Abstract
The ubiquitin ligase Cbl mediates ubiquitination of activated receptor tyrosine kinases (RTKs) and interacts with endocytic scaffold complexes, including CIN85/endophilins, to facilitate RTK endocytosis and degradation. Several mechanisms regulate the functions of Cbl to ensure the fine-tuning of RTK signalling and cellular homeostasis. One regulatory mechanism involves the binding of Cbl to Sprouty2, which sequesters Cbl away from activated epidermal growth factor receptors (EGFRs). Here, we show that Sprouty2 associates with CIN85 and acts at the interface between Cbl and CIN85 to inhibit EGFR downregulation. The CIN85 SH3 domains A and C bind specifically to proline–arginine motifs present in Sprouty2. Intact association between Sprouty2, Cbl and CIN85 is required for inhibition of EGFR endocytosis as well as EGF-induced differentiation of PC12 cells. Moreover, Sprouty4, which lacks CIN85-binding sites, does not inhibit EGFR downregulation, providing a molecular explanation for functional differences between Sprouty isoforms. Sprouty2 therefore acts as an inducible inhibitor of EGFR downregulation by targeting both the Cbl and CIN85 pathways.
Keywords: Cbl, CIN85, EGFR, endocytosis, Sprouty2
Introduction
Activation of receptor tyrosine kinases (RTKs) by growth factors induces intracellular signalling pathways that determine whether the cell proliferates, differentiates or undergoes apoptosis. Activated receptors also initiate negative signalling events that modulate the positive signals to precisely regulate the biological outcome (Dikic & Giordano, 2003). Many of the key molecules involved in negative receptor signalling control receptor endocytosis. The Cbl family of ubiquitin ligases has an important role in this process, mediating multiple monoubiquitination of activated RTKs, which targets them for lysosomal degradation (Haglund et al, 2003a, 2003b; Mosesson et al, 2003). Moreover, Cbl recruits endocytic complexes, including the Cbl-interacting protein of 85 kDa (CIN85)/endophilins, to activated RTKs to facilitate endocytosis (Petrelli et al, 2002; Soubeyran et al, 2002). Several recent findings indicate that RTK signalling can be finely adjusted by regulating the functions of Cbl. At least three different mechanisms of Cbl regulation have been described: (i) sequestration of Cbl away from activated epidermal growth factor receptors (EGFRs) by means of phosphorylated Sprouty2 or activated Cdc42 in complex with βPix (Christofori, 2003; Wu et al, 2003; Kim & Barsagi, 2004); (ii) proteasome-mediated degradation of Cbl (Bao et al, 2003); and (iii) interference with ubiquitin-dependent sorting of activated receptors in the endosome by means of Sts-1 and Sts-2 (Kowanetz et al, 2004).
One of the best-studied inhibitors of Cbl is Sprouty2, an inducible regulator that becomes phosphorylated on a conserved tyrosine residue (Y55) after EGFR activation. This phosphorylated tyrosine functions as a docking site for the SH2 domain of Cbl, and competes with pY1045 in the activated EGFR (Egan et al, 2002; Fong et al, 2003; Hall et al, 2003; Rubin et al, 2003). Thus, Sprouty2 removes Cbl from activated EGFRs and blocks Cbl-mediated EGFR ubiquitination and degradation, which leads to sustained receptor signalling (Wong et al, 2002; Fong et al, 2003; Hall et al, 2003; Rubin et al, 2003). However, other Sprouty isoforms, such as Sprouty3 and Sprouty4, do not interfere with EGFR downregulation, in spite of containing a Cbl-binding motif (Wong et al, 2002). Differences between the individual Sprouty family members are not yet fully understood, but findings presented in this manuscript could help in explaining this apparent anomaly. We show how Sprouty2 associates with CIN85 and acts at the interface between Cbl and CIN85 to inhibit EGFR ubiquitination and downregulation.
Results
Sprouty2 and CIN85 form a complex in mammalian cells
Recently, we have shown that the three SH3 domains of CIN85 bind to atypical proline–arginine motifs (PxxxPR) present in the carboxyl termini of Cbl and Cbl-b. In this way, CIN85 clusters Cbl molecules, which is crucial for efficient EGFR endocytosis and degradation (Kowanetz et al, 2003). As Sprouty2 associates with Cbl, we were interested in analysing whether Sprouty2 affects the function of CIN85 during EGFR endocytosis. Interestingly, endogenous Sprouty2 associated with CIN85 in C2C12 cells (Fig 1A). When coexpressed in cells, Sprouty2 wild type as well as its separate amino- and carboxy-terminal parts readily co-precipitated with full-length CIN85 (Fig 1B; supplementary Fig 1 online). Moreover, the three SH3 domains of CIN85 bound to Sprouty2 in glutathione-S-transferase (GST) pull-down assays, whereas its proline-rich region and coiled-coil domain did not (data not shown). Furthermore, the isolated SH3 domains A and C of CIN85 associated with Sprouty2, indicating that both domains specifically recognized their consensus sequences in Sprouty2, whereas SH3 domain B did not (Fig 1C).
