Abstract
Mitochondria are the main sites of biological energy generation in eukaryotes. These organelles are remnants of a bacterial endosymbiont that took up residence inside a host cell over 1,500 million years ago. Comparative genomics studies suggest that the mitochondrion is monophyletic in origin. Thus, the original mitochondrial endosymbiont has evolved independently in anaerobic and aerobic environments that are inhabited by diverse eukaryotic lineages. This process has resulted in a collection of morphologically, genetically and functionally heterogeneous organelle variants that include anaerobic and aerobic mitochondria, hydrogenosomes and mitosomes. Current studies aim to determine whether a central common function drives the retention of mitochondrial organelles in different eukaryotic organisms.
Keywords: mitochondrial evolution, anaerobic eukaryotes, remnant organelles, amitochondrial organisms, eukaryotic evolution
Introduction
A wide range of biochemical reactions—such as pyruvate oxidation, the citric acid cycle, electron transport, oxidative phosphorylation and ATP generation—takes place in the mitochondria of aerobic eukaryotes. Mitochondria also have key roles in buffering cytosolic calcium, fatty acid oxidation, haem biosynthesis and the biosynthesis of iron–sulphur (FeS) clusters. The importance of mitochondria in cellular metabolism has been emphasized by the discovery of an increasing number of human diseases that are caused either by defects in mitochondrial proteins encoded in the nucleus or by point mutations or large deletions in mitochondrial DNA (Scheffler, 2000).
The discovery of highly degenerate mitochondrion-related organelles such as hydrogenosomes (Cerkasovová et al, 1973; Lindmark & Müller, 1973) and mitosomes (Mai et al, 1999; Tovar et al, 1999) in eukaryotes that lack typical mitochondria has revived interest in the origins and evolutionary aspects of the endosymbiosis-derived organelles of eukaryotes. Known as hydrogenosomes or mitosomes, depending on whether or not they generate molecular hydrogen as a metabolic end-product, these organelles are found in a range of microbial eukaryotes. Some, such as Cryptosporidium parvum and Neocallimastix patriciarum are related to mitochondrion-containing organisms, whereas others, such as Giardia intestinalis and Trichomonas vaginalis, are considered basal because of their apparent early branching in phylogenetic reconstructions. In this short review, we look at the multifaceted morphological and biochemical nature of mitochondria and review recent progress in the characterization of mitochondrial remnant organelles.
Mitochondrial heterogeneity
Mitochondria are genetically diverse. The mitochondrial genome is a remnant of the original bacterial endosymbiont that gave rise to current mitochondria. The original interaction between the endosymbiont and host resulted in the transfer of endosymbiont genes to the host nucleus and the irreversible loss of redundant genes (Adams & Palmer, 2003; Timmis et al, 2004). The observation that mitochondrial genomes vary enormously in size and gene content suggests that gene transfer might be dependent on some environmental conditions. Reclimonas americana, a freshwater protozoan, contains the least derived mitochondrial genome with the largest coding capacity and encodes 97 genes in a genome that is just over 69 kb in size. At the other end of the spectrum, Plasmodium falciparum, the causative agent of malaria, contains the smallest reported mitochondrial genome, which encodes just five genes in a 6-kb element. Under certain conditions, this reductive tendency may result in the eventual displacement of all mitochondrial genes from the organelle, as suggested by the apparent absence of an organellar genome in diverse mitochondrion-related organelles (Abrahamsen et al, 2004; León-Avila & Tovar, 2004; Turner & Müller, 1983; van der Giezen et al, 1997).
Two main theories to explain the presence of mitochondrial genomes have been advanced: the hydrophobicity theory (von Heijne, 1986) and the co-location for redox regulation (CORR) hypothesis (Allen, 1993). The former suggests that mitochondria retained certain genes because the hydrophobicity and membrane topologies of their products would not allow their import into mitochondria if translated in the cytoplasm. The latter states that mitochondria retained a genome because this enables them to regulate expression tightly depending on the redox state of the organelle. Because the mitochondrial genes that exist today mostly encode hydrophobic proteins involved in bioenergetic processes that are absent in mitochondrial remnant organelles, both hypotheses could explain the absence of a genome in these organelles.
