Abstract
Cucumber mosaic virus (CMV), the type member of the genus Cucumovirus (family Bromoviridae), is transmitted by aphids in a nonpersistent manner. Mutagenesis experiments identified the βH-βI loop of the capsid subunit as a potential key motif responsible for interactions with the insect vector. To further examine the functional characteristics of this motif, we generated monoclonal antibodies that bound to native virions but not to βH-βI mutants. Fab fragments from these antibodies were complexed with wild-type CMV and the virus-Fab structure was determined to 12-Å resolution by using electron cryomicroscopy and image reconstruction techniques. The electron density attributed to the bound antibody has a turret-like appearance and protrudes from each of the 12 fivefold axes of the icosahedral virus. Thus, the antibody binds only to the pentameric clusters (pentons) of A subunits of the T=3 quasisymmetric virus and does not appear to bind to any of the B and C subunits that occur as hexameric clusters (hexons) at the threefold (quasi-sixfold) axes. Modeling and electron density comparisons were used to analyze the paratope-epitope interface and demonstrated that the antibody binds to three βH-βI loops in three adjacent A subunits in each penton. This antibody can discriminate between A and B/C subunits even though the βH-βI loop adopts the same structure in all 180 capsid subunits and is therefore recognizing differences in subunit arrangements. Antibodies with such character have potential use as probes of viral assembly. Our results may provide an additional rationale for designing synthetic vaccines by using symmetrical viral particles.
Cucumber mosaic virus (CMV), the type member of the genus Cucumovirus (family Bromoviridae), infects more than 800 plant species and causes economically important diseases of many crops worldwide (26). CMV is transmitted by aphids in a nonpersistent manner. The virus does not circulate or replicate in the aphid and is quickly acquired from and transmitted to a host during feeding. Virus interacts with the anterior portion of the alimentary tract (food canal to foregut), from which it can be subsequently inoculated by egestion. Unlike some other plant viruses that are transmitted in a nonpersistent manner, CMV does not require helper proteins for transmission. Hence, the aphid recognition motifs must reside on the capsid itself. The ∼24-kDa coat protein also appears to play a role in normal cell-to-cell and systemic movement independent of particle formation (17, 29).
The structures of CMV and cowpea chlorotic mottle virus (CCMV), another member of the family Bromoviridae, were both determined by using cryoelectron microscopy (37, 39) and X-ray crystallography (32, 37). Though the CMV and CCMV capsid proteins share only 19% amino acid sequence identity, they have highly homologous central β-barrel structures. Despite this, a number of significant differences distinguish these T=3, icosahedral viruses. First, CMV is transmitted by aphids, whereas bromoviruses are transmitted by beetles via a fundamentally different mechanism (14). Second, the N termini of the B and C subunits in CCMV form a β-barrel domain similar to that found in other plant viruses but, in CMV, the N termini form a unique hexameric bundle of amphipathic helices that penetrate into the RNA core. Third, CCMV has a cluster of acidic residues at the quasi-threefold interface that may play a role in metal and pH dependent swelling of the capsid (37). In CMV, which does not exhibit swelling behavior, these residues are replaced by ionically complementary acids and bases. Fourth, the diameter of CMV is ∼12 Å larger than that of CCMV, which may be a consequence of longer βF-βG and βF-αEF loops. Finally, whereas genomic RNA in CCMV interacts with the internal residues of the capsid subunits around the quasi-threefold axes (relating A, B, and C subunits), similar interactions were not observed in CMV (32).
The X-ray crystal structure of CMV revealed an exposed βH-βI loop with several unique properties (32). In contrast to the predominant neutral or basic charge character of the capsid surface, the βH-βI loop is highly acidic. Close inspection of the electron density near this loop indicated that a metal ion might be chelated by this cluster of acidic residues. The likelihood of an important role for these residues is additionally supported by the observation that six of the eight amino acids that comprise this loop are highly conserved among strains of CMV and other cucumoviruses (20). Furthermore, mutations in several of the loop residues (D191, D192, L194, and E195) had no significant effect on virion formation or stability but they did reduce or eliminate aphid transmission (20). Hence, to better understand the molecular basis for virus transmission by insects, we targeted this region for further studies.
Here we report results of an electron cryomicroscopy (cryoTEM) and three-dimensional (3D) image reconstruction and pseudo-atomic modeling study of a CMV-Fab complex. The monoclonal antibody used was selected based on its inability to bind to several of the βH-βI loop mutants and was therefore believed to recognize the βH-βI loop. The results of the present study demonstrate that this antibody binds to the pentons (the terms pentons and hexons used here have been borrowed from studies describing similar features on much larger viruses) formed by the A capsid subunits at the fivefold axes but not to the chemically identical B and C subunits at the threefold axes. Nearly all residues in contact with the antibody are charged, a finding consistent with a number of other antibody and/or antigen studies (23, 36).
MATERIALS AND METHODS
CMV production.
CMV was propagated in Nicotiana clevelandii and purified by differential centrifugation, followed by sucrose gradient fractionation as previously described (22, 32).
Monoclonal antibody production.
The preparation and screening of the monoclonal antibodies will be described elsewhere (K. L. Perry et al., unpublished data). Briefly, BALB/c mice were injected with native CMV, and spleen cells from the mice were fused with Sp2 myeloma cells by using standard procedures (12). Antibodies from hybridoma cell lines were screened for their ability to bind intact virions in a triple antibody sandwich enzyme-linked immunosorbent assay. The antibody used for these studies, 3A8-5C10, reacted with native virions and some of the βH-βI mutants but not with D191A, D191K, D192A, and E195K mutants (20).
Fab and virus-Fab complex preparation.