Figure 1.
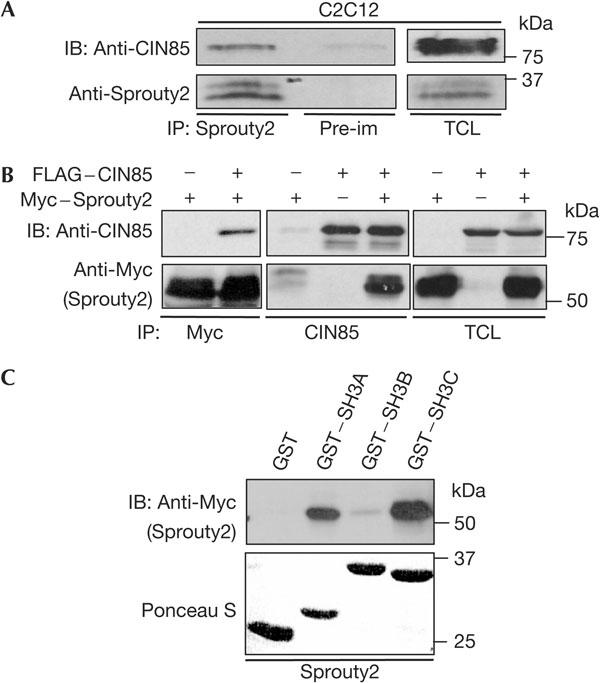
Complex between Sprouty2 and CIN85 in mammalian cells. (A) Endogenous Sprouty2 from C2C12 cell lysates was immunoprecipitated (IP) with antisprouty2 antibodies. Immunoblotting (IB) was performed using specific antibodies against Sprouty2 and CIN85. Pre-immune serum (Pre-im) was used as a negative control. TCL, total cell lysate. (B) Immunoprecipitates of Myc–Sprouty2 and FLAG–CIN85 were separated by SDS–polyacrylamide gel electrophoresis and analysed by immunoblotting with antibodies against the Myc epitope and CIN85. (C) Glutathione-S-transferase (GST) pull-down of Myc–Sprouty2 with isolated SH3 domains of CIN85 (SH3A, SH3B, SH3C) or GST alone. Levels of GST fusion protein were determined by Ponceau staining.
Identification of the CIN85-binding motifs in Sprouty2
Interestingly, Sprouty2 contains two PxxxPR motifs (PTVVPR-64 and PTVPPR-309; Fig 2A). Mutation of arginine to alanine abolishes the binding of the CIN85 SH3 domains to such motifs (Kowanetz et al, 2003). Indeed, the association of Sprouty2-R309A with CIN85sH3A was strongly reduced, whereas Sprouty2-R64A showed slightly decreased binding (supplementary Fig 2A online). The Sprouty2-R64,309A double mutant did not interact with CIN85-SH3A (Fig 2B), indicating that both motifs contribute to the association with CIN85sH3A in vitro. However, as this double mutant was still associated with CIN85-SH3C, a further binding site was assumed. Two potential sites were located in the N-terminal part of Sprouty2, GEPDPR-31 and LKPAPR-72, both of which lack the first proline of the PxxxPR motif, but have a proline at position three (Fig 2A). The fact that Sprouty2-R72A, unlike Sprouty2-R31A, lost its ability to associate with CIN85sH3C indicated that the LKPAPR-72 motif constitutes the second principal binding site (Fig 2B; supplementary Fig 2B online). As expected, the triple mutant Sprouty2-R64,72,309A was not able to associate with either of the SH3 domains of CIN85 (Fig 2B). Taken together, these binding studies showed that CIN85sH3A preferentially binds to the PTVPPR (R309) motif, whereas CIN85-SH3C associates with the LKPAPR (R72) motif. Moreover, the PTVVPR (R64) motif partially contributed to the binding of Sprouty2 to CIN85-SH3A in vitro.
Figure 2.