Mitochondria are also biochemically diverse. Traditionally, these organelles are seen as energy-generating entities that produce ATP through a complex set of biochemical reactions involving pyruvate decarboxylation, proton-pumping electron transport and oxidative phosphorylation. During aerobic respiration, oxygen acts as the final acceptor of electrons from NADH and FADH2, but many eukaryotic organisms contain mitochondria that can produce ATP without consuming oxygen. Instead, these facultative anaerobic organelles use terminal oxidases that are capable of using endogenous organic compounds, such as fumarate or inorganic nitrate, as final electron acceptors (Tielens et al, 2002). Facultative anaerobic mitochondria are found not only in some obscure unicellular protists that live in micro-aerophilic or anaerobic environments, but also in 'higher' multicellular eukaryotes, such as mussels, snails and parasitic helminths. The biochemical diversity of anaerobic mitochondria and the biochemical range of aerobic mitochondria illustrate the biochemical heterogeneity of this organelle.
Mitochondria are also morphologically diverse. They are surrounded by a limiting double membrane, but the traditional baffle-like cristae model of the inner mitochondrial membrane (Fig 1A) is a simplified reflection of the true dynamics of mitochondrial morphology. Their morphology depends on the physiological state of the cell and varies significantly between different cell types. Mitochondria can be present as independent organelles in some cells, but in others, they are a highly dynamic continuous network. The morphological heterogeneity of the organelle is clearly illustrated by the cristae-less mitochondria of human testicular somatic cells and by the marked stagespecific morphological changes of the single mitochondrion of the African trypanosome (Priest & Hajduk, 1994). In addition, yeast mitochondria lose their cristae under anoxic conditions, a phenotype that can be reversed by aeration (Lloyd, 1974). These observations indicate that a standard mitochondrion, with invariable genetic, biochemical and morphological properties, does not exist.
Figure 1.
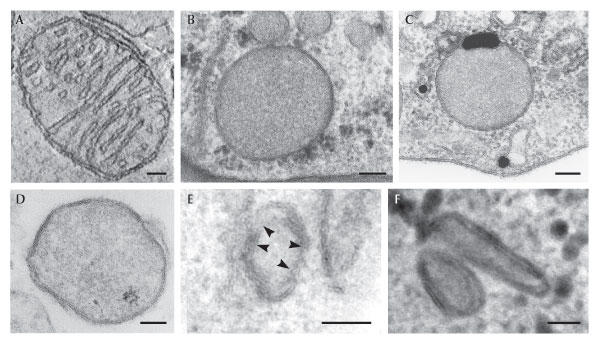
Electron micrographs of different mitochondrial manifestations. (A) Mitochondrion from chicken cerebellum. (B,C) Hydrogenosomes from (B) the anaerobic fungus Neocallimastix patriciarum and (C) the cattle parasite Tritrichomonas foetus. (D–F) Mitosomes from (D) the intestinal parasite Entamoeba histolytica, (E) the microsporidian Trachipleistophora hominis and (F) the diplomonad Giardia intestinalis. Scale bars: (A–D) 100 nm and (E,F) 50 nm. (A) Kindly provided by T.G. Frey, San Diego State University, CA, USA, and G.A. Perkins, University of California, San Diego, CA, USA; (B,C) kindly provided by M. Benchimol, Universidade Santa Ursula, Brazil; (D) reproduced with permission from the American Society for Microbiology; (E,F) reproduced with permission from Nature Publishing Group, Macmillan Limited.
Mitochondrion-related remnant organelles
Mitochondrion-related organelles, such as hydrogenosomes and mitosomes, are found in eukaryotic organisms that lack 'typical' mitochondria (Fig 1B–F). The human parasites Trichomonas vaginalis, Giardia intestinalis, Entamoeba histolytica, Encephalitozoon cuniculi and Trachipleistophora hominis were all once thought to be the direct descendents of the proto-eukaryotic cell that is hypothesized to have hosted the original mitochondrial endosymbiont (Cavaliersmith, 1983). However, the basal placement of these organisms has been shown to be an artefact of phylogenetic analyses (Embley & Hirt, 1998). This, plus the fact that mitochondrion-related organelles have been found in all anaerobic microbial eukaryotes that have been investigated in sufficient molecular detail thus far, provides a strong argument against the idea that these organisms are primitive amitochondriates (Embley et al, 2003a; Gray, 2005; Hrdy et al, 2004).
