Abstract
DRD1 is a SNF2-like protein previously identified in a screen for mutants defective in RNA-directed DNA methylation of a seed promoter in Arabidopsis. Although the initial study established a role for DRD1 in RNA-directed DNA methylation, it did not address whether DRD1 is needed for de novo or maintenance methylation, or whether it is required for methylation of other target sequences. We show here that DRD1 is essential for RNA-directed de novo methylation and acts on different target promoters. In addition, an unanticipated role for DRD1 in erasure of CG methylation was shown when investigating maintenance methylation after segregating away the silencing trigger. DRD1 is unique among known SNF2-like proteins in facilitating not only de novo methylation of target sequences in response to RNA signals, but also loss of methylation when the silencing inducer is withdrawn. The opposing roles of DRD1 could contribute to the dynamic regulation of DNA methylation.
Keywords: demethylation, de novo methylation, maintenance methylation, RNA-directed DNA methylation, SNF2-like protein
Introduction
RNA-directed DNA methylation has been documented in plants (Mathieu & Bender, 2004) and in human cells (Kawasaki & Taira, 2004; Morris et al, 2004). Genetic analyses in Arabidopsis have identified the DNA methyltransferases that catalyse de novo cytosine (C) methylation in response to RNA signals (Cao et al, 2003; Aufsatz et al, 2004) and maintain symmetrical C(N)G methylation (N is A, T or C) in cooperation with histone-modifying enzymes (Jones et al, 2001; Aufsatz et al, 2002a; Jackson et al, 2002; Malagnac et al, 2002). A plantspecific SNF2-like putative chromatin-remodelling protein, DRD1, was identified in a screen for mutants impaired in RNA-directed DNA methylation and silencing of the seed-specific α′ promoter (Kanno et al, 2004). From the initial genetic screen, it was unclear whether drd1 mutants are deficient in de novo methylation or in maintenance methylation. In the case of RNA-directed DNA methylation, de novo methylation leads to the modification of Cs in all sequence contexts within the region of RNA–DNA sequence identity, whereas maintenance methylation refers to perpetuation of symmetrical C(N)G methylation during DNA replication in the absence of the RNA trigger (Matzke et al, 2004). To further understand the role of DRD1 in RNA-directed DNA methylation, we examined separately the effects of a drd1 mutation on RNA-directed de novo methylation and on maintenance methylation. We report here that DRD1 influences both of these processes in a manner that facilitates the dynamic regulation of DNA methylation.
Results and Discussion
Transgenic systems to study RNA-mediated silencing and methylation of the seed-specific α′ promoter (Kanno et al, 2004) and the constitutive nopaline synthase (NOS) promoter (Aufsatz et al, 2002b) have been described previously. These systems rely on a doublestranded RNA—encoded at a silencer H complex (hygromycin resistance)—that is homologous to the respective target promoter sequence. The double-stranded RNAs are processed to short RNAs, 21–24 nucleotides in length, which are thought to trigger de novo methylation and silencing of the homologous target promoter at an unlinked target K complex (kanamycin resistance; supplementary Fig 1 online). In the absence of the silencer, the respective target promoter is active and unmethylated (Fig 1A: left, α′ promoter; right, NOS promoter), whereas it is repressed and methylated in the presence of the silencer (Fig 1B: left, α′ promoter; right, NOS promoter).
Figure 1.