Hybridomas (3A8-5C10) were cultured by using a Cellmax Quad-4 System (Cellco Corp., Germantown, Md.) and a microcapillary cartridge with a 10-kDa molecular mass cutoff. The cells were removed from the suspension by centrifugation at 10,000 × g for 10 min. Antibody was precipitated from the supernatant by the addition of 349 g of ammonium sulfate per liter of antibody solution. The solution was incubated overnight at 4°C, and the precipitate collected by centrifugation at 10,000 × g for 10 min. The precipitate was pooled and dialyzed against 50 mM sodium phosphate buffer (pH 7.0) containing 1 mM sodium azide. Debris was then removed from the dialyzed precipitate by centrifugation, and antibody was purified by using protein G affinity chromatography (Amersham Biosciences, Piscataway, N.J.). The antibody was digested at a concentration of ∼0.3 mg/ml in the presence of 25 mM β-mercaptoethanol and at a papain/antibody ratio of 1:1,000. The reaction was incubated at 37°C for 2 h and quenched by the addition of 75 mM iodoacetamide (final concentration). The digestion solution was then dialyzed against 20 mM Tris buffer (pH 7.6) and concentrated ∼5-fold by using Centricon-10 concentrators (Millipore Corp., Bedford, Mass.). The samples were then passed through a 0.45-μm (pore-size) filter and loaded onto a mono-Q anion-exchange column that had been equilibrated with 10 mM Tris buffer (pH 7.0). At this pH, the Fab fragments elute in the void volume, whereas the Fc fragments and intact antibodies bind to the matrix. The Fab fractions were pooled and concentrated to ∼0.5 mg/ml. To 1 ml of the Fab solution at 0.5 mg/ml, 2 ml of CMV at 0.8 mg/ml was added and sodium chloride was added to yield a final concentration of 100 mM. This mixture was then concentrated ∼10-fold with a Centricon-10. This mixture was incubated overnight at 4°C, and the unbound Fab fragments were removed via size exclusion chromatography (Superose 6; Amersham Biosciences) with 20 mM Tris (pH 7.2) containing 100 mM sodium chloride as buffer. The virus-Fab complex eluted in the void volume and was concentrated back to its original volume with a Centricon-10, yielding a virus concentration of ∼10 mg/ml.
Cryoelectron microscopy and 3D image reconstruction.
Small (∼3.5-μl) aliquots of purified CMV-Fab samples were adhered to holey carbon-coated electron microscope grids and vitrified in liquid ethane as described previously (3). Micrographs were recorded in a Philips CM300 field emission gun transmission electron microscope on Kodak SO-163 film under low-dose conditions (∼24 e/Å2) and at a calibrated magnification of 47,000. Eight micrographs with defocus values ranging between 1.7 and 3.6 μm underfocus were digitized on a Zeiss SCAI scanner by using a 7-μm step size. The digitized data were twofold bin averaged to give a 14-μm step size corresponding to a final pixel size of 2.98 Å at the specimen.
The defocus level of each micrograph was determined by using the X-windows program RobEM, which fits theoretical microscope contrast transfer functions (CTF) to the incoherent sum of the Fourier transforms of all particle images from each micrograph. The orientation and center of each boxed particle was initially determined by means of the model-based polar-Fourier transform method (2), with the previously determined, 23-Å CMV image reconstruction (39) used as the starting model and with each image modified by a filter function (FILTER3; see below) designed to partially compensate for the effects of the microscope CTF. Orientation refinement was monitored with real and reciprocal space correlation coefficients (3).
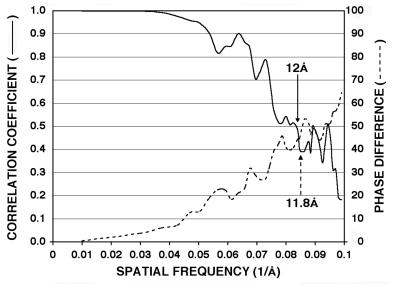
The final 3D reconstruction was computed from 1,667 particle images to a resolution cutoff of 10 Å (Fig. 1). The actual resolution of the density map was estimated by randomly splitting the image data into two roughly equal sets, computing independent 3D reconstructions, and comparing the structure factors calculated independently for each reconstruction (Fig. 1). This analysis demonstrated good agreement to better than 12-Å resolution between the independent data as measured both by phases (<45°) and correlation coefficients (>0.5).
FIG. 1.
Resolution assessment of CMV-Fab 3D reconstruction. The final set of boxed CMV-Fab images was subdivided in half, and two independent reconstructions were computed. Correlation coefficient (solid line) and phase difference (dashed line) comparisons were computed from the structure factors derived from the two reconstructions. Based on conservative estimates (CC, >0.5; phase differences, <45°), these plots demonstrate that the data are reliable to at least 12-Å resolution, and significant Fourier terms are present to ca. 10.5 Å.
In computing the 3D reconstruction with Fourier Bessel procedures (3), the Fourier transform of each image was multiplied by a pair of filters (FILTER1 and FILTER2) designed to compensate in part for the phase and amplitude effects of the CTF. The first filter (FILTER1 = 1.0/[|CTF| + δ1∗(1.0 − |CTF|)]) was imposed on Fourier data up to the first peak of the CTF (see reference 3 for definition of CTF, for which a value of 0.05 was assumed for amplitude contrast), and the second filter (FILTER2 = |CTF|/[CTF2 + δ2∗(1.0 − |CTF|)]) was imposed on all Fourier data beyond the first CTF peak. The variables δ1 and δ2, which are set to small values in a range between 0.01 and 0.2, are used to suppress noise amplification in the data near the nodes of the CTF. For the present study, δ1 and δ2 were set to 0.05 and 0.1, respectively, for all images. The differential treatment of the data before the first CTF peak is similar in principle to that described by Conway and Steven (8). The combined effect of the two filters is to avoid noise amplification near the nodes of the CTF, as well as to restore contrast at the lowest spatial frequencies so that densities for each of the different components such as the solvent and protein subunits are rendered more uniform. Except for a slight gain in computational efficiency, the filter used during the model-based refinement of particle centers and orientations (FILTER3 = |CTF|/[CTF2 + δ3], with δ3 = 0.2 in the present study) acts in a manner analogous to that provided when two filters are used to correct the image data.
For display purposes and to render the data comparable to the X-ray models, the final reconstruction map was sharpened by multiplying the structure factors with an inverse temperature factor [= exp(B/4dhkl2)], in which B = 500 Å2 and dhkl is the spacing in angstroms for each structure factor (13).