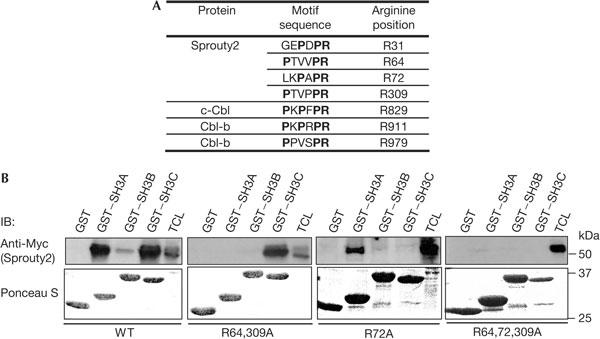
Identification of CIN85-binding sites in Sprouty2. (A) List of proline–arginine motifs in Sprouty2 and c-Cbl/Cbl-b. The amino-acid position of the conserved arginine is indicated. (B) Glutathione-S-transferase (GST) pull-down of Myc-tagged Sprouty2 or Sprouty2 mutants, performed as in Fig 1C. IB, immunoblotting; TCL, total cell lysate; WT, wild type.
To test the significance of the identified motifs in vivo, we analysed the binding between the Sprouty2 mutants and CIN85 in mammalian cells. The association between Sprouty2 and CIN85 was independent of EGF stimulation (Fig 3A). Importantly, Sprouty2-R72A, Sprouty2-R309A and Sprouty2-R72,309A, which lacked the principal binding sites, associated with CIN85 to a much lesser extent than either Sprouty2 wild type or Sprouty2-R64A (Fig 3A). Moreover, no association was detected between CIN85 and Sprouty2-R64,72,309A or Sprouty2-R64,72,309A,Y55F (Fig 3A). In conclusion, the PTVPPR-309 and LKPAPR-72 motifs are essential for the association between CIN85 and Sprouty2 in mammalian cells.
Figure 3.
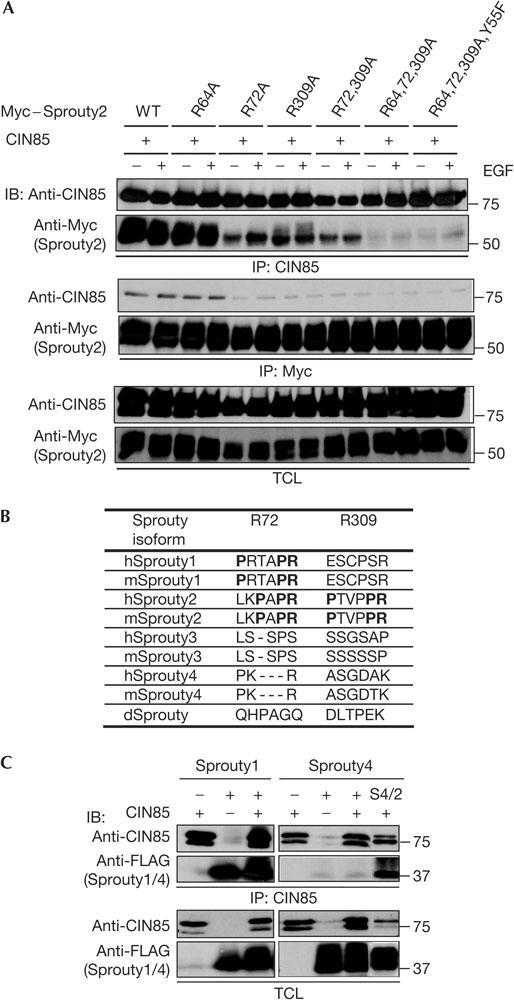
CIN85-binding motifs in Sprouty in vivo. (A) Serumstarved human embryonic kidney (HEK) 293T cells overexpressing CIN85 and Myc-tagged Sprouty2 or indicated Sprouty2 mutants were mock treated or stimulated with 50 ng/ml epidermal growth factor (EGF) for 10 min. Levels of CIN85, Sprouty2 and its mutants in immunoprecipitates (IP) and total cell lysate (TCL) were determined by immunoblotting (IB). WT, wild type. (B) Table listing the CIN85-binding motifs in the Sprouty isoforms in different species. d, Drosophila; h, human; m, mouse. (C) Immunoprecipitation of CIN85 to analyse its interaction with FLAG-tagged Sprouty1, Sprouty4 or the Sprouty4/2 chimaera (S4/2). Sprouty1 is found in complex with endogenous CIN85 that is only detected on longer exposures.
