM 32P-labelled tRNAGlu with recombinant SIG2 or BSA instead of recombinant RPOT3. (D) EMSA was performed after incubation of 32P-labelled tRNAGlu (20 nM) in the absence (lane 1) or presence of 60 nM RPOT3 either without (lane 2) or with 1 μl of the anti-His-tag antibody (lane 3). (E) EMSAs were performed after incubation of 32P-labelled tRNAGlu (20 nM) in the presence of 60 nM RPOT3 either without (lane 1) or with unlabelled tRNAGlu, tRNAVal, tRNAGly or tRNATrp, as indicated, at 20, 200 or 2000 nM (lanes 2–4, respectively).
Two distinct types of RNA polymerase are sequentially activated during infection with certain bacteriophages, including T7 phage in Escherichia coli. In the initial stage of phage propagation, E. coli RNA polymerase mediates the transcription of early genes, including that for the phage-encoded RNA polymerase. During the middle and late stages of phage propagation, however, phage genes are transcribed predominantly by T7 RNA polymerase, with the activity of the E. coli enzyme being repressed through the action of the proteins encoded by T7 gene 2 or gene 0.7 (Nechaev & Severinov, 2003). We have shown the operation of an analogous RNA polymerase switch during chloroplast development in higher plants (Fig 4). In the early stage of plastid differentiation, plastid housekeeping genes such as accD and rpoB are transcribed by NEP. Transcription of photosynthesis-related genes by PEP does not occur at this time, presumably because of the lack of nuclear-encoded sigma factors (Fig 4A). The subsequent light-dependent expression of sigma factors, including SIG2, results in the activation of PEP and the conversion of proplastids to photosynthetic chloroplasts. The PEP holoenzyme that contains SIG2 (PEP–SIG2) mediates the transcription of several tRNA genes, the products of one of which, tRNAGlu, directly interacts with NEP and represses its activity during the late stage of chloroplast development (Fig 4B). Such repression of NEP-dependent gene transcription does not occur in mutants that lack either SIG2 (Fig 4C) or core subunits of PEP (Fig 4D) because the chloroplasts of these plants do not show PEP–SIG2-dependent accumulation of tRNAGlu. The abundance of certain NEP-dependent transcripts was recently found to remain constant throughout chloroplast development in maize as a result of compensatory regulation of mRNA stability and synthesis rate (Cahoon et al, 2004), suggesting that the regulatory mechanisms of chloroplast gene expression might differ between dicot and monocot plants.
Figure 4.
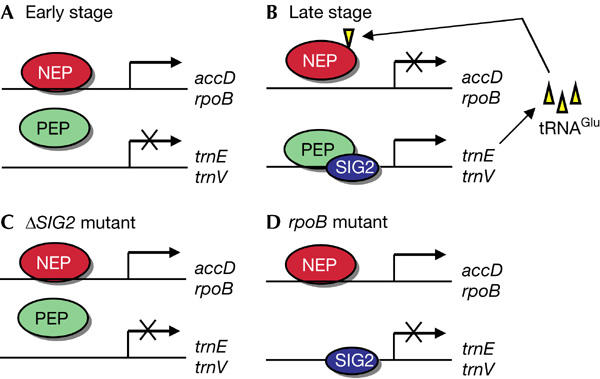
Proposed role for PEP–SIG2-dependent expression of tRNAGlu in the control of NEP RNA polymerase activity during chloroplast development. Transcription from NEP- or PEP-dependent promoters is shown for early (A) and late (B) stages of chloroplast development in wild-type plants as well as for ΔSIG2 (C) and ΔrpoB (D) mutants.
In the chloroplasts of higher plants as well as in various other organisms, tRNAGlu is required not only for translation, but also for the synthesis of δ-aminolevulinic acid, a precursor of tetrapyrroles including haeme and chlorophyll (Schön et al, 1986). We have shown yet another role for tRNAGlu: regulation of the NEP RNA polymerase during chloroplast development. Chloroplast tRNAGlu is thus a multifunctional molecule having roles in translation, tetrapyrrole biosynthesis and regulation of the switch between the two types of plastid RNA polymerase, which render it indispensable for chloroplast development.