Pseudo-atomic modeling.
To model the CMV-Fab complex the program “O” (16) was used to place a single antibody Fab into the turret-like feature in the electron density map. Since steric hindrance precluded the binding of a second Fab to this site, an occupancy of 0.2 was assigned to all of the atoms in the Fab, and an additional 59 copies of the structure were generated by imposing icosahedral symmetry. The programs SFALL and FFT (1) were then used to compute an electron density map from the resulting pseudo-atomic CMV-Fab model, and the map was viewed with the program “O” (16). This process was repeated several times to adjust the model to give optimum agreement between the calculated pseudo-atomic model and 3D image reconstruction maps.
RESULTS
Image reconstruction of the CMV-Fab complex.
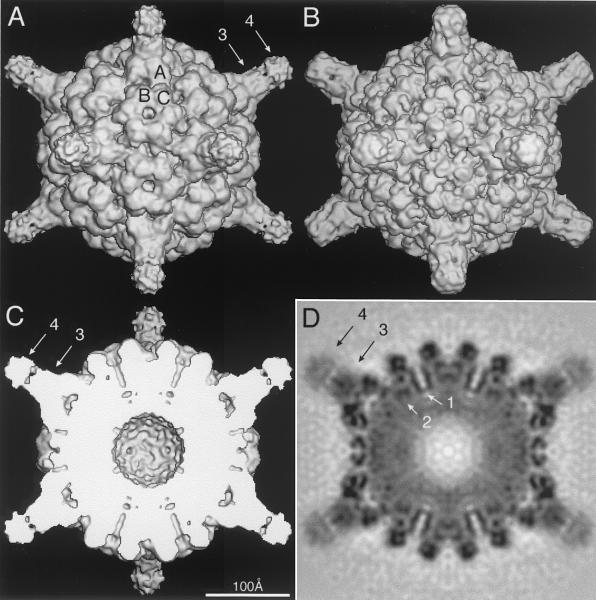
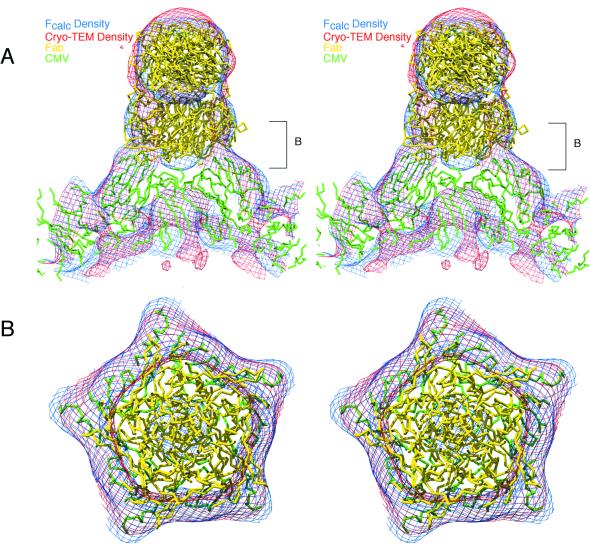
The 3D image reconstruction of the vitrified CMV-Fab complex clearly shows the site of attachment of the Fab to the surface of CMV (Fig. 2). Although unexpected because the T=3 capsid consists of 180 chemically identical subunits, the antibody only binds to the A subunits that form the 12 pentons about the icosahedral fivefold axes. Even when the image reconstruction is rendered at much lower contour levels, no density appears associated at or near the B or C subunits that form the hexons at the 20 quasi-sixfold axes. In contrast with the present study, previous image reconstructions of other virus-Fab complexes showed electron densities for the bound Fab molecules as characteristic, bilobed extensions that could be clearly correlated with the known four domain structure for a Fab molecule (34-36). However, the electron density of the Fab molecule bound to CMV exhibits a bilobed, turret morphology, which is a consequence of the antibody binding quite close to a symmetry axis (in this instance the fivefold). In previous virus-Fab studies, the Fabs bound away from symmetry axes and in orientations such that no portion of the Fab crossed an axis of symmetry. The CMV Fab exhibits a clear separation between the variable and constant domains (Fig. 2, arrows 3 and 4, respectively). In addition, the constant domains have lower densities than the variable domains (Fig. 2D), a finding which likely reflects an inherent increased mobility of the constant domains owing to flexibility conferred at the elbow region of the Fab.
FIG. 2.
Shaded-surface (A to C) and equatorial section (D) representations of the CMV-Fab complex, all viewed along a twofold axis of symmetry. The arrows highlight aspects of the cryoTEM 3D reconstruction that are depicted in greater detail in subsequent figures. (A) 3D reconstruction of the CMV-Fab complex with the approximate locations of the three quasiequivalent (chemically identical) A, B, and C subunits labeled. The large, turret-like projections at the icosahedral vertices represent the bound Fab molecules, and the variable and constant domains of one Fab are identified by arrows 3 and 4, respectively. (B) Pseudo-atomic model of CMV-Fab complex computed to 10-Å resolution; β factors were applied to match the 12-Å cryo-TEM data, using the X-ray crystal structures of CMV (32) and Fab17 (19). (C) Same as panel A with the front half of the structure removed to visualize internal features of the reconstruction. (D) Density projection of the central section of the CMV-Fab reconstruction, with the blackest regions representing the highest density regions in the map. Two key internal features are the radially directed density representing the six-helical bundle at the quasi-sixfold axes (arrow 1) and density that lies adjacent to and projects away from the helical bundle (arrow 2). The lower density for the Fabs (especially the constant domains [arrow 4]) is consistent with the hypothesis that only a single Fab binds to each of the 12 pentons at the vertices of the icosahedral virus.
RNA interactions with the CMV capsid.