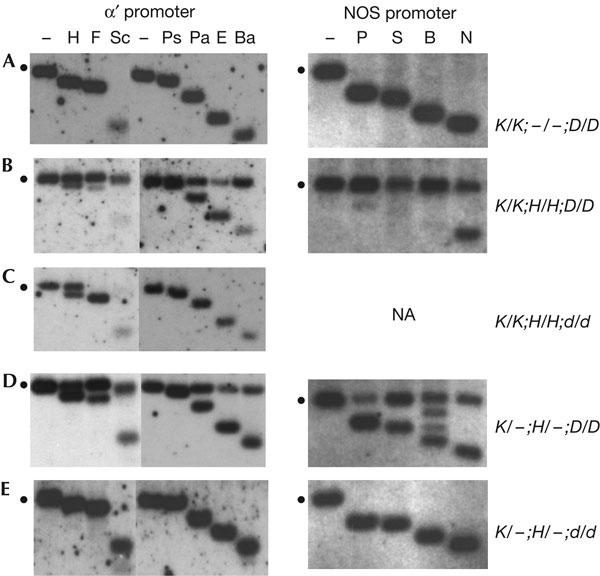
Analysis of de novo methylation. Methylation of the target α′ promoter was analysed using restriction enzymes diagnostic for non-CG methylation (F, Sc, Ps, Pa, E, Ba) or CG methylation (H). Methylation of the nopaline synthase (NOS) promoter was analysed using restriction enzymes diagnostic for non-CG methylation (N), CG methylation (P, B) or both (S). The respective methylationsensitive enzymes were added after a standard digest with non-methylation-sensitive restriction enzymes (‘−' lanes). The position of the methylated fragment is denoted by the dots to the left of each blot. Shifts to the smaller fragment(s) indicate no methylation at the site(s) tested. Maps of the promoters showing positions of restriction enzyme sites and the probes used for hybridization are depicted in supplementary Fig 1 online. Genotypes of the plants analysed are shown to the right; dashes indicate a hemizygous transgene locus. Bold letters to the left represent boxed genotypes shown in the breeding schemes in Fig 2. Abbreviations: K, target complex; H, silencer complex; d, drd1-6 mutant; D, wild type; NA, not applicable. Abbreviations of enzymes and their recognition sequences (sensitivity to C methylation indicated by the superscript ‘m'): α′ promoter: B, BstUI (mCGmCG); Ba, BamHI (GGATmCmC); E, EcoT22I (ATGmCAT); F, Fnu4HI (GmCmNGmC, if N is C); H, HpyCH4IV (AmCGT); NOS promoter: N, NheI (GCTAGmC); P, Psp1406I (AAmCGTT); Pa, PagI (TmCATGA); Ps, PstI (mCTGmCAG); S, SacII (mCmCGmCGG); Sc, ScrF1 (CmCmNGG (if N is C).
In drd1 mutants, non-CG methylation of the target α′ promoter is greatly reduced, as evidenced by complete digestion with methylation-sensitive restriction enzymes abbreviated F, Sc, Ps, Pa, E and Ba (Fig 1C, left: data are for third-generation drd1-6 plants). By contrast, substantial CG methylation is retained, as indicated by the persistent double band generated by the restriction enzyme abbreviated H (Fig 1C, left). Similar findings for drd1-1 led to the suggestion that DRD1 is important primarily for non-CG methylation (Kanno et al, 2004).
To analyse whether DRD1 is needed for RNA-directed de novo methylation of target sequences, crosses were made to generate F1 plants in which a naive target K complex was combined with the silencer H complex in either wild-type (D/D; Fig 2D) or homozygous drd1-6 (d/d) plants (Fig 2E). Methylation of the target promoter was then examined in the resulting F1 progeny.
Figure 2.
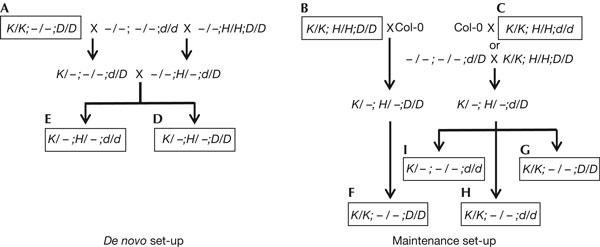
Breeding schemes. De novo methylation analysis: wild-type plants that were homozygous for either the target K complex (K/K;−/−;D/D) or the silencer H complex (−/−;H/H;D/D) were crossed with a transgene-free, homozygous drd1-6 mutant (−/−;−/−;d/d), producing respective heterozygous drd1-6 progeny (K/−;−/−;d/D and −/−;H/−;d/D). Heterozygous drd1-6 plants containing either the target K complex or silencer H complex were crossed and F1 progeny that were either drd1-6 mutant (K/−;H/−;d/d) or wild type (K/−;H/−;D/D) were identified by genotyping. Maintenance methylation analysis: α′ promoter system: segregating the target K complex from the silencer H complex in wild-type (K/K;H/H;D/D) or homozygous drd1-6 plants (K/K;H/H;d/d) required an initial backcross to untransformed wild-type plants (Col-0). This produced progeny that were hemizygous for the K and H complexes in either wild-type (K/−;H/−;D/D) or heterozygous drd1-6 (K/−;H/−;d/D) backgrounds. The nopaline synthase (NOS) promoter system: the K/K;H/H;D/D plants were likewise crossed to Col-0 to produce wild-type progeny (K/−;H/−;D/D). To generate the K/−;H/−;d/D genotype, wild-type plants that were homozygous for the respective target K complex and silencer H complex (K/K;H/H;D/D) were crossed with a transgene-free, heterozygous drd1-6 mutant (−/−;−/−;d/D). For both silencing systems, wild-type plants (K/−;H/−;D/D) were selfed and the resulting F2 progeny genotyped to identify those containing K and lacking H (K/K;−/−;D/D). The heterozygous drd1-6 plants (K/−;H/−;d/D) were also selfed and the resulting F2 progeny were genotyped to identify those containing K and lacking H in wild-type (K/K;−/−;D/D) and homozygous drd1-6 mutant (K/(K);−/−;d/d) backgrounds. The boxed and lettered genotypes correspond to the lettered blots in Figs 1 and 4.
