Abstract
The human cytomegalovirus IRS1 and TRS1 open reading frames encode immediate-early proteins with identical N-terminal domains and divergent C-terminal regions. Both proteins have been shown previously to activate reporter genes in transfection assays in cooperation with other viral gene products. We have constructed two viruses carrying substitution mutations within either the IRS1 or TRS1 open reading frame. ADsubIRS1 failed to produce the related IRS1 and IRS1263 proteins, but it replicated with normal kinetics to produce a wild-type yield in human fibroblasts. The addition in trans of the IRS1263 protein, which antagonizes the ability of IRS1 and TRS1 proteins to activate reporter genes, did not inhibit the growth of the mutant virus. ADsubTRS1 failed to produce the TRS1 protein, and it generated an ∼200-fold-reduced yield of infectious virus in comparison to its wild-type parent. Viral DNA accumulated normally, as did a set of viral mRNAs that were monitored in ADsubTRS1-infected cells. However, two tegument proteins were partially mislocalized and infectious virus particles did not accumulate to normal levels within ADsubTRS1-infected cells. Further, infectious ADsubTRS1 particles sedimented abnormally in a glycerol-tartrate gradient, indicating that the structure of the mutant particles is aberrant. Our analysis of the ADsubTRS1 phenotype indicates that the TRS1 protein is required, either directly or indirectly, for efficient assembly of virus particles.
Human cytomegalovirus (HCMV) is a ubiquitous member of the beta-herpesvirus family. Although HCMV infection usually is asymptomatic in healthy children and adults, it is a leading viral cause of birth defects and it is a major cause of morbidity and mortality in immunocompromised individuals (16).
The HCMV genome is large, ∼230,000 bp, and comprises two unique domains (unique long [UL] and unique short [US]) bracketed by repeated sequences (12). The IRS1 and TRS1 open reading frames (22, 26) encode proteins of 846 and 788 amino acids, respectively, and span from repeated domains (c′ and c) into the US domain (see Fig. 1A). Expression of their mRNAs is controlled by identical promoters located within the repeated domain that are active during the immediate-early phase of the virus replication cycle. The two encoded proteins, pIRS1 and pTRS1, are identical over their N-terminal 549 amino acids, which are encoded by the repeated domains, and the proteins diverge in their US-encoded C-terminal regions. A promoter that directs the expression of an mRNA corresponding to the 3′ region of IRS1 resides within the IRS1 coding region (19). This mRNA encodes pIRS263, which is identical to the C-terminal 263 amino acids of pIRS1 (19).
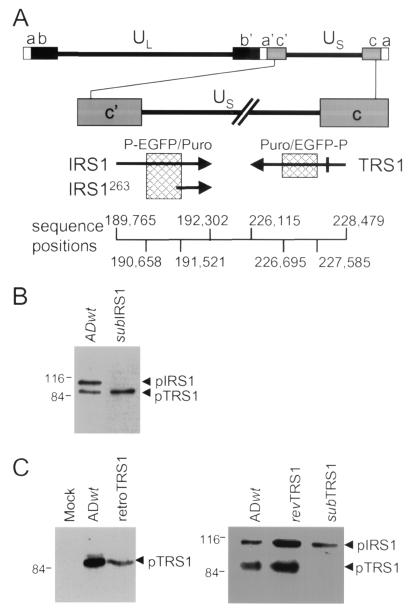
FIG. 1.
Locations of alterations in mutant and revertant viruses and effect of mutations on protein expression within infected cells. (A) Diagram of IRS1 and TRS1 coding regions depicting the locations of mutations. The top portion displays the entire viral genome with unique (UL and US) and repeated (a, b, and c) domains. The region containing the IRS1, IRS1263, and TRS1 open reading frames (arrows) is expanded and displayed below. Cross-hatched boxes, regions deleted when producing the substitution mutations in ADsubIRS1 and ADsubTRS1. Above the boxes the inserted marker cassette is indicated (P, SV40 early promoter; EGFP, EGFP coding region; Puro, puromycin resistance gene). Bar on TRS1 arrow, single base pair change (C to A) introduced into ADrevTRS1. The sequence boundaries of the open reading frames and deletions are indicated at the bottom. (B) Western blot analysis of pIRS1 and pTRS1 expression after infection at a multiplicity of infection of 5 PFU/cell with ADwt or ADsubIRS1. (C) (Left) Western blot assay of pTRS1 expression after infection at a multiplicity of infection of 5 PFU/cell with ADwt or at a multiplicity of infection of 3 transducing units/cell with retroTRS1. (Right) Western blot assay of pIRS1 and pTRS1 expression in retroTRS1-containing cells infected with ADsubTRS1 or ADrevTRS1 at a multiplicity of infection of 5 PFU/cell.