The presence of mitochondrion-related organelles is not restricted to these discredited 'primitive' lineages; these organelles are also found in diverse groups of protists, fungi and ciliates that have close mitochondrial relatives, such as Cryptosporidium parvum, Neocallimastix patriciarum and Dasytricha ruminantium (Fig 2; Embley et al, 2003a; Riordan et al, 2003). Although more research is necessary to characterize these organelles in full, evidence suggests that mitochondrion-related organelles are a collection of biochemically and morphologically diverse organelles that have independent evolutionary histories but that share a common origin, similar to mitochondria.
Figure 2.
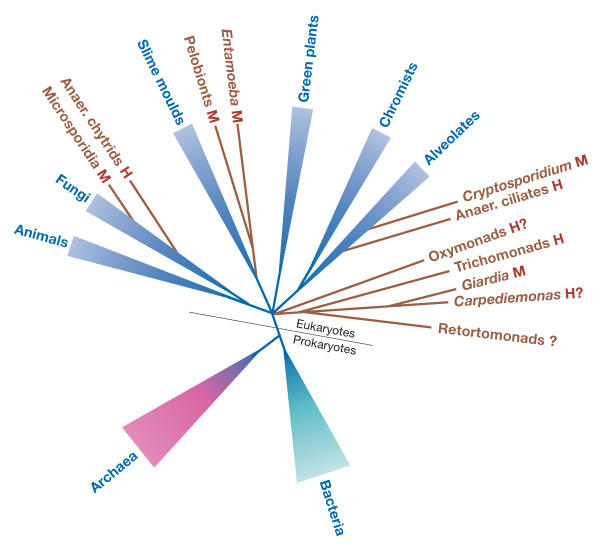
Schematic representation of the distribution of anaerobic microbial eukaryotes on a universal tree of life. The presence of mitochondrial-derived organelles is indicated: H, hydrogenosome; M, mitosome; ?, unconfirmed or suspected. Anaerobic microbial eukaryotes are shown in brown.
Organelle evolution
Exactly how mitochondrion-related organelles originated and how they have evolved to their present state is a matter of scientific debate. Their presence in diverse groups of organisms that have close mitochondrial relatives indicates that both hydrogenosomes and mitosomes have evolved independently on several occasions (Fig 2). Detailed phylogenetic analyses of mitochondrial and plastid remnant genomes have established unambiguously the independent endosymbiotic origins of mitochondria and chloroplasts (Gray et al, 1999). However, until recently, no organellar DNA had been detected in most mitochondrion-related organelles (Abrahamsen et al, 2004; León-Avila & Tovar, 2004; Turner & Müller, 1983; van der Giezen et al, 1997), which hampered the direct genetic demonstration of their evolutionary ancestry. This has now changed. Phylogenetic analysis of the remnant hydrogenosomal genome of the anaerobic ciliate Nyctotherus has provided unequivocal evidence that these mitochondrion-related organelles are indeed modified mitochondria (Boxma et al, 2005). The recent identification of DNA in the mitosomes of Blastocystis (Nasirudeen & Tan, 2004) should allow the genetic probing of the evolutionary history of mitosomes as well.
Two alternative hypotheses to explain the origins of mitochondrion-related organelles have been put forward (Fig 3). The first proposes vertical descent from the original mitochondrial endosymbiont (Embley et al, 2003b; Martin & Müller, 1998; Tielens et al, 2002; van der Giezen & Tovar, 2004). The second invokes two serial endosymbiotic events: one that led to the acquisition of the original mitochondrial endosymbiont, and the other, subsequent to the loss of the original mitochondrial endosymbiont, that led to the formation of a chimerical organelle that evolved into hydrogenosomes and/or mitosomes (Dyall et al, 2004a,b).
Figure 3.