Interestingly, the Cbl-binding motif including Y55 is found in all four Sprouty homologues (Guy et al, 2003), whereas principal CIN85-binding motifs are found only in Sprouty2 and Sprouty1 (Fig 3B). Consistent with this, Sprouty1 associated with CIN85, whereas Sprouty4 did not (Fig 3C). Moreover, the CIN85-binding motifs are conserved in the mammalian Sprouty homologues, but are not found in the Drosophila orthologues (Fig 3B). To analyse the requirement of CIN85 binding for the functions of Sprouty, we engineered a Sprouty4/2 chimaera by mutating amino acids in the Sprouty4 sequence and introducing the two principal CIN85-binding sites from Sprouty2. This yielded a chimaera, which associated with CIN85 (Fig 3C), thereby underpinning the significance of the identified proline–arginine motifs for the association between Sprouty2 and CIN85.
Sprouty2 acts on both Cbl and CIN85
To investigate the functional relevance of the CIN85/Sprouty2 association, we tested whether Sprouty2 mutants not binding to CIN85 affected EGFR ubiquitination and downregulation. In accordance with previous studies, Sprouty2 wild type blocked the binding between EGFR and Cbl and thereby Cbl-mediated EGFR ubiquitination, whereas Sprouty2-Y55A, which is unable to associate with Cbl, did not (Fig 4A). Surprisingly, the CIN85 nonbinding mutant Sprouty2-R64,72,309A did not inhibit Cbl-mediated ubiquitination of EGFR (Fig 4A), although the Cbl-binding motif (Y55) was still intact, phosphorylated and bound to Cbl on EGF stimulation (supplementary Fig 3 online). The fact that this mutant bound less efficiently to Cbl than Sprouty2 wild type indicates that CIN85 binding to Sprouty2 is required for its efficient complex formation with Cbl.
Figure 4.
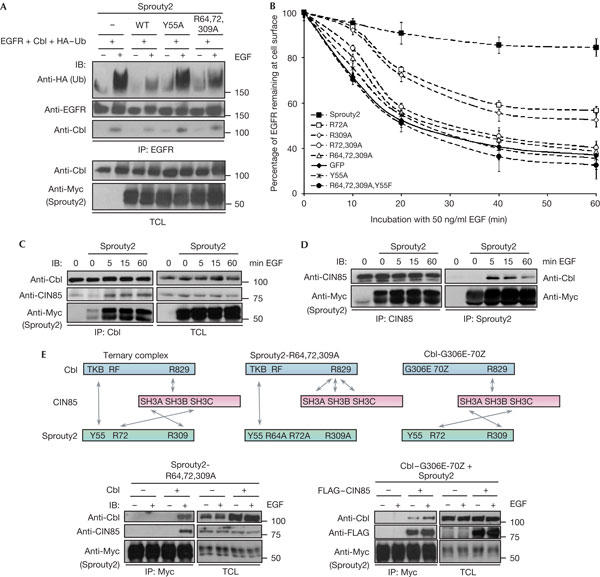
Sprouty2 blocks epidermal growth factor receptor (EGFR) downregulation by acting on both Cbl and CIN85. (A) Serumstarved human embryonic kidney (HEK) 293T cells overexpressing EGFR, Cbl, haemagglutinin (HA)–ubiquitin (Ub), alone or in combination with Sprouty2 wild type (WT), Sprouty2-Y55A or Sprouty2-R64,72,309A, were mock treated or stimulated with 50 ng/ml EGF for 10 min. Immunoprecipitated EGFR was assayed for ubiquitination and association with Cbl. IB, immunoblotting; IP, immunoprecipitation; TCL, total cell lysate. (B) Chinese hamster ovary (CHO) cells were transfected with constructs for EGFR, Cbl and Sprouty2, Sprouty2-R72A, Sprouty2-R309A, Sprouty2-R72,309A, Sprouty2-R64,72,309A, Sprouty2-Y55A, Sprouty2-R64,72,309A,Y55F or green fluorescent protein (GFP) as a control and subjected to an EGFR downregulation assay, as described in the Methods. (C) CHO–EGFR cells were untransfected or transfected with Myc–Sprouty2 as indicated, serum starved and stimulated with 50 ng/ml EGF for 0, 5, 15 or 60 min. Total cell lysates (TCL) and immunoprecipitates of endogenous Cbl were analysed as shown. (D) CHO–EGFR cells were transfected with Myc–Sprouty2 and stimulated as in (C). Immunoprecipitates of endogenous CIN85 and overexpressed Sprouty2 were analysed by immunoblotting, as indicated. (E) In a ternary complex, Cbl interacts in an EGF-inducible way with both Sprouty2 and CIN85 by means of the tyrosine kinase-binding (TKB) domain and the PKPFPR motif (R829), respectively. Simultaneously, CIN85 associates constitutively with Sprouty2 by means of its SH3A and SH3C domains. Sprouty2-R64,72,309A is impaired in binding to CIN85, but interacts with Cbl. Conversely, Cbl-G306E-70Z does not bind to Sprouty2, but still retains the ability to interact directly with CIN85. Bottom left panel: HEK 293T cells were transfected with Myc–Sprouty2-R64,72,309A, alone or together with Cbl, as indicated. Bottom right panel: HEK 293T cells were transfected with HA–Cbl-G306E-70Z and Myc–Sprouty2, alone or together with FLAG–CIN85. Cells were serum starved and mock treated or stimulated for 5 min with 50 ng/ml EGF. Total cell lysates and immunoprecipitates of Myc–Sprouty2 were analysed by immunoblotting. RF, ring finger domain.