In wild-type F1 plants, the target α′ promoter showed increased methylation in CGs and in non-CGs after introducing the silencer H complex (Fig 1D, left). The level of methylation observed in wild-type F1 plants approximated that seen in plants in which the target complex and silencer complex had been together in the same genome for several generations (Fig 1B, left). The similar methylation patterns indicate that the maximum attainable level of RNA-directed methylation of the target α′ promoter is essentially reached in the first generation containing both transgene complexes. By contrast, the target α′ promoter did not acquire detectable methylation after being combined with the silencer H complex in homozygous drd1-6 plants (Fig 1E, left), which showed a hybridization pattern identical to that of the unmethylated target α′ promoter (Fig 1A, left). The lack of methylation was not because of inadequate production of RNA signals, as indicated by the continued presence of α′ promoter short RNAs in drd1-6 plants (Fig 3A).
Figure 3.
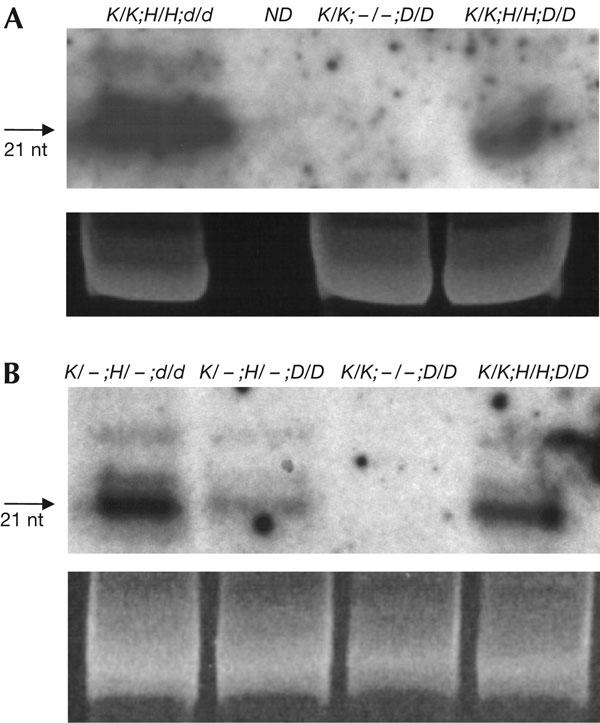
Short RNA analysis. (A) Northern blot analysis of α′ promoter short RNAs in wild-type plants containing only the target complex (K/K;−/−;D/D) or both the target and silencer complexes (K/K;H/H;D/D), or in the drd1-6 mutant (K/K;H/H;d/d). (B) Northern blot analysis of nopaline synthase (NOS) promoter short RNAs in F1 progeny from the ‘de novo' cross (Fig 2) that were homozygous for the drd1-6 mutation (K/−;H/−;d/d) or wild type (K/−;H/−;D/D), and in wild-type plants that were either homozygous for the target K complex only (K/K;−/−;D/D) or double homozygous for the target K complex and silencer H complex (K/K;H/H;D/D). In wild-type plants, NOS promoter short RNAs are approximately twice as abundant in homozygous H/H as in hemizygous H/− plants. The quantities of short RNAs are increased in drd1-6 mutants, perhaps because they are not used and turned over efficiently in these plants. Ethidium bromide staining of the principal RNA species in the samples is shown as a loading control. ND indicates that α′ promoter short RNAs were not tested in the genetype K/−;H/−;D/D.