Methods
Isolation of total cellular RNA and northern blot analysis. Seeds of A. thaliana Ws (Wassilewskija) and the sig2-1 mutant (Kanamaru et al, 2001) were sterilized with a solution containing 70% ethanol and 3% sodium hypochlorite before sowing on ashless paper filters soaked in MS medium. After stratification at 4°C for 24 h in the dark, the seeds were grown at 23°C under continuous white light. Total RNA was extracted and purified from whole seeds and young seedlings with the use of the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Digoxigenin-labelled singlestranded DNA probes for accD and rpoB transcripts were prepared as described (Kanamaru et al, 2001). Genespecific DNA fragments were amplified with the following primer sets: accD, 5′-TTCAAGCTTTCATATCGGATGACAC-3′ and 5′-TGGGATCCGCAGAGACC-3′; rpoB, 5′-TCAAAGCTTATGCTTGGGGATGAAAAAG-3′ and 5′-TTCCTTACATAAGGATTCAG-3′. Then, identical antisense-strand oligonucleotides of the two genes were used as primers for producing digoxigenin-labelled probes by 15–20 cycles of one-side polymerase reaction with 1 pmol of the purified PCR products. Specific probes for plastid tRNAs were prepared by end-labelling with digoxigenin of the oligonucleotides 5′-GCTGCCTCCTTGAAAGAGAGAT-3′ for trnE (UUC), 5′-GACCTGCTCGGTGTAAACGAGGT-3′ for trnV (UAC), 5′-GTCTTCTCCTTGGCAAGGAGAA-3′ for trnG (GCC) and 5′-GACATTGGGTTTTGGAGACCCAC-3′ for trnW (CCA). Total RNA (5 μg for accD and rpoB transcripts, 10 μg for tRNAs) was subjected to electrophoresis on a denaturing gel containing 1.2% (accD, rpoB) or 1.6% (tRNAs) agarose. The RNA molecules were then transferred under vacuum to a positively charged nylon membrane. Hybridization with the specific probes was performed for 12 h at 55°C (accD, rpoB) as described (Kanamaru et al, 2001) or for 14 h at 45°C (tRNAs) in Easy Hyb solution (Roche Diagnostics, Basel, Switzerland). Ribosomal RNA on the membrane was stained with methylene blue.
Preparation of proplastid extracts. Suspension-cultured cells derived from root callus tissue of A. thaliana (ecotype Col-0) were maintained as described (Umeda et al, 1998). Protoplasts were isolated from the cultured cells, essentially as described (Tamura et al, 2002). Isolation of proplastids and preparation of a transcriptionally active proplastid extract were performed in a manner similar to that described for tobacco (Kapoor & Sugiura, 1999).
Synthesis of tRNAs in vitro. General molecular biological techniques were performed according to standard protocols (Sambrook et al, 1989). The synthesis of tRNAs was achieved by in vitro transcription with the use of a T7 RiboMAX kit (Promega, Madison, WI, USA). For preparation of template DNA, a DNA fragment corresponding to each full-length tRNA was amplified by PCR from A. thaliana chloroplast P1 clones (Sato et al, 1999) with primers that contained a T7 promoter sequence at the 5′ end. The amplified DNA fragments were purified by electrophoresis before being subjected to in vitro transcription. The primer sequences were as follows:
trnE (UUC), 5′-CAGAGATGCATAATACGACTCACTATAGCCCC CATCGTCTAGTGGTT-3′ and 5′-TACCCCCAGGGGAAGTCGAATCCCCGCTGC-3′;
trnV (UAC), 5′-CAGAGATGCATAATACGACTCACTATAAGGGA TATAACTCAGCGGTA-3′ and 5′-TAGGGATAATCAGGCTCGAACTGATGACTT-3′;
trnG (GCC), 5′-CAGAGATGCATAATACGACTCACTATAGCGGA TATAGTCGAATGGTA-3′ and 5′-GGCGGATAGCGGGAATCGAACCCGCGTCTT-3′; and
trnW (CCA), 5′-CAGAGATGCATAATACGACTCACTATAACGCT CTTAGTTCAGTTCGG-3′ and 5′-CACGCTCTGTAGGATTTGAACCTACGACAT-3′.