Next, the importance of CIN85 binding was tested in EGFR downregulation assays. As previously shown, Sprouty2 wild type blocked EGFR downregulation, whereas the Cbl nonbinding mutant (Sprouty2-Y55A) did not (Fig 4B). Interestingly, the CIN85 nonbinding mutant (Sprouty2-R64,72,309A) was also unable to inhibit Cbl-mediated EGFR downregulation (Fig 4B), which is consistent with its inability to block Cbl-mediated EGFR ubiquitination (Fig 4A). Thus, the loss of either the Cbl-binding site or the CIN85-binding sites in Sprouty2 had identical effects on EGFR downregulation. Moreover, the combined loss of the Cbl- and CIN85-binding sites (Sprouty2-R64,72,309A,Y55F) failed to block EGFR endocytosis, similarly to the loss of the Cbl-binding site (Sprouty2-Y55A) or the CIN85-binding sites (Sprouty2-R72,309A or Sprouty2-R64,72,309A; Fig 4B). This indicates that the two binding events depend on each other and are required to mediate the inhibitory effect of Sprouty2 on Cbl. Conversely, Sprouty2-R72A or Sprouty2-R309A, both of which were individually mutated in the two principal CIN85-binding motifs, were partially able to block EGFR downregulation (Fig 4B), suggesting that a residual association between Sprouty2 and CIN85 allows the mutants to interfere with EGFR endocytosis. Accordingly, the CIN85-binding Sprouty4/2 chimaera, but not Sprouty4 wild type, blocked the disappearance of EGFR from the cellular surface to an extent similar to that of either Sprouty2 or Sprouty1 (supplementary Fig 4 online). Taken together, these data suggest that Sprouty2 requires binding to both Cbl and CIN85 to efficiently inhibit Cbl-mediated EGFR ubiquitination and downregulation.
We investigated further the interaction dynamics between Cbl, CIN85 and Sprouty2 on EGF stimulation. Both CIN85 and Sprouty2 were recruited to Cbl after short EGF stimulation and remained in the complex for prolonged EGF stimulation time (Fig 4C), which is in agreement with previous reports (Soubeyran et al, 2002; Fong et al, 2003; Hall et al, 2003; Rubin et al, 2003). This suggests that ternary complexes between Cbl, CIN85 and Sprouty2 can form after EGF stimulation. However, this reasoning does not exclude the simultaneous formation of pools of dimeric Sprouty2/CIN85 and Sprouty2/Cbl complexes. As shown in Fig 4D, CIN85 constitutively interacts with Sprouty2, whereas Cbl associates with Sprouty2 on EGF stimulation. To show the existence of ternary complexes between Cbl, CIN85 and Sprouty2, we set up an approach using mutants of Sprouty2 (Sprouty2-R64,72,309A) and Cbl (Cbl-G306E-70Z) that are unable to interact with CIN85 or Sprouty2, respectively (Fig 3A; Wong et al, 2001; Fong et al, 2003). We reasoned that if ectopic overexpression of Cbl could rescue the binding between Sprouty2-R64,72,309A and CIN85, this would be due to Cbl-mediated recruitment of CIN85 to the Sprouty2 mutant (Fig 4E, model). Indeed, Sprouty2-R64,72,309A was able to associate with CIN85 only in the presence of high amounts of Cbl subsequent to EGF stimulation (Fig 4E, left panel). Similarly, Cbl-G306E-70Z was detected in complexes with Sprouty2 on addition of exogenous CIN85, which suggests that CIN85 was able to connect Sprouty2 and the Cbl mutant (Fig 4E, model and right panel). These data indicate that ternary complexes between Cbl, CIN85 and Sprouty2 can form in the cell.