The target NOS promoter also becomes methylated de novo at CGs and non-CGs in wild-type plants after introducing the respective silencer H construct (Fig 1D, right; Aufsatz et al, 2004), but it failed to acquire measurable methylation in the drd1-6 mutant (Fig 1E, right). Bisulphite sequencing confirmed that no detectable methylation was induced at the target NOS promoter in drd1-6 mutants, whereas methylation was observed at Cs in all sequence contexts within the region of RNA–DNA sequence identity in wild-type plants (supplementary Fig 2 online). NOS promoter short RNAs were detected in the F1 plants (Fig 3B), indicating that RNA signals were available but unable to induce methylation in the drd1-6 mutants. In contrast to the α′ promoter, the level of NOS promoter methylation was less in F1 progeny than in plants in which the target and silencer had been together for several generations (compare Fig 1D, right, with Fig 1B, right). Despite this difference, the results show that DRD1 is indispensable for RNA-directed de novo methylation of two distinct promoters.
The efficiency of maintenance methylation was examined in wild-type and drd1-6 plants after crossing out the respective silencer H complexes to remove the source of the RNA signals. In wild-type F2 progeny descended from DRD1 wild-type parents (Fig 2F), the target α′ promoter lost both CG and non-CG methylation after segregating away the silencer complex (Fig 4F, left), resulting in a hybridization pattern that is indistinguishable from the unmethylated α′ promoter (Fig 1A, left). An identical pattern of methylation (Fig 4G, left) was observed in wild-type F2 progeny descended from the drd1-6 mutant (Fig 2G). Bisulphite sequencing confirmed the loss of non-CG methylation and showed only some residual methylation at two CG dinucleotides (supplementary Fig 3A online). Thus, in the α′ promoter silencing system, almost all methylation is lost in wild-type progeny when the source of the RNA signal is withdrawn. Unexpectedly, however, in drd1-6 progeny lacking the silencer H complex (Fig 2H,I), substantial CG methylation was detected, even though non-CG methylation was lost. Retention of CG methylation is again exemplified by the persistent double band generated by the restriction enzyme abbreviated H. This double band was observed in drd1-6 progeny that are homozygous (Fig 4H, left) and hemizygous (Fig 4I, left) for the target K complex, indicating no dependence on dosage of the target promoter. Bisulphite sequencing confirmed the enhanced maintenance of CG methylation in the drd1-6 mutant (supplementary Fig 3B online) relative to wild-type plants (supplementary Fig 3A online).
Figure 4.
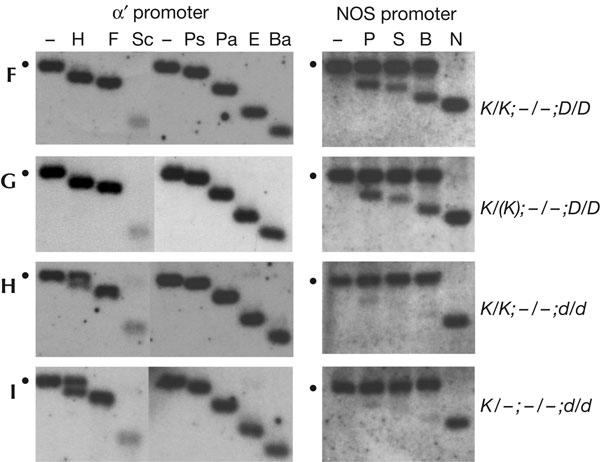
Maintenance methylation analysis. Methylation of the target promoter at the respective K complex (α′ promoter (left) and nopaline synthase (NOS) promoter (right)) after segregating away the respective silencer H complex was studied using methylationsensitive restriction enzymes as described in the legend to Fig 1. The genotypes of the plants analysed are shown to the right. The bold letters to the left represent the boxed genotypes shown in the breeding scheme in Fig 2. ‘K/(K)' denotes both homozygous K/K and hemizygous K/− plants. The plants shown in (G,H,I) are siblings.
A comparable result was obtained when analysing maintenance methylation in the NOS promoter system. In contrast to the α′ promoter, the target NOS promoter retains significant methylation in the absence of the silencer H complex in wild-type plants: although non-CG methylation seems to be lost (exemplified by complete digestion with the enzyme abbreviated N (NheI)), considerable CG methylation remains, as shown by only partial digestion with restriction enzymes abbreviated P (Psp1406I) and B (BstUI) (Fig 4F, right). Similarly to the α′ promoter, however, the NOS promoter shows increased CG methylation in drd1-6 progeny. This is evidenced by poorer digestion with enzymes P and B in drd1-6 mutants (Fig 4H,I, right) than in wild-type plants (Fig 4F,G, right). By contrast, cleavage by enzyme N, which is diagnostic of non-CG methylation, is equivalent in both drd1-6 mutants and wild-type plants (Fig 4F–I, right). Collectively, these findings show a previously unsuspected role for DRD1 in the complete erasure of CG methylation after segregating away the silencer H complex that encodes the RNA trigger.