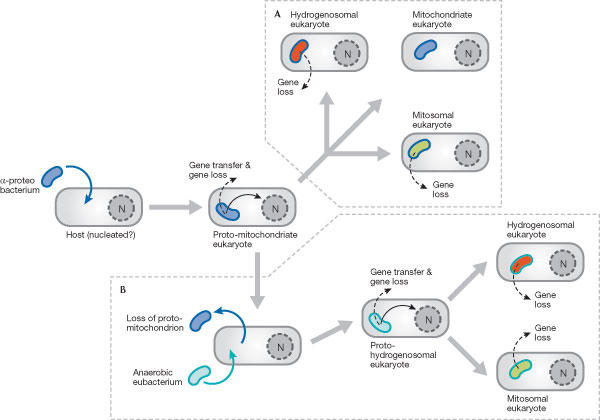
Evolution of mitochondria and mitochondrion-related organelles. (A) Vertical descent from the original mitochondrial endosymbiont. (B) Chimeric origins of mitosomes and hydrogenosomes from sequential endosymbionts. See text for details. Mitochondria/α-proteobacteria are shown in blue, hydrogenosomes in red, mitosomes in lime and anaerobic eubacterium in turquoise. N, nucleus.
The first proposal (Fig 3A) is the most parsimonious hypothesis and is consistent with most of the experimental data available. Direct evolution from the mitochondrial endosymbiont explains the presence of the limiting double membrane that surrounds mitochondrion-related organelles and the functionally conserved mechanisms of protein import into hydrogenosomes, mitosomes and mitochondria. It also explains the mutually exclusive distribution of mitochondria and mitochondrion-related organelles, and the independent evolution of mitochondrion-related organelles in a range of unrelated eukaryotic lineages, whether or not they are related to known mitochondrial groups.
The second proposal is less parsimonious and although it might help to explain the presence and phylogenetic affinities of the anaerobic enzymes hydrogenase and pyruvate:ferredoxin oxidoreductase in hydrogenosomes, it does not account for the multiple independent origins of mitochondrion-related organelles in unrelated eukaryotic lineages (Fig 3B). To account for the latter under this scheme, two assumptions are required: first, that the loss of the original mitochondrial endosymbiont occurred several times independently in different eukaryotic lineages, presumably at different geological times; and second, that in each case, independent anaerobic eubacterial endosymbionts subsequently became established and evolved into diverse, highly derived mitochondrion-related organelles. Moreover, one unfulfilled prediction in this scenario is the existence of bona fide secondarily amitochondrial lineages related to those that are known to harbour mitochondrion-related organelles (that is, the descendents of secondarily amitochondrial cells that did not participate in the second endosymbiotic interaction). Because no confirmed secondarily amitochondrial eukaryotes have yet been identified, the evidence for this proposal is at present tenuous.
A common function for mitochondrial organelles?
Understanding the functions of mitochondrion-derived genes and organelles is now a priority. Oxidative phosphorylation-dependent ATP biosynthesis, which is a major function of mitochondria, has not been observed in mitochondrion-related organelles. Isolated hydrogenosomes take up ADP from the medium and excrete equimolar amounts of ATP (Müller, 1993). ATP is synthesized in these organelles by substrate-level phosphorylation through the catalytic conversion of succinyl-CoA to succinate by succinate thiokinase. Mitosome-containing organisms, conversely, are known to synthesize ATP by substrate-level phosphorylation in the cytosol, with no direct involvement of their mitochondrion-related organelles. If energy metabolism is not the unifying functional feature between mitochondria and its related organelles, what is?