Finally, the functional consequences of the expression of Sprouty2 mutants in PC12 cells were investigated. Nerve growth factor (NGF) stimulation promotes sustained mitogen-activated protein kinase (MAPK) activation and neuronal differentiation, whereas transient MAPK activation triggered by EGF increases proliferation of PC12 cells. However, by increasing the strength and duration of EGFR signalling, either by EGFR overexpression or inhibition of EGFR endocytosis, PC12 cells form neurites (Wong et al, 2002). Indeed, overexpression of Sprouty2 wild type promoted EGF-induced PC12 cell differentiation (Fig 5A,B). Conversely, mutants lacking the CIN85-binding sites showed significantly reduced neurite outgrowth. In agreement with the EGFR downregulation assays, the loss of either of the principal CIN85-binding sites (Sprouty2-R72A and Sprouty2-R309A) did not completely block neurite outgrowth, whereas combined mutations (Sprouty2-R72,309A or Sprouty2-R64,72,309A) had a much stronger effect (Fig 5A,B). Moreover, both Sprouty2-R64,72,309A-Y55F, deficient in binding to both CIN85 and Cbl, and Sprouty2-Y55A, which is unable to bind to Cbl, failed to induce differentiation of PC12 cells (Fig 5A,B). Consequently, Sprouty2 inhibits EGFR endocytosis, promotes sustained EGFR signalling and induces neurite outgrowth in response to EGF by associating with both Cbl and CIN85.
Figure 5.
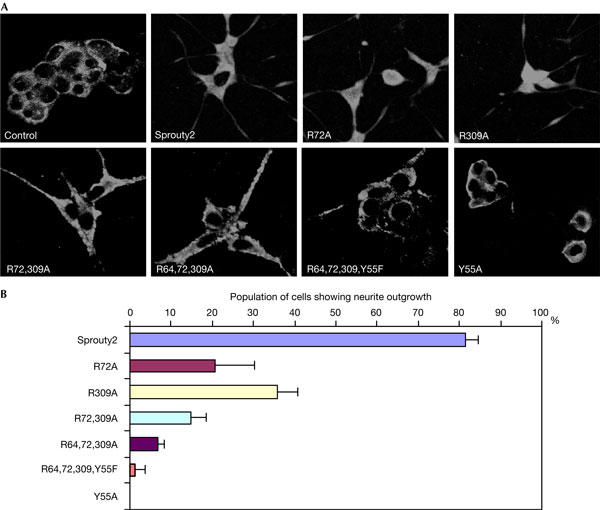
Sprouty2 requires binding to both Cbl and CIN85 to promote neurite outgrowth. (A) PC12 cells grown on poly-D-lysine-coated coverslips were transfected with Myc-tagged plasmids encoding Sprouty2, Sprouty2-R72A, Sprouty2-R309A, Sprouty2-R72,309A, Sprouty2-R64,72,309A, Sprouty2-R64,72,309A,Y55F or Sprouty2-Y55A. Cells were fixed after incubation for 4 days in lowserum medium without added growth factor (Sprouty2 ‘Control') or with 100 ng/ml epidermal growth factor. Cells were subjected to immunofluorescence, as described in the Methods. (B) Experimental sets in (A) were quantified in three independent experiments. Cells with neurite extensions of one to two times the length of the cell body were scored as neurite-containing cells. The graph shows the average percentage of cells showing neurite outgrowth from the three experiments. The error bars represent the standard deviation.
Discussion
Here, we report that the inhibitory effect of Sprouty2 on EGFR endocytosis depends on its interference with the functions of both Cbl and CIN85. Ligand stimulation of EGFRs leads to the recruitment of Cbl and CIN85 to activated receptors, inducing their endocytosis and degradation (Dikic & Giordano, 2003). EGF stimulation also leads to synthesis of Sprouty2, its targeting to the receptor complexes and sequestration of Cbl away from activated EGFRs. Our data indicate that Sprouty2 can constitutively interact with two SH3 domains of CIN85 (Figs 1, 2 and 3), whereas the third SH3 domain of CIN85 can still associate with Cbl on cell activation (Fig 4; Kowanetz et al, 2003). This allows Sprouty2 to block CIN85-mediated clustering of Cbl molecules, a step that is important for stabilization of Cbl–EGFR interactions and efficient ubiquitination and downregulation of EGFR (Kowanetz et al, 2003). Moreover, in this way, CIN85 may contribute to the translocation of Sprouty2 to Cbl/EGFR complexes, where phosphorylated Sprouty2 can compete with activated EGFRs for binding of the SH2 domain of Cbl. In such a scenario, Sprouty2 requires binding to both CIN85 and Cbl to inhibit EGFR endocytosis. Consistent with this hypothesis, the loss of either CIN85 or Cbl binding to Sprouty2 allows EGFR downregulation to occur normally in the presence of the mutant Sprouty2 molecules (Fig 4).