Either pIRS1 or pTRS1 is required for complementation of HCMV oriLyt-dependent DNA replication in a transfection assay (14, 15), but as yet there is no evidence that the proteins act directly in the process of DNA replication. They likely influence replication by facilitating expression of other viral genes in the assay (8). Both pIRS1 and pTRS1 have been shown to activate the expression of reporter genes in transfection assays, working in cooperation with other immediate-early proteins (8, 10, 19, 23). The two activators are packaged into the virion (18), so they would be available in cells to help activate the immediate-early class of HCMV genes. pIRS1263 antagonizes the activation function of pIRS1 and pTRS1 (19).
To explore the function of pIRS1, pIRS1263, and pTRS1 within infected cells, we constructed two mutant viruses. ADsubIRS1 contains a substitution mutation in the central domain of the IRS1 open reading frame, and it fails to produce pIRS1 and pIRS1263. ADsubTRS1 contains a substitution mutation in the central domain of the TRS1 open reading frame, and it cannot produce pTRS1. Whereas ADsubIRS1 does not exhibit a growth defect, ADsubTRS1 replicates slowly, producing a reduced yield in human fibroblasts. The pTRS1-deficient virus exhibits a defect late in the replication cycle, suggesting that the protein is directly or indirectly involved in the assembly of virus particles.
MATERIALS AND METHODS
Viruses and cells.
ADwt is a plaque-purified derivative of the AD169 strain (20) of HCMV, and it was propagated in primary human foreskin diploid fibroblasts. ADsubIRS1 and ADsubTRS1 were derived from ADwt by recombination within infected cells. Recombination vectors that contained a marker cassette surrounded by the HCMV regions flanking the segment to be deleted were prepared. The marker cassette (17) comprises the simian virus 40 (SV40) early promoter controlling expression of a bicistronic mRNA containing the enhanced green fluorescent protein (EGFP) coding region, an internal ribosomal entry site, and the puromycin-N-acetyltransferase coding region. HCMV DNA segments flanking the viral sequences to be deleted were prepared by PCR amplification and cloned on either side of the marker cassette. For construction of ADsubIRS1, these segments included bp 189646 to 190658 and 191521 to 192320, and, for construction of ADsubTRS1, they included bp 189646 to 190658 and 226695 to 225740 (sequence numbers from reference 3). The same DNA segment was used as the 5′ flank to target the two mutations because this region resides in a repeated region located upstream of both IRS1 and TRS1. To produce recombinant viruses carrying substitution mutations, ADwt DNA (2 μg), a linearized recombination vector (5 μg), and a plasmid expressing pUL82 (1 μg), which enhances the infectivity of viral DNA (1), were introduced into fibroblasts (2.5 × 106 cells in 0.25 ml) by electroporation (260 mV, 960 μF). Puromycin (2 μg/ml) was added to cultures from 24 to 48 h after electroporation to select for recombinant virus with the puromycin resistance marker. To produce ADrevTRS1, a recombination plasmid with a wild-type TRS1 coding region was prepared. The coding region encoded a leucine-to-valine substitution at amino acid 190, so that the revertant could be distinguished from a wild-type contaminant. An enhanced cyan fluorescent protein (ECFP) coding region, whose expression was controlled by the SV40 early promoter, was inserted downstream of the 3′-noncoding region of the TRS1 locus in the recombination vector. The linearized recombination vector, ADsubTRS1 DNA, and the pUL82 expression plasmid were introduced into fibroblasts by electroporation. Blue fluorescent colonies were formed at high frequency after recombination to the 5′ side of the mutation within the TRS1 coding region and to the 3′ side of the mutation between the EGFP coding sequence in ADsubTRS1 DNA and the highly related ECFP coding sequence in the recombination plasmid. These recombinant viral genomes were unstable and produced progeny lacking the ECFP marker since the marker was bracketed by two copies of the TRS1 3′-noncoding region. Nonfluorescent plaques containing ADrevTRS1 were obtained in the second round of plaque purification. Stocks of ADsubIRS1 and ADrevTRS1 were prepared after three sequential plaque purifications on human fibroblasts, and a stock of ADsubTRS1 was made after three sequential plaque purifications on pTRS1-expressing human fibroblasts.