FeS proteins have important roles in electron transfer, enzymatic catalysis and metabolic regulation and are universally distributed in prokaryotic and eukaryotic organisms. Several genes encoding proteins that participate in the biosynthesis of FeS clusters have been identified in the genomes of protists that harbour mitochondrion-related organelles, including G. intestinalis, T. vaginalis, C. parvum, E. histolytica and E. cuniculi (Abrahamsen et al, 2004; Ali et al, 2004; Katinka et al, 2001; LaGier et al, 2003; Sutak et al, 2004; Tachezy et al, 2001; Tovar et al, 2003; van der Giezen et al, 2004). Furthermore, Giardia mitosomes and trichomonad hydrogenosomes have been shown to function in FeS cluster biosynthesis and in FeS protein maturation (Lill & Kispal, 2000; Sutak et al, 2004; Tovar et al, 2003), similar to mitochondria. Thus, evidence suggests that the biosynthesis of critical FeS moieties might be a unifying functional feature of mitochondria and mitochondrion-related organelles. Such a crucial observation has prompted the suggestion that the requirement for FeS proteins might have had a central role in securing the establishment and retention of the original mitochondrial endosymbiont (Embley et al, 2003b; Martin & Müller, 1998; Tachezy et al, 2001; Tovar et al, 2003; van der Giezen & Tovar, 2004).
Despite the crucial nature of FeS cluster biosynthesis, several FeS proteins are known to be dispensable under a variety of environmental conditions—for example, glutamate synthase, aconitase, subunits of respiratory complexes I, II, and III, pyruvate:ferredoxin oxidoreductase and hydrogenase. Investigating precisely why FeS metabolism is essential in organisms that harbour mitochondrion-related organelles may help to explain why degenerate mitochondria have been retained in different eukaryotic lineages. Or more precisely, which FeS protein(s) could be so crucial that the whole FeS machinery has been retained? The most promising candidate so far is Rli1, the only essential FeS protein in yeast that is not involved in FeS assembly itself (Balk et al, 2004). Its universal presence in eukaryotes, including those that contain mitochondrial remnants, and its involvement in tRNA and/or rRNA maturation (Dong et al, 2004; Gabaldon & Huynen, 2004) hint at an ancestral essential role for this protein.
Conclusions and future directions
Much remains to be investigated about the physiology and metabolic range of mitosomes and hydrogenosomes. Have other mitochondrial functions, such as amino-acid and fatty-acid metabolism, calcium homeostasis and even apoptosis, been retained? What is the level of functional heterogeneity among mitochondrion-related organelles? With the genomes of several protozoa completed (Katinka et al, 2001; Loftus et al, 2005; Abrahamsen et al, 2004) or nearly completed (T. vaginalis, G. intestinalis), we might soon be in a position to understand the role that mitochondrion-related organelles have in the overall biology of 'amitochondrial' organisms, some of which are human parasites of major medical importance.
Similar to mitochondria, hydrogenosomes and mitosomes contain ATP-dependent molecular chaperones Cpn60 and/or mitochondrial heat-shock protein mtHsp70 (Mai et al, 1999; Tovar et al, 1999; Williams et al, 2002). ATP/ADP transport occurs in hydrogenosomes and members of the mitochondrial carrier family of transporters have been identified in trichomonad and fungal hydrogenosomes (Dyall et al, 2003; Müller, 1993; Tjaden et al, 2004; van der Giezen et al, 2002; Voncken et al, 2002). How ATP is imported into mitosomes is not known, but the presence of ATP-consuming chaperones suggests that nucleoside transport across their limiting membranes must also take place in vivo. Recent data indicate that Entamoeba mitosomes use an unusual member of the mitochondrial carrier family to transport ATP across their membranes (Chan et al, 2005).
Investigating how proteins are targeted to mitosomes will help to elucidate the degree of degeneracy of the protein import apparatus. Not all mitosomal proteins have obvious mitochondrial targeting presequences, and the scant distribution of identifiable protein translocators in organisms that harbour degenerate mitochondria is all too apparent. For example, the microsporidian genome only encodes one translocator for the outer membrane (tom70) and one for the inner membrane (tim22), in contrast with the multiplicity of proteins that is required for protein import in yeast (Katinka et al, 2001; Truscott et al, 2003). Studying the import systems of these degenerate mitochondrial organelles might help to define the minimum molecular set that is required for protein translocation across a double-membrane-bound organelle.