Furthermore, our results provide more clues to the opposing actions of different Sprouty family members. Whereas all isoforms in mammals and Drosophila contain the conserved Cbl-binding tyrosine residue Y55, only mammalian Sprouty2 and Sprouty1 are able to inhibit EGFR endocytosis (Wong et al, 2002; Fong et al, 2003; Hall et al, 2003; Rubin et al, 2003). The functional differences could be explained by the differential recruitment of distinct Sprouty isoforms to the EGFR/Cbl complex. In accordance, our findings show that the ability of Sprouty isoforms to bind to CIN85 correlates with their ability to inhibit EGFR endocytosis. For example, binding between Sprouty2 and CIN85 is required for Sprouty2-mediated inhibition of EGFR downregulation and induction of PC12 cell differentiation (Fig 4). Conversely, Sprouty4, which does not bind to CIN85 (Fig 3), fails to block EGFR downregulation (supplementary Fig 4 online) and is thus unable to promote neurite outgrowth in response to EGF (Wong et al, 2002).
The general significance of the mechanisms described here is underlined by our recent findings that another EGF-induced feedback regulator of Cbl, βPix, also targets the Cbl/CIN85 interface to inhibit Cbl-mediated EGFR downregulation (M.H.H.S. and I.D., unpublished results). In conclusion, these results, along with several recent reports (Hall et al, 2003; Rubin et al, 2003; Wu et al, 2003), point to the crucial role of Sprouty2 and βPix in blocking the Cbl/CIN85 interface and fine-tuning receptor endocytosis, which is implicated in both physiological and pathological conditions.
Methods
Reagents, antibodies and plasmids. EGF was from Intergen (Oxford, UK) and 125I-EGF was from ICN Biomedicals (Irvine, CA, USA). Antibodies against CIN85, EGFR and Cbl have been described previously (Soubeyran et al, 2002). Antibodies against Myc (9E10) were from Babco (Princeton, NJ, USA), FLAG (M2) from Sigma Aldrich (St Louis, MO, USA), haemagglutinin (HA; 12CA5) from Boehringer Mannheim (Mannheim, Germany) and phosphotyrosine (pY99) from Santa Cruz Biotech (Santa Cruz, CA, USA). Antibodies against Sprouty2 were a gift from Hong Joo Kim, Dafna Barsagi, Chanan Rubin and Yosef Yarden. Human Sprouty2 and Sprouty2-Y55A in pcDNA3–6 × Myc were provided by Atsuo Sasaki and Akihiko Yoshimura (Kyushu University, Fukuoka, Japan). HA–G306E-70Z-Cbl in pBABEpuro was provided by Wallace Y. Langdon (The University of Western Australia, Nedlands, Australia). Mouse Sprouty1 and Sprouty4 and human Sprouty2-CT (residues 178–315) in pXJ40–FLAG have been described previously (Wong et al, 2001, 2002). pRK5–EGFR, pRK5–Cbl, pcDNA3–HA–ubiquitin, pcDNA3–FLAG–CIN85 and GST fusion proteins of SH3 domains A, B and C of CIN85 have been described previously (Soubeyran et al, 2002; Haglund et al, 2003b).
Site-directed mutagenesis. Sprouty2 arginine to alanine mutants were generated by site-directed mutagenesis. pcDNA3–6 × Myc–Sprouty2-NT was constructed by inserting a stop codon at the position of amino acid 178. The pXJ40–Sprouty4/2 chimaera was engineered by PCR by introducing the CIN85-binding sites (LKPAPR-72 and PTVPPR-309) at corresponding sites in the Sprouty4 sequence. Oligonucleotide sequences are available on request.
Cell culture and biochemical studies. Cell culture, transfections, cell lysis, immunoprecipitation, GST pull-down assays, SDS–polyacrylamide gel electrophoresis and western blotting were performed as described previously (Wong et al, 2002; Haglund et al, 2003b, 2004). C2C12 cells were provided by Hong Joo Kim and Dafna Barsagi and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 5% calf serum and antibiotics. For EGFR downregulation assays, Chinese hamster ovary (CHO) cells were transfected with EGFR, Cbl and Sprouty2, various Sprouty2 mutants, Sprouty1, Sprouty4, the Sprouty4/2 chimaera or GFP as a control. The amount of EGFR molecules remaining at the cellular membrane was determined by measuring surface-bound 125I-labelled EGF after stimulation with EGF for the indicated durations, as described (Kowanetz et al, 2004). The percentage of surface receptor was calculated by comparing the values of EGFstimulated cells and nonstimulated cells. Each time point was measured in triplicate and the experiments were repeated twice.