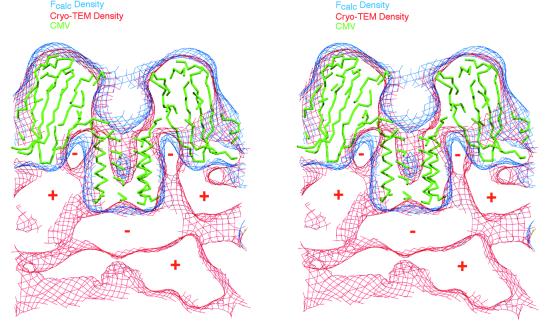
The image reconstruction of the virus alone is entirely consistent with the X-ray structure and also reveals some unique features. Hexameric bundles that lie along the icosahedral threefold axes at the inner surface of the capsid subunits (32) are prominent features in the image reconstruction (arrow 1, Fig. 2D). The architecture of internal components not observed in the X-ray crystal structure can be seen in the image reconstruction. For example, a strong positive density streams off from the sides of the hexameric helical bundle (Fig. 2, arrow 2, and Fig. 3). The highly concentrated positive charge attributed to the basic residues in this region was previously suggested to facilitate interactions with genomic RNA (32). Because the first 27 residues of this N-terminal region in CMV are disordered in the X-ray structure, this density in the reconstruction is likely comprised of RNA and protein. However, strong negative density occurs at the inner surface in the regions around the quasi-threefold axes where RNA was observed in close contact with the capsid in CCMV (37). Therefore, RNA-capsid interactions appear to be quite distinct in CMV and CCMV.
FIG. 3.
Stereo cross-section view of CMV density maps and X-ray model in region near a quasi-sixfold axis. The 3D reconstruction and X-ray map (rendered at a 15-Å resolution) are depicted in red and blue contours, respectively. The C-α backbone of the CMV model is represented by thin green tubes. Selected regions of positive (high) and negative (low or zero) cryoTEM electron density are denoted by “+” and “−” signs for clarity. Two large regions of positive density stream away from the sides of the helical bundle (see also arrow 2, Fig. 2). Negative density, centered directly beneath the helical bundle, occurs in a region that contains positive density in CCMV and which is attributed to organized, genomic RNA (37). Figures 3 to 5 and 7 were made by using the program MolView (http://www.danforthcenter.org/smith/molview.htm [31]).
Quasi-equivalent differences and construction of the pseudo-atomic model.
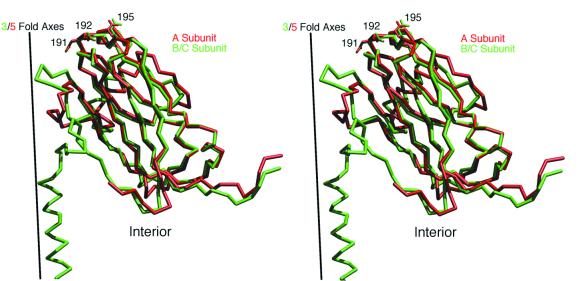
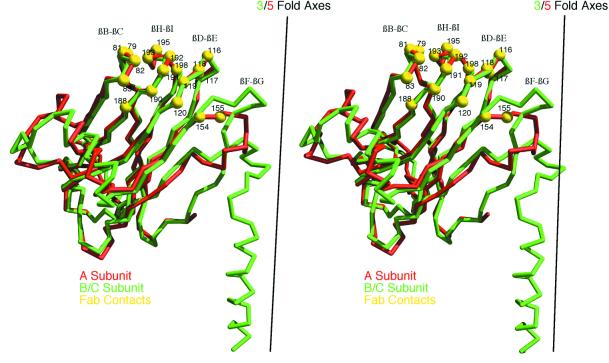
The selectivity of the CMV antibody for the A versus B and C subunits could reflect structural differences in the subunits arising from quasi-equivalence in the T=3 icosahedron. This explanation seems unlikely, however, since the majority of the exposed loops adopt essentially identical structures (Fig. 4). In particular, this antibody is sensitive to mutations at 191, 192, and 195 on the βH-βI loop, yet even the side chain conformations of these residues are nearly identical among the A, B, and C subunits. The primary structural difference among the three quasiequivalent subunits lies in the βF-βG loop, but this loop does not appear to contribute significantly to the antibody contact.
FIG. 4.
Stereo view comparison of the tertiary structures of the A (red) and B/C (green) subunits in CMV. The subunits are aligned with the icosahedral three- and fivefold axes superimposed. The RNA core of the virus (not shown) starts at the bottom of the panel. The side chains for residues 191, 192, and 195 are depicted to illustrate the conservation of main and side chain conformations, especially at the viral surface of the quasiequivalent subunits.
The footprint of the antibody on the penton was examined by constructing a pseudo-atomic model of the CMV-Fab complex from the X-ray crystal structures of CMV (32) and Fab17 (19). It was immediately clear during these modeling studies that this CMV-Fab complex could not be interpreted by placing a single Fab structure in a unique orientation onto the virion surface as has been done in several other studies (6, 34-36). The unusual turret shape, therefore, was likely a consequence of the spatial averaging that occurs when the icosahedrally symmetric reconstruction is computed and only a single Fab binds to each penton. To simulate this mode of binding, a pseudo-atomic model was constructed by placing one Fab in the density at the penton, assigning a relative occupancy of 0.2 to it, and then applying icosahedral symmetry to generate an additional 59 Fab molecules. The electron density map generated from this model gave excellent correlation with the 3D reconstruction (Fig. 3B and 5), and it was evident that the Fab variable domains assumed a pentameric profile as a consequence of the averaging process (Fig. 5B). Further refinement of the Fab orientation was performed until an optimum fit was obtained between the calculated and cryoTEM maps. Though such modeling experiments do not offer positive proof for the suggested mode of binding, they do strongly support our contention that one Fab binds per penton. In addition, the model indicates that the spherical nature of the electron density for the constant domains likely results from the Fabs binding in an orientation that allows them to intersect the fivefold axes.
FIG. 5.