The results of the experiments reported here, which have examined separately de novo and maintenance methylation, necessitate a revision of the original proposal that DRD1 is primarily important for non-CG methylation (Kanno et al, 2004). DRD1 is required for RNA-directed de novo methylation of Cs in all sequence contexts, including methylation of CG dinucleotides (Fig 5). However, DRD1 is also necessary for efficient loss of methylation, particularly CG methylation, once the source of the RNA signal is removed. The latter requirement is more evident for the target α′ promoter, which loses almost all CG methylation in wild-type plants after segregating away the silencer complex but shows elevated CG methylation in the absence of the silencer in the drd1-6 mutant. Therefore, the methylation pattern observed in third-generation drd1 plants can be explained as an additive defect caused by inadequate erasure of CG methylation coupled to failed de novo methylation, which affects predominantly non-CG nucleotide groups (Fig 5). Whether the remaining CG methylation in drd1 mutants is due to more robust maintenance or to a deficiency in either passive loss or active removal of CG methylation is not known. Passive loss occurs when there is a failure to maintain methylation during successive rounds of DNA replication. By contrast, active demethylation requires a demethylase activity and can take place in nondividing cells (Santos et al, 2002). In animal systems, DNA glycosylases, which are normally involved in base excision repair, have been reported to be involved in active demethylation of CG dinucleotides (Kress et al, 2001). In Arabidopsis, two large proteins that contain DNA glycosylase domains, DEMETER (Xiao et al, 2003; Kinoshita et al, 2004) and REPRESSOR OF SILENCING 1 (Gong et al, 2002), have been implicated in the removal of CG methylation and the re-activation of silenced genes. Additional work is required to clarify how DRD1 contributes to the loss of CG methylation.
Figure 5.
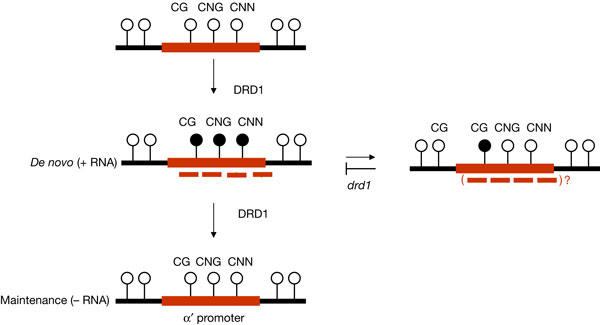
Dual role of DRD1 in the dynamic regulation of RNA-directed DNA methylation. DRD1 is required for RNA-directed de novo methylation of Cs in all sequence contexts for the target promoters tested here. DRD1 is also needed for the complete erasure of methylation after segregation of the source of the RNA signal, as indicated by enhanced retention of CG methylation in drd1 mutants. This is particularly clear for the target α′ promoter, which normally loses most methylation after withdrawing the silencing trigger. Methylation is indicated by filled circles, the target promoter by the red line and short RNAs by red dashed lines.
Although DRD1 seems to be a common requirement for RNA-directed DNA methylation, the two target promoters tested here differ in the rate of their response to the introduction or withdrawal of RNA signals in wild-type plants. The target α′ promoter is very responsive, acquiring a full level of methylation in the first generation that combines the target and silencer complexes and losing most methylation in the first generation after crossing out the silencer complex. By contrast, the target NOS promoter does not attain the full level of RNA-directed methylation in the F1 generation and retains substantial CG methylation after removing the source of the trigger RNA. The reasons for these differences in response time are not known. In both systems, the respective short RNAs seem to be present in comparable amounts. Variations in the sequence composition, structure or chromatin condensation state of the two target complexes might alter their sensitivity to RNA signals (Matzke et al, 2004). For example, there are several copies of the NOS promoter at the corresponding target K complex (Aufsatz et al, 2002b), whereas only a single copy of the α′ promoter is present at the corresponding target K complex (Kanno et al, 2004). The higher density of CG dinucleotides in the NOS promoter (supplementary Fig 2 online) may contribute to more efficient maintenance of CG methylation at this promoter. By contrast, the α′ promoter, which is comparatively deficient in CG dinucleotides (supplementary Fig 3 online), is not repressed by CG methylation but instead seems to be silenced by non-CG methylation (Kanno et al, 2004). As shown here, this non-CG methylation must be established de novo in each generation by a DRD1-dependent, RNA-directed pathway. The fact that the α′ promoter is silenced by non-CG methylation probably allowed us to identify the drd1 mutant in the corresponding genetic screen. By contrast, proteins required for CG methylation, the DNA methyltransferase MET1 (Aufsatz et al, 2002a) and histone deacetylase 6 (Aufsatz et al, 2002b), were identified in a genetic screen carried out on the NOS promoter silencing system.