Immunofluorescence. PC12 cells were seeded on poly-D-lysine-coated coverslips and transfected using the GenePORTER™ Transfection Reagent (Gene Therapy Systems, San Diego, CA, USA). At 24 h after transfection, cells were subjected to either low serum or 100 ng/ml EGF for 4 days. Cells were fixed with 3% paraformaldehyde, and stained with a polyclonal antibody directed against the c-Myc epitope tag (Transduction Laboratories, Lexington, KY, USA) followed by fluorescein isothiocyanate-conjugated AffiniPure goat anti-rabbit IgG (Jackson Immuno-Research Laboratories, West Grove, PA, USA). Images were visualized and captured by confocal laser scanning microscopy (Zeiss, Oberkochen, Germany).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank A. Sasaki, A. Yoshimura, H.J. Kim, D. Bar-Sagi, W.Y. Langdon, C. Rubin and Y. Yarden for providing reagents. We are grateful to D.-H. Lao, who advised us to create a Sprouty4/2 chimaera, and to P. Soubeyran, I. Marinovic and K. Husnjak for help in the early phases of this project. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG DI 93112) and Boehringer Ingelheim Fonds to I.D. M.H.H.S. is a fellow of the European Molecular Biology Organization (EMBO, ALTF 881-2003).
References
- Bao J, Gur G, Yarden Y. (2003) Src promotes destruction of c-Cbl: implications for oncogenic synergy between Src and growth factor receptors. Proc Natl Acad Sci USA 100: 2438–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G (2003) Split personalities: the agonistic antagonist Sprouty. Nat Cell Biol 5: 377–379 [DOI] [PubMed] [Google Scholar]
- Dikic I, Giordano S (2003) Negative receptor signalling. Curr Opin Cell Biol 15: 128–135 [DOI] [PubMed] [Google Scholar]
- Egan JE, Hall AB, Yatsula BA, Barsagi D (2002) The bimodal regulation of epidermal growth factor signaling by human Sprouty proteins. Proc Natl Acad Sci USA 99: 6041–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CW, Leong HF, Wong ES, Lim J, Yusoff P, Guy GR (2003) Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J Biol Chem 278: 33456–33464 [DOI] [PubMed] [Google Scholar]
- Guy GR, Wong ES, Yusoff P, Chandramouli S, Lo TL, Lim J, Fong CW (2003) Sprouty: how does the branch manager work? J Cell Sci 116: 3061–3068 [DOI] [PubMed] [Google Scholar]
- Haglund K, Di Fiore PP, Dikic I (2003a) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci 28: 598–603 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I (2003b) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Haglund K, Ivankovic-Dikic I, Shimokawa N, Kruh GD, Dikic I (2004) Recruitment of Pyk2 and Cbl to lipid rafts mediates signals important for actin reorganization in growing neurites. J Cell Sci 117: 2557–2568 [DOI] [PubMed] [Google Scholar]
- Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Barsagi D (2003) hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr Biol 13: 308–314 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Barsagi D (2004) Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol 5: 441–450 [DOI] [PubMed] [Google Scholar]
- Kowanetz K, Szymkiewicz I, Haglund K, Kowanetz M, Husnjak K, Taylor JD, Soubeyran P, Engstrom U, Ladbury JE, Dikic I (2003) Identification of a novel proline–arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J Biol Chem 278: 39735–39746 [DOI] [PubMed] [Google Scholar]
- Kowanetz K, Crosetto N, Haglund K, Schmidt MH, Heldin CH, Dikic I (2004) Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J Biol Chem 279: 32786–32795 [DOI] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y (2003) Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem 278: 21323–21326 [DOI] [PubMed] [Google Scholar]
- Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S (2002) The endophilin–CIN85–Cbl complex mediates ligand-dependent downregulation of c-Met. Nature 416: 187–190 [DOI] [PubMed] [Google Scholar]
- Rubin C, Litvak V, Medvedovsky H, Zwang Y, Lev S, Yarden Y (2003) Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr Biol 13: 297–307 [DOI] [PubMed] [Google Scholar]
- Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I (2002) Cbl–CIN85–endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature 416: 183–187 [DOI] [PubMed] [Google Scholar]
- Wong ES, Lim J, Low BC, Chen Q, Guy GR (2001) Evidence for direct interaction between Sprouty and Cbl. J Biol Chem 276: 5866–5875 [DOI] [PubMed] [Google Scholar]
- Wong ES, Fong CW, Lim J, Yusoff P, Low BC, Langdon WY, Guy GR (2002) Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. EMBO J 21: 4796–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WJ, Tu S, Cerione RA (2003) Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell 114: 715–725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