Stereo view showing fit of the CMV (green) and Fab (yellow) X-ray models into the cryoTEM electron density map (red contours) and the electron density calculated from the pseudo-atomic model (blue contours). A single Fab was fit into the reconstructed density envelope, and then icosahedral fivefold symmetry was applied. Since the density envelope could only accommodate one Fab at each penton, an occupancy of 0.2 was applied to each of the fivefold-related Fab molecules. Therefore, although all five Fabs are shown in this figure to demonstrate how the cryoTEM electron density is filled by the Fab structures, in reality only one Fab molecule is bound at a time. (A) Close-up view of the CMV-Fab model and density maps, with the viral RNA core at the bottom of the figure and a fivefold axis aligned in a vertical direction. (B) Same as panel A but viewed down a fivefold axis and including just a portion of the “turret” corresponding to the region demarked by brackets labeled “B” in panel A. Agreement between the calculated and observed maps is excellent. Also, the averaged variable domains of the Fabs form a pentameric structure docked to the surface of the A subunit penton.
Antibody footprinting.
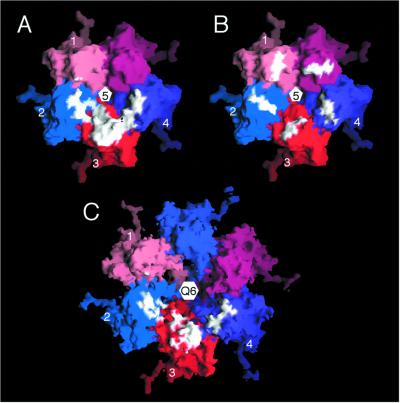
The results of the pseudo atomic modeling enabled us to investigate specific contacts made by the Fab on the virus. The program MS (7) was used to identify the contact between a single Fab and a penton as depicted in the pseudo-atomic model. Though any data derived in this way from low-resolution cryoTEM results must be viewed with caution, previous results with human rhinovirus 14 (HRV14) have demonstrated that the modeling can be sufficiently accurate to approximate contact surfaces at near-atomic resolution (33, 36). The Fab contacts three of the five A subunits of a penton and buries a total of 673 Å2 of the CMV surface upon binding. These contacts are summarized in Tables 1 to 3 and are illustrated in Fig. 6 and 7. Furthermore, it appears that one Fab is capable of simultaneously binding to three “antigenic” βH-βI loops via both its heavy and light-chain hypervariable regions (Table 1 and Fig. 6B). Consistent with how this particular monoclonal antibody was selected, contacts with the βH-βI loops are predominant, although there are significant contacts also with the immediately adjacent βB-βC and βD-βE loops (Table 2). Only minimal contact (∼3%) is made with the βF-βG loop and, in view of the above-mentioned caution with which pseudo-atomic model data should be interpreted, may not be reliable. Therefore, we conclude that the antibody appears to bind to sets of loops, with the individual loop structures being nearly identical in the A subunits of pentons and the B and C subunits in hexons (Fig. 7). The nature of the contacts is particularly interesting in that nearly all of the capsid residues (∼84% of the total contact surface) that contact the antibody are charged.
TABLE 1.
Antibody contacts
| Subunit | Loop | Residue(s) | Area (Å2) | Area (%) |
|---|---|---|---|---|
| 1 | βB-βC | 82 | 2.9 | 0.43 |
| 2 | βB-βC | 82 | 38.0 | 5.60 |
| βD-βE | 118-120 | 30.0 | 4.46 | |
| βF-βG | 154 | 4.5 | 0.67 | |
| βH-βI | 190-192 | 71.5 | 10.70 | |
| 3 | βB-βC | 79, 81-83 | 130.8 | 19.41 |
| βD-βE | 117-120 | 90.9 | 13.43 | |
| βF-βG | 154-155 | 20.7 | 2.60 | |
| βH-βI | 188, 190-193, 195 | 163.0 | 24.20 | |
| 4 | βB-βC | 82 | 23.0 | 3.40 |
| βD-βE | 116, 118 | 66.0 | 9.80 | |
| βH-βI | 191-192, 198 | 33.3 | 4.89 |
TABLE 3.
Summary of types of chemical contacts
| Type | Main chain (%) | Side chain (%) |
|---|---|---|
| Acid (D, E) | 12.0 | 37.0 |
| Base (H, K, R) | 4.5 | 30.0 |
| Polar (N, Q, S, T) | 4.3 | 3.2 |
| Small (A, G) | 2.4 | 1.5 |
| Aromatic N-P (F) | 0.6 | 1.2 |
| Aromatic P (W, Y) | 0.4 | 2.7 |
FIG. 6.
Molecular surface representations of CMV penton (A and B) and hexon (C) with Fab contact area and location of mutations depicted. (A) Contact area (white) of a single Fab on a penton with individual subunits distinguished by different colors and the four subunits noted in Table 1 are labeled 1 to 4. The view is along the fivefold axis toward the virus center. A single Fab contacts significant regions of three different A subunits. (B) Same as panel A but with white coloring to denote the locations and surface formed by residues, 191, 192, and 195. Based on panel A, the Fab is seen to contact some or all of these residues in penton subunits 2, 3, and 4. (C) Mapping of contact residues seen in panel A onto the hexon B and C subunits demonstrates that the hypothetical hexon contact is discontinuous compared to the contact observed in the penton. This figure was made by using the program GRASP (24).
FIG. 7.
Stereo view of aligned A (red) and B/C (green) subunit Cα backbones with contact residues (yellow) within a penton mapped onto a single A subunit. The three major contact loops (βB-βC, βH-βI, and βD-βE) have nearly identical structures in the quasiequivalent subunits (see also Fig. 4). Some contact with the βF-βG loop, which bends away from the fivefold axis in the A subunit, may occur, but this loop only contributes ∼3% of the total virus-Fab contact area and therefore may not represent an actual contact in the real virus-antibody complex.
TABLE 2.