Determining the protein complement of various mitochondrion-related organelles will be useful in mapping their evolutionary paths. However, a direct demonstration of their precise origins can only come from the sequencing and phylogenetic analyses of their genomes. In this respect, the recent discovery of organellar DNA in the mitosomes of B. hominis is a promising development (Nasirudeen & Tan, 2004). Most importantly, the genetic demonstration that Nyctotherus hydrogenosomes are modified mitochondria (Boxma et al, 2005) offers unqualified support to the hypothesis that mitochondrion-related organelles arose by vertical descent from the original mitochondrial endosymbiont. As such, they should be considered degenerate mitochondria.


References
- Abrahamsen MS et al. (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304: 441–445 [DOI] [PubMed] [Google Scholar]
- Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 [DOI] [PubMed] [Google Scholar]
- Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T (2004) An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron–sulfur cluster assembly under anaerobic conditions. J Biol Chem 279: 16863–16874 [DOI] [PubMed] [Google Scholar]
- Allen JF (1993) Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol 165: 609–631 [DOI] [PubMed] [Google Scholar]
- Allen JF (2003) The function of genomes in bioenergetic organelles. Philos Trans R Soc Lond B Biol Sci 358: 19–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Pierik AJ, Netz DJ, Muhlenhoff U, Lill R (2004) The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron–sulphur proteins. EMBO J 23: 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxma B et al. (2005) An anaerobic mitochondrion that produces hydrogen. Nature 434: 74–79 [DOI] [PubMed] [Google Scholar]
- Cavaliersmith T (1983) A 6 kingdom classification and a unified phylogeny. In Schwemmler W, Schenk HEA (eds) Endocytobiology. II. pp 1027–1034. Berlin, Germany: De Gruyter [Google Scholar]
- Cerkasovová A, Lukasová G, Cerkasòv J, Kulda J (1973) Biochemical characterization of large granule fraction of Tritrichomonas foetus (strain KV1). J Protozool 20: 525 [Google Scholar]
- Chan KW et al. (2005) A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr Biol 15: 737–742 [DOI] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Dyall SD, Lester DC, Schneider RE, Delgadillo-Correa MG, Plumper E, Martinez A, Koehler CM, Johnson PJ (2003) Trichomonas vaginalis Hmp35, a putative pore-forming hydrogenosomal membrane protein, can form a complex in yeast mitochondria. J Biol Chem 278: 30548–30561 [DOI] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ (2004a) Ancient invasions: from endosymbionts to organelles. Science 304: 253–257 [DOI] [PubMed] [Google Scholar]
- Dyall SD, Yan W, Delgadillo-Correa MG, Lunceford A, Loo JA, Clarke CF, Johnson PJ (2004b) Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature 431: 1103–1107 [DOI] [PubMed] [Google Scholar]
- Embley TM, Hirt RP (1998) Early branching eukaryotes? Curr Opin Genet Dev 8: 624–629 [DOI] [PubMed] [Google Scholar]
- Embley TM, van der Giezen M, Horner DS, Dyal PL, Foster P (2003a) Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Philos Trans R Soc Lond B Biol Sci 358: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, van der Giezen M, Horner DS, Dyal PL, Bell S, Foster P (2003b) Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life 55: 387–395 [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Huynen MA (2004) Prediction of protein function and pathways in the genome era. Cell Mol Life Sci 61: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW (2005) The hydrogenosome's murky past. Nature 434: 29–31 [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283: 1476–1481 [DOI] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Embley TM (2004) Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432: 618–622 [DOI] [PubMed] [Google Scholar]
- Katinka MD et al. (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453 [DOI] [PubMed] [Google Scholar]
- LaGier MJ, Tachezy J, Stejskal F, Kutisova K, Keithly JS (2003) Mitochondrial-type iron–sulfur cluster biosynthesis genes (IscS and IscU) in the apicomplexan Cryptosporidium parvum. Microbiology 149: 3519–3530 [DOI] [PubMed] [Google Scholar]