Several other SNF2-like proteins, including DDM1 (Jeddeloh et al, 1999), Lsh (Dennis et al, 2001) and ATRX (Gibbons et al, 2000), can regulate DNA methylation. However, DRD1 is the only one of these factors that has been implicated both in RNA-directed de novo methylation of cytosines in all sequence contexts and in erasure of CG methylation. The drd1 alleles identified in the original genetic screen contain mutations in functionally implicated regions of the SWI2/SNF2 ATPase domain (Kanno et al, 2004), suggesting that DRD1 functions as a chromatin-remodelling protein to disrupt histone–DNA contacts and/or displace nucleosomes. One possibility is that DRD1 is specialized to allow RNA signals to access homologous target DNA in a chromatin context. Depending on the availability of RNA signals and various DNA-modifying enzymes in different cell types, DRD1 activity could facilitate RNA-guided de novo methylation catalysed by DNA methyltransferases or demethylation of CG dinucleotides catalysed by DNA glycosylases. In this model, DRD1 is a pivotal factor in the dynamic regulation of promoter activity, which might contribute to developmental plasticity, epigenetic reprogramming or adaptation of plants to the environment. Future research will have to focus on determining the full scope of DRD1 activity by using genomics approaches to identify endogenous targets in the genome.
Speculation
In view of the proposed role of DRD1 in opening chromatin to RNA signals to create a substrate for de novo methylation, one can ask whether loss of CG methylation, which also requires DRD1, occurs at DNA sequences targeted by short RNAs (Fig 5, short RNAs in parentheses). Although the silencing complex that encodes the trigger RNA was segregated away in plants used to assess maintenance methylation, we cannot exclude the possibility that parental RNA signals are carried over into cells of the next generation, providing a short time period early in development when RNA-guided demethylation could occur. For example, DEMETER acts only in the central cell of the female gametophyte to remove CG methylation (Xiao et al, 2003; Kinoshita et al, 2004). Thus, whether RNA-directed de novo methylation or RNA-guided demethylation occurs would be governed by the differential cell-typespecific availability of either DNA methyltransferases or DNA glycosylases, respectively.
Methods
Plant material and genotyping. All experiments were performed on drd1-6 mutant (Kanno et al, 2004) or wild-type Arabidopsis thaliana (ecotype Columbia-0). The genotypes of plants used for molecular analyses were determined by PCR-based screens, as described in the supplementary information online.
DNA methylation analysis. DNA was isolated from genotyped plants and analysed for cytosine methylation, using methods described previously (Aufsatz et al, 2002a, 2002b, 2004; Kanno et al, 2004). Southern data results were reproduced for at least two plants of each genotype. For the drd1-6 mutant shown in Fig 1C (left), DNA was isolated from plants of the M3 generation (genotype K/K;H/H;d/d).
Short RNA analysis. NOS promoter and α′ promoter short RNAs were isolated and analysed by northern blotting, as described previously (Aufsatz et al, 2002a, 2002b, 2004; Kanno et al, 2004).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We acknowledge financial support from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (Grant P-15611-B07) and the European Union (contract no. HPRN-CT-2002-00257).
References
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJM (2002a) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double stranded RNA. EMBO J 21: 6832–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke AJM, Matzke M (2002b) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA 99: 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, Matzke AJM, Matzke M (2004) The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol Biol 54: 793–804 [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q, Muegge K (2001) Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev 15: 2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR (2000) Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet 24: 368–371 [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu J-K (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth A, Cao X, Jacobsen SE (2002) Control of CpNpG methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Jones L, Ratcliff F, Baulcombe DC (2001) RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 11: 747–757 [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJM (2004) Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 14: 801–805 [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K (2004) Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431: 211–217 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Kress C, Thomassin H, Grange T (2001) Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS Lett 494: 135–140 [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J (2002) An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Bender J (2004) RNA-directed DNA methylation. J Cell Sci 117: 4881–4888 [DOI] [PubMed] [Google Scholar]
- Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJM (2004) Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta 1677: 129–141 [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SWL, Jacobsen SE, Looney DJ (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292 [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241: 172–182 [DOI] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL (2003) Imprinting of the MEA polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5: 891–901 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