Summary of loop contacts
| Loop | Area (Å2) | Area (%) |
|---|---|---|
| βB-βC | 195 | 29 |
| βD-βE | 187 | 28 |
| βF-βG | 22 | 3 |
| βH-βI | 268 | 40 |
How can the antibody distinguish pentons from hexons when the loops in the A, B, and C subunits are the same? To help answer this apparent paradox, we mapped the atoms contacted in the pentons onto the hexons (Fig. 6C). This simple experiment demonstrated that the putative hexon footprint was discontinuous and dispersed relative to the penton footprint. The more extended hexon footprint is likely a consequence of two geometric constraints dictated by the differences between pentameric versus hexameric arrangements of the capsid subunits. The footprint residues in the pentons roughly map onto a circle of smaller radius than that for the imaginary hexon footprint and so the penton footprint is both more curved and less extensive than that for the hexon. Therefore, the antibody appears to acquire penton specificity simply by virtue of the quaternary differences in subunit arrangements in pentons and hexons (Fig. 6) but not because of quasiequivalent differences that distinguish the intersubunit contacts within pentons and hexons (Fig. 7). These results have potential impact on the design of virus-based synthetic vaccines since the location of the foreign epitope on the virion surface could greatly impact the repertoire of the elicited antibodies.
DISCUSSION
Antibody-virus interactions.
Our cryoTEM and modeling results clearly demonstrate that antibody 3A8-5C10 binds to the βH-βI loop of CMV, a region that has been implicated in aphid transmission (20, 32). Unlike in other virus-antibody complexes studied to date, this antibody binds immediately adjacent to an axis of icosahedral symmetry and only one Fab binds per penton. The nature of the CMV-Fab contact is reminiscent of the lysozyme-HYHEL-10 complex in which the antibody and antigen interact in a blunt-faced manner (25). This contrasts with the mode of binding adopted by other antibodies such as Fab17, in which a protruding antigenic loop on HRV14 binds into the cleft between the heavy- and light-chain variable domains (33).
The area of CMV in contact with the antibody is almost entirely comprised of charged amino acids. This is somewhat surprising since it has been suggested that the energy to desolvate charged residues in the interface is greater than the energy gained by the electrostatic interaction of oppositely charged species (15). It was further suggested that the thermodynamic driving force for most processes in aqueous solutions results from nonpolar interactions and that the ion pairs and hydrogen bonds are rearranged to minimize the free-energy cost of desolvation. However, in HRV14-antibody complexes, columbic interactions at the paratope-epitope interface were found to be crucial for antibody binding (6, 33, 36). Mutations that negate or reverse the charge of residues in and around the NIm-IA epitope on HRV14 had a profound effect on antibody binding (6). More detailed analysis of a number of immune complexes has shown that there is a higher degree of complementarity in the electrostatic fields about the antigen and antibody contacts regions than individual hydrogen bonds and salt bridges (23). CMV-Fab interactions may likewise be dominated by electrostatic fields. In addition, antibody binding may not require the desolvation of the entire interface upon binding.
Antibodies are known to be able to bind simultaneously to multiple viral subunits (see, for example, reference 30). However, the CMV-Fab results appear to represent the first example in which an antibody bridges the same regions of two or more identical and adjacent capsid subunits. The binding of Fabs to cowpea mosaic virus (27) is similar in one respect to the CMV-Fab binding. In cowpea mosaic virus, the Fab binds to different portions of two adjacent large subunits. However, the contact regions in these two subunits were not identical, as is the case in this CMV-Fab complex. This antibody is centered about the βH-βI loop that is thought to interact with the receptor molecule in the aphid. This loop is immediately adjacent to and facing toward axes of icosahedral symmetry. Therefore, it may be that the aphid receptor, like the antibody 3A8-5C10, also exhibits quasiequivalent specificity and may interact only with pentons or only with hexons.
Recognition of quasiequivalence differences.
Although the CMV-Fab result is atypical of most virus-antibody interactions studied to date, some instances have been reported in which proteins that bind to capsid proteins are capable of distinguishing differences in quasiequivalently related subunits. However, in these studies a number of mechanisms were proposed to explain how these quasiequivalent subunits could be discerned by the ligating proteins. Knowledge of the atomic structures of components of the CMV-Fab complex allows some of these possible mechanisms to be eliminated.
In herpes simplex virus type 1 (HSV-1), one copy of the 12-kDa VP26 capsid protein binds to each VP5 subunit in the 150 hexons of the nucleocapsid, whereas VP26 does not bind to any of the VP5 subunits in the 12 pentons (40). VP26 appears to form a continuous ring on the outer tip of the hexons (41), whereas in solution it exists in equilibrium as monomers and dimers (40). Based on such observations, it was concluded that VP26 is not predisposed to form hexamers that could then bind specifically to hexons, but instead HSV-1 pentons are unable to bind VP26 (40). Binding specificity in this example could arise from structural differences in VP5 that occur when it assembles into pentamers or hexamers. Alternatively, specificity could merely reflect differences in the spatial arrangement of structurally identical subunits.
Recent studies of papillomavirus-antibody interactions (4) indicate a possible close correlation with the present CMV-Fab results. Two different antibodies to L1, the major coat protein of papillomaviruses, were examined by using cryoTEM and image reconstruction methods and antibody 5B6 was shown to bind to the side of each of the five L1 molecules in the hexavalent capsomers but to none of the five L1 molecules in the pentavalent capsomers. Binding specificity in this example was attributed to several factors. Close proximity of the tips of the pentavalent capsomers to the neighboring hexavalent capsomers might sterically hinder antibody binding to the pentavalent but not hexavalent capsomeres. It was also suggested that the different coordination of the pentavalent and hexavalent capsomeres could yield conformational differences that are detected by the antibody. Finally, the presence of the L2 capsid protein solely within the pentavalent capsomers might alter the L1 structure in those capsomers in a way that would preclude antibody binding. It is noteworthy that the five L1 sites on the hexavalent capsomers, which are in distinct rather than quasiequivalent environments, do not exhibit identical occupancies. This likely reflects differences in steric restrictions imposed by the tips of the neighboring pentavalent capsomers.