In vitro transcription and primer extension assay. In vitro transcription was performed as described (Kapoor & Sugiura, 1999). The synthesized RNAs were subjected to hybridization with the accD A (5′-GCTTTACTTAGCTCACCTCTG-3′), accD B (5′-TTTCCATAGAGCTTCTGGCCTC-3′) and accD C (5′-GAGGTCGACGGTATCGATAAGC-3′) primers labelled at their 5′ end with [γ-32P]ATP, and were analysed by primer extension as described (Hanaoka et al, 2003). The 5′ ends of the extension products were determined by comparison with complementary DNA sequences generated from the same primers with the use of a LI-COR sequencing kit (Epicentre, Madison, WI, USA). For preparation of the transcription template, a DNA fragment encompassing the promoter region of accD (nucleotides −900 to +100 relative to the translation initiation codon) was amplified by PCR from A. thaliana genomic DNA with accD DN (5′-GTACTGCAGCCCTTGGGGAAATGCACCGGG-3′, PstI site underlined) and accD UP (5′-GGCGAATTCCAGGAGCAAAACTATCCATTG-3′, EcoRI site underlined) primers. The PCR product was cloned into pBluescript SK+ (Stratagene, La Jolla, CA, USA) with the use of the restriction sites included in the PCR primers.
Cloning of RPOT3 cDNA. Two RPOT3 cDNA fragments were amplified by PCR from an A. thaliana cDNA library (Tanaka et al, 1997) with the primer pairs Pt1 (5′-GGACTCGAGGAATTCGCAAGCTTGATTGAGGA GC-3′) and Pt2 (5′-TTTCTAGCCACAGAAAGTTTG-3′) or Pt3 (5′-TTAACAGAGAGAGGCATTCTC-3′) and Pt4 (5′-TGAATTCAGATCTCAGTTGAAGAAGTACTGTG -3′). The amplified fragments were each cloned into pBluescript II KS+ (Stratagene) at the SmaI site, generating pPT5′ and pPT3′α, respectively. The 1.4-kb SacI fragment of pPT5′ was then cloned into the SacI site of pPT3′α to generate pPTα, which contains a 2.7-kb contiguous RPOT3 cDNA.
Expression and purification of RPOT3 and SIG2. For expression of RPOT3 in E. coli, a DNA fragment was amplified from pPTα by PCR with the primers RPOT3 DN (5′-GACTCGAGGCAAGCTTGATTGAGGAGCTTG-3′, XhoI site underlined) and RPOT3 UP (5′-GGCTCGAGTCAGTTGAAGAAGTACTGTGAT-3′, XhoI site underlined). The amplified fragment was digested with XhoI and inserted in-frame into pET-15b (Novagen, La Jolla, CA, USA) to generate pET-RPOT3, which encodes amino acids 102–993 of RPOT3 with six histidine residues added at the amino terminus (the N-terminal region of RPOT3, which is required for targeting to chloroplasts, was omitted). The recombinant protein was purified, on a HIS-Select nickel affinity column (Sigma, St Louis, MO, USA), from the soluble fraction of a lysate of E. coli BL21 (DE3) that had been transformed with pET-RPOT3. A fraction containing single RPOT3 protein was obtained by washing with 40 mM imidazole and eluting with 50 mM imidazole. Protein concentration was estimated using the BCA protein assay kit (Pierce, Erembodegem, Belgium). Proteins were separated on 10% polyacrylamide gels, and analysed by Coomassie brilliant blue staining and western blotting using the antibody for His tag (MBL, Nagoya, Japan) or T7 RNA polymerase (Novagen). Expression and purification of the recombinant SIG2 protein was performed as described previously (Hanaoka et al, 2003).
EMSA analysis. A 10–80 nM portion of recombinant RPOT3, recombinant SIG2 or BSA (Wako, Osaka, Japan) was incubated for 30 min at 30°C with 20 nM 32P-labelled tRNA in 20 μl of a solution containing 10 mM Tris–HCl (pH 8.0), 50 mM KCl, 1 mM EDTA, 5 mM MgCl2 and 5 mM dithiothreitol in the presence or absence of 1 μl of His-tag antibody or non-labelled tRNA as a competitor. The tRNA–protein complexes were then separated by electrophoresis on a native 5% polyacrylamide gel and detected with a BAS1000 image analyser (Fujix, Tokyo, Japan). The tRNA probes were prepared by in vitro transcription and labelled by incubation with T4 polynucleotide kinase (TaKaRa, Kyoto, Japan) and [γ-32P]ATP. Labelled tRNA probes were purified from nonreacted [γ-32P]ATP with Micro Bio-spin column 30 (Bio-Rad, Hercules, CA, USA). In advance of binding reaction, tRNA probes were denatured at 70°C for 10 min and renatured by gradually cooling down to 30°C.
Acknowledgments
We thank M. Umeda for providing the suspension culture of A. thaliana cells. This work was supported by Grants-in-Aid for Scientific Research to K.K., to H.T. and to KT (14340248 and 16GS0304) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. M.H. was a Research Fellow of the Japan Society for the Promotion of Science.
References
- Allison LA (2000) The role of sigma factors in plastid transcription. Biochimie 82: 537–548 [DOI] [PubMed] [Google Scholar]
- Cahoon AB, Harris SM, Stern DB (2004) Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep 5: 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rudiger W, Koop HU, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J 18: 477–489 [DOI] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG (2000) Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6: 269–279 [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Nagashima A, Kanamaru K, Tanaka K, Takahashi H (2000) Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase sigma factors in Arabidopsis thaliana. FEBS Lett 481: 47–52 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Takahashi H, Tanaka K (2003) Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res 31: 7090–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277: 809–811 [DOI] [PubMed] [Google Scholar]
- Hess WR, Börner T (1999) Organellar RNA polymerases of higher plants. Int Rev Cytol 190: 1–59 [DOI] [PubMed] [Google Scholar]
- Hess WR, Prombona A, Fieder B, Subramanian AR, Börner T (1993) Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J 12: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H (1997) Leafspecifically expressed genes for polypeptides destined for chloroplasts with domains of sigma70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 14948–14953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K, Nagashima A, Fujiwara M, Shimada H, Shirano Y, Nakabayashi K, Shibata D, Tanaka K, Takahashi H (2001) An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol 42: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Kapoor S, Sugiura M (1999) Identification of two essential sequence elements in the nonconsensus type II PatpB-290 plastid promoter by using plastid transcription extracts from cultured tobacco BY-2 cells. Plant Cell 11: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Mizutani A, Nishimura M, Iba K (1997) A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J 12: 1241–1250 [Google Scholar]
- Liere K, Kaden D, Maliga P, Börner T (2004) Overexpression of phage-type RNA polymerase RpoTp in tobacco demonstrates its role in chloroplast transcription by recognizing a distinct promoter type. Nucleic Acids Res 32: 1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Troxler RF (1996) Molecular characterization of a positively photoregulated nuclear gene for a chloroplast RNA polymerase sigma factor in Cyanidium caldarium. Proc Natl Acad Sci USA 93: 3313–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Severinov K (2003) Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol 57: 301–322 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. New York, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 6: 283–290 [DOI] [PubMed] [Google Scholar]
- Schön A, Krupp G, Gough S, Berry-Lowe S, Kannangara CG, Söll D (1986) The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature 322: 281–284 [DOI] [PubMed] [Google Scholar]
- Shirano Y et al. (2000) Chloroplast development in Arabidopsis thaliana requires the nuclear-encoded transcription factor sigma B. FEBS Lett 485: 178–182 [DOI] [PubMed] [Google Scholar]
- Sugita M, Sugiura M (1996) Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H (2002) Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA doublestrand breaks. Plant J 29: 771–781 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Oikawa K, Ohta N, Kuroiwa H, Kuroiwa T, Takahashi H (1996) Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science 272: 1932–1935 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Tozawa Y, Mochizuki N, Shinozaki K, Nagatani A, Wakasa K, Takahashi H (1997) Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett 413: 309–313 [DOI] [PubMed] [Google Scholar]
- Umeda M, Bhalerao RP, Schell J, Uchimiya H, Koncz C (1998) A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 5021–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]