- León-Avila G, Tovar J (2004) Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organellar genome. Microbiology 150: 1245–1250 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Lindmark DG, Müller M (1973) Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem 248: 7724–7728 [PubMed] [Google Scholar]
- Lloyd D (1974) The mitochondria of microorganisms. London, UK: Academic
- Loftus B et al. (2005) The genome of the protist parasite Entamoeba histolytica. Nature 433: 865–868 [DOI] [PubMed] [Google Scholar]
- Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J (1999) Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol 19: 2198–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Müller M (1998) The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- Müller M (1993) The hydrogenosome. J Gen Microbiol 139: 2879–2889 [DOI] [PubMed] [Google Scholar]
- Nasirudeen AM, Tan KS (2004) Isolation and characterization of the mitochondrion-like organelle from Blastocystis hominis. J Microbiol Methods 58: 101–109 [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL (1994) Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J Bioenerg Biomembr 26: 179–191 [DOI] [PubMed] [Google Scholar]
- Riordan CE, Ault J, Langreth SG, Keithly JS (2003) Cryptosporidium parvum Cpn60 targets a relict organelle. Curr Genet 44: 138–147 [DOI] [PubMed] [Google Scholar]
- Scheffler IE (2000) A century of mitochondrial research: achievements and perspectives. Mitochondrion 1: 3–31 [DOI] [PubMed] [Google Scholar]
- Sutak R, Dolezal P, Hrdy I, Dancis A, Delgadillo M, Johnson PJ, Müller M, Tachezy J (2004) Mitochondrial-type assembly of iron–sulfur centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc Natl Acad Sci USA 101: 10368–10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy J, Sánchez LB, Müller M (2001) Mitochondrial type iron–sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol Biol Evol 18: 1919–1928 [DOI] [PubMed] [Google Scholar]
- Tielens AGM, Rotte C, van Hellemond JJ, Martin W (2002) Mitochondria as we don't know them. Trends Biochem Sci 27: 564–572 [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Tjaden J, Haferkamp I, Boxma B, Tielens AG, Huynen M, Hackstein JH (2004) A divergent ADP/ATP carrier in the hydrogenosomes of Trichomonas gallinae argues for an independent origin of these organelles. Mol Microbiol 51: 1439–1446 [DOI] [PubMed] [Google Scholar]
- Tovar J, Fischer A, Clark CG (1999) The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol 32: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Tovar J, León-Avila G, Sánchez L, Sutak R, Tachezy J, van der Giezen M, Hernández M, Müller M, Lucocq JM (2003) Mitochondrial remnant organelles of Giardia function in iron–sulphur protein maturation. Nature 426: 172–176 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Brandner K, Pfanner N (2003) Mechanisms of protein import into mitochondria. Curr Biol 13: R326–R337 [DOI] [PubMed] [Google Scholar]
- Turner G, Müller M (1983) Failure to detect extranuclear DNA in Trichomonas vaginalis and Tritrichomonas foetus. J Parasitol 69: 234–236 [PubMed] [Google Scholar]
- van der Giezen M, Tovar J (2004) Hydrogenosomes, mitosomes and mitochondria: variations on a theme? In Horner DS, Hirt RP (eds) Organelles, Genomes and Eukaryote Phylogeny: An Evolutionary Synthesis in the Age of Genomics pp 289–308. Boca Raton, FL, USA: CRC Press [Google Scholar]
- van der Giezen M, Sjollema KA, Artz RR, Alkema W, Prins RA (1997) Hydrogenosomes in the anaerobic fungus Neocallimastix frontalis have a double membrane but lack an associated organelle genome. FEBS Lett 408: 147–150 [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Slotboom DJ, Horner DS, Dyal PL, Harding M, Xue G-P, Embley TM, Kunji ER (2002) Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J 21: 572–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Giezen M, Cox S, Tovar J (2004) The iron–sulfur cluster assembly genes IscS and IscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol Biol 4: 7. 10.1186/1471-2148-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G (1986) Why mitochondria need a genome. FEBS Lett 198: 1–4 [DOI] [PubMed] [Google Scholar]
- Voncken F et al. (2002) Multiple origins of hydrogenosomes: functional and phylogenetic evidence from the ADP/ATP carrier of the anaerobic chytrid Neocallimastix sp. Mol Microbiol 44: 1441–1454 [DOI] [PubMed] [Google Scholar]
- Williams BA, Hirt RP, Lucocq JM, Embley TM (2002) A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418: 865–869 [DOI] [PubMed] [Google Scholar]