Our analysis of the CMV-Fab complex permits clarification of some ambiguities in previous virus-antibody studies. The atomic structure of CMV clearly demonstrates that most of the antibody contact region is conserved in the three quasiequivalent yet chemically identical subunits. Contrary to the papillomavirus example, the antigenic site in CMV is equally exposed in all subunits. Also, unlike the HSV-1 example, in which VP26 differentiates between VP5 in hexons and pentons, in CMV there is but one capsid protein. Hence, our hypothesis that each antibody binds to three or more subunits in each CMV penton seems well supported by the evidence. While all 180 CMV capsid subunits have the same antigen binding loops, the antibody only recognizes a specific pattern of loops that only occurs in pentons.
RNA influence on capsid assembly.
The CCMV crystal structure analysis showed that the highest density of RNA occurs at the B-C coat protein interface and extends toward the quasi-threefold junction (11, 37). In the absence of RNA, the CCMV coat protein can form empty capsids that appear to have the same structure as authentic virions, but these capsids only form at low pH and in the presence of high-ionic-strength buffers. Presumably, RNA bound at the B-C interfaces helps direct capsid formation under physiological conditions by inducing curvature in the capsid and neutralizing basic residues (37). CMV appears to assemble via a very different pathway and the mode of interaction between the RNA genome and the capsid is distinct from that observed in CCMV. Such differences might arise from the extensive interactions that coalesce the N-terminal helices at the quasi-sixfold axes. It is also notable that successful reassembly of empty CMV virions has yet to be reported. Thus, the amphipathic helices may dominate early assembly events and thereby abrogate empty capsid formation. Since CCMV is highly homologous to CMV with the major exception being that the CCMV has a β-annulus structure at the quasi-sixfold axis rather than the hexameric helical bundle observed in CMV, these two viruses make an excellent test case for viral assembly modeling (see, for example, reference 42).
Conclusions.
The results reported here have at least two significant implications. First, the development of specific antibodies that can distinguish quasi-equivalent sites is now possible, and such antibodies may be useful for monitoring both in vivo and in vitro assembly of icosahedral particles. For example, the assembly of CMV cannot be examined in vitro. However, with antibodies that could discern pentons and hexons, it would be possible to monitor CMV assembly intermediates in vivo. Such studies would also determine whether CCMV follows the same or similar assembly pathways in vivo and in vitro or whether cellular factors are required.
Second, our results demonstrate that the location of an antigenic loop on a viral surface is an important criteria in determining how best to engineer hybrid viruses for use as vaccines. Using the CMV structure as an example, the βH-βI loop might represent a logical position into which a foreign antigenic peptide could be introduced. However, because this loop is directed toward the fivefold icosahedral symmetry axis and the typical footprint of a bound antibody is large relative to this area of the penton (i.e., ca. 600 to 1,000 Å2), is seems that many antigens inserted in this loop would likely constrain elicited antibodies to simultaneously contact several subunits. This might therefore result in the selection of antibodies that recognize the targeted epitope in a context that does not properly mimic its native form. Hence, a more fruitful approach might be to place the antigenic determinant into a region that will help assure it is presented in a more exposed, native form and also which would be less likely to yield antibodies with paratopes that bind more than one epitope at a time. In this sense, for example, the NIm-IA site on HRV14 is a good candidate for designing hybrid virions (6, 36) since it is on a ridge that points away from the fivefold axis, whereas the NIm-IB is a poor place for an insertion since it is closer to and points toward the fivefold axis. Indeed, the sites homologous to the NIm-IA site, or the NIm-IA site itself, have been used for successful antigen insertions in the cases of poliovirus (5, 9), cowpea mosaic virus (18, 21, 28, 38), and HRV14 (10).
Acknowledgments
We thank R. Ashmore for dedicated programming efforts and C. Xiao for use of his EMRESOL program to compare independent image reconstructions.
This work was supported by NIH grants to T.J.S. (GM-10704) and T.S.B. (AI-45976 and GM-33050) and by USDA-NRI grants to K.L.P. (1999-02511 and 2002-00647). We also thank the Keck Foundation and Purdue University for instrumentation and reinvestment grant support, respectively, provided to the Purdue Structural Biology Group.
REFERENCES
- 1.Bailey, S. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 3.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booy, F. P., R. B. S. Roden, H. L. Greenstone, J. T. Schiller, and B. L. Trus. 1998. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 Å resolution. J. Mol. Biol. 281:95-106. [DOI] [PubMed] [Google Scholar]
- 5.Burke, K. L., G. Dunn, M. Ferguson, P. Minor, and J. W. Almond. 1988. Antigen chimeras of poliovirus as potential vaccines. Nature 332:81-82. [DOI] [PubMed] [Google Scholar]
- 6.Che, Z., N. H. Olson, D. Leippe, W.-M. Lee, A. Mosser, R. R. Rueckert, T. S. Baker, and T. J. Smith. 1998. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryo-electron microscopy and X-ray crystallography of virus-Fab complexes. J. Virol. 72:4610-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connolly, M. L. 1981. Protein surfaces and interiors. Ph.D. thesis. University of California, Berkeley.