Two recombinant retroviruses, retroIRS1263 and retroTRS1, were prepared by cloning the appropriate HCMV open reading frame into pRetro-EBNA. Retroviruses were propagated by transfecting (7 μl of Lipofectamine reagent/μg of DNA) the plasmids into Phoenix Ampho cells (11). Retroviruses were used to infect Polybrene (Sigma)-treated (4 μg/ml) human fibroblasts. Polybrene was maintained in the culture for 24 h after addition of the retrovirus, and then cells were washed twice and fed with fresh medium. Immunofluorescence assays were used to monitor expression of HCMV proteins in human fibroblasts infected with recombinant retroviruses and to determine the transducing titers of the retroIRS1263 and retroTRS1 stocks. Retrovirus-infected human fibroblasts were maintained in culture for at least 4 days before infection with HCMV because we observed that residual Polybrene inhibited HCMV infection.
HCMV particles were purified (2, 7, 25) from the medium of infected cultures when ∼50% of the cells were detached from the plate. The medium was layered over a sorbitol cushion (20% d-sorbitol, 50 mM Tris [pH 7.4], 1 mM MgCl2, 100 μg of bacitracin/ml), and virus was pelleted by centrifugation (20,000 rpm, 1 h, 4°C, Beckman SW28 rotor). Bacitracin was included to inhibit aggregation of virus particles (24). This crude preparation of virus particles was resuspended in buffer containing 50 mM Tris, pH 7.4, and 100 mM NaCl. To further purify virus particles, the crude preparation was supplemented with bacitracin (100 μg/ml) and urea (0.5 M) and the solution was sonicated for 15 s, layered onto a glycerol-tartrate gradient (0 to 30% glycerol, 35 to 15% potassium tartrate, 50 mM Tris [pH 7.4], 100 mM NaCl), and subjected to centrifugation (40,000 rpm, 15 min, 4°C, Beckman SW41 rotor). Three fractions were removed from the gradient (noninfectious enveloped particles [NIEPs], virions, and dense bodies [DBs]), diluted in buffer (50 mM Tris [pH 7.4], 100 mM NaCl), pelleted by centrifugation, and resuspended in the same buffer.
Analysis of viral nucleic acids.
Viral DNA present in total-cell-DNA preparations was analyzed by Southern blot assay. After digestion with restriction enzymes, DNA was separated on an 0.8% agarose gel, blotted to a nylon membrane, and probed with [32P]dCTP-labeled DNA prepared by random priming of the entire HCMV genome. Specific viral mRNAs in total cellular RNA isolated with the TRIzol reagent (Gibco-BRL) were analyzed by Northern blot assay. RNAs were separated by electrophoresis for 3 h at 100 V in a formaldehyde-containing gel and blotted to a nylon membrane, and viral mRNAs were detected by hybridization to [32P]dCTP-labeled DNA probes prepared by random priming of viral sequences that were purified from recombinant plasmids.
Analysis of viral proteins.
The steady-state levels of individual proteins in preparations of purified virus particles or lysates of infected cells were determined by Western blot assay. Protein samples were separated by electrophoresis in a sodium dodecyl sulfate-containing 8 or 10% polyacrylamide gel and transferred to a nitrocellulose membrane. Blots were probed with antibodies and then developed with the ECL reagent (Amersham). An antibody (MAb810) recognizing the common domain of pUL122 and pUL123 was purchased (Chemicon). Antibodies to pUL32 (13), pUL55 (13), pUL69 (5), pUL83 (13), pUL86 (27), pUL99 (13), pIRS1 (19), and pTRS1 (4) have been described previously, and an antibody to the Golgi marker mannosidase II was a gift from G. Waters (Princeton University).
The locations of viral proteins within infected cells were determined by indirect immunofluorescence. Cells were fixed in paraformaldehyde (4%) and treated with blocking solution (phosphate-buffered saline containing 0.5% bovine serum albumin, 5% goat serum, 0.5% Tween 20, 1% Triton X-100). Cells were incubated with the primary antibody in blocking solution for 1 h and with the secondary antibody in blocking solution for 30 min and washed with blocking solution three times after each incubation. Cells were then washed twice with distilled water, incubated with YOYO-1 (Molecular Probes) or Hoechst 33342 (Sigma) to stain DNA, washed twice in distilled water, and mounted onto slides.
Pulse-chase experiments were performed to monitor