- 8.Conway, J. F., and A. C. Steven. 1999. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128:106-118. [DOI] [PubMed] [Google Scholar]
- 9.Crabbe, M., D. Evans, and J. Almond. 1990. Modelling of poliovirus HIV-1 antigen chimeras. FEBS Lett. 271:194-198. [DOI] [PubMed] [Google Scholar]
- 10.Ding, J., A. D. Smith, S. C. Geisler, X. Ma, G. F. Arnold, and E. Arnold. 2002. Crystal structure of a human rhinovirus that displays part of the HIV-1 V3 loop and induces neutralizing antbodies. Structure 10:999-1011. [DOI] [PubMed] [Google Scholar]
- 11.Fox, J. M., G. Wang, J. A. Speir, N. H. Olson, J. E. Johnson, T. S. Baker, and M. J. Young. 1998. Comparison of the native CCMV virion with in vitro assembled CCMV virions by cryoelectron microscopy and image reconstruction. Virology 244:212-218. [DOI] [PubMed] [Google Scholar]
- 12.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Havelka, W. A., R. Henderson, and D. Oesterhelt. 1995. Three-dimensional structure of halorhodopsin at 7 Å resolution. J. Mol. Biol. 247:726-738. [DOI] [PubMed] [Google Scholar]
- 14.Hobbs, H. A., and J. P. Fulton. 1979. Beetle transmission of cowpea chlorotic mottle virus. Phytopathology 69:255-256. [Google Scholar]
- 15.Honig, B., and A. Nicholls. 1995. Classical electrostatics in biology and chemistry. Science 268:1144-1149. [DOI] [PubMed] [Google Scholar]
- 16.Jones, T. A., J.-Y. Zou, and S. W. Cowan. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, I. B., L. Zhang, and P. Palukaitis. 1998. Characterization of cucumber mosaic virus. V. Cell-to-cell movement requires capsid protein but not virions. Virology 246:221-231. [DOI] [PubMed] [Google Scholar]
- 18.Lin, T., C. Porta, G. Lomonossoff, and J. Johnson. 1996. Structure-based design of peptide presentation on a viral surface: the crystal structure of a plant/animal virus chimera at 2.8 Å resolution. Fold Des. 1:179-187. [DOI] [PubMed] [Google Scholar]
- 19.Liu, H., T. J. Smith, W. M. Lee, A. Mosser, R. R. Rueckert, N. H. Olson, R. H. Cheng, and T. S. Baker. 1994. Structure determination of an fab fragment that neutralizes human rhinovirus 14 and analysis of the Fab-virus complex. J. Mol. Biol. 240:127-137. [DOI] [PubMed] [Google Scholar]
- 20.Liu, S., X. He, G. Park, C. Josefsson, and K. L. Perry. 2002. A conserved capsid protein surface domain of cucumber mosaic virus is essential for aphid vector transmission. J. Virol. 76:9756-9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomonossoff, G. P., and J. E. Johnson. 1995. Viral expression systems for peptides. Semin. Virol. 6:257-267. [Google Scholar]
- 22.Lot, H., J. Marrou, J. B. Quiot, and C. Esvan. 1972. Contribution a l'etude de virus de la mosaique du concombre (CMV). II. Methode de purification rapide du virus. Ann. Phytopathol. 4:25-38. [Google Scholar]
- 23.McCoy, A. J., V. Chandana Epa, and P. M. Colman. 1997. Electrostatic complementarity at protein/protein interfaces. J. Mol. Biol. 268:570-584. [DOI] [PubMed] [Google Scholar]
- 24.Nicholls, A. 1993. GRASP: graphical representation and analysis of surface properties. Columbia University, New York, N.Y.
- 25.Padlan, E. A., W. W. Silverton, S. Sheriff, G. H. Cohen, S. J. Smith-Gill, and D. R. Davies. 1989. Stucture of an antibody-antigen complex: crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc. Natl. Acad. Sci. USA 86:5938-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41:281-348. [DOI] [PubMed] [Google Scholar]
- 27.Porta, C., R. H. Cheng, Z. Chen, T. S. Baker, and J. E. Johnson. 1994. Direct imaging of interactions between an icosahedral virus and conjugate Fab fragments by cryoelectron microscopy and X-ray crystallography. Virology 204:777-788. [DOI] [PubMed] [Google Scholar]
- 28.Porta, C., V. Spall, J. Loveland, J. Johnson, P. Barker, and G. Lomonossoff. 1994. Development of cowpea mosaic virus as a high-yielding system for the presentation of foreign peptides. Virology 202:949-955. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz, I., and A. L. N. Rao. 1998. Deletions in the conserved amino-terminal basic arm of cucumber mosaic virus coat protein disrupt virion assembly but do not abolish infectivity and cell-to-cell movement. Virology 248:323-331. [DOI] [PubMed] [Google Scholar]
- 30.Sherry, B., A. G. Mosser, R. J. Colonno, and R. R. Rueckert. 1986. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J. Virol. 57:246-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, T. J. 1995. MolView: a program to analyze and display atomic structures on the Macintosh personal computer. J. Mol. Graphics 13:122-125. [DOI] [PubMed] [Google Scholar]
- 32.Smith, T. J., E. Chase, T. J. Schmidt, and K. Perry. 2000. The structure of cucumber mosaic virus and comparison to cowpea chlorotic mottle virus. J. Virol. 74:7578-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, T. J., E. S. Chase, T. J. Schmidt, N. H. Olson, and T. S. Baker. 1996. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature 383:350-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, T. J., R. H. Cheng, N. H. Olson, P. Peterson, E. Chase, R. J. Kuhn, and T. S. Baker. 1995. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 92:10648-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, T. J., N. H. Olson, R. H. Cheng, E. S. Chase, and T. S. Baker. 1993. Structure of a human rhinovirus-bivalently bound antibody complex: implications for virus neutralization and antibody flexibility. Proc. Natl. Acad. Sci. USA 90:7015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, T. J., N. H. Olson, R. H. Cheng, H. Liu, E. Chase, W. M. Lee, D. M. Leippe, A. G. Mosser, R. R. Ruekert, and T. S. Baker. 1993. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J. Virol. 67:1148-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speir, J. A., S. Munshi, G. Wang, T. S. Baker, and J. E. Johnson. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryoelectron microscopy. Structure 3:63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Usha, R., J. Rohll, V. Spall, M. Shanks, A. Maule, J. Johnson, and G. Lomonossoff. 1993. Expression of an animal virus antigenic site on the surface of a plant virus particle. Virology 197:366-374. [DOI] [PubMed] [Google Scholar]
- 39.Wikoff, W. R., C. J. Tsai, G. Wang, T. S. Baker, and J. E. Johnson. 1997. The structure of cucumber mosaic virus: cryoelectron microscopy, X-ray crystallography, and sequence analysis. Virology 232:91-97. [DOI] [PubMed] [Google Scholar]
- 40.Wingfield, P. T., S. J. Stahl, D. R. Thomsen, F. L. Homa, F. P. Booy, B. L. Trus, and A. C. Steven. 1997. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J. Virol. 71:8955-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]
- 42.Zlotnick, A., J. M. Johnson, P. W. Wingfield, S. J. Stahl, and D. Endres. 1999. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry 38:14644-16652. [DOI] [PubMed] [Google Scholar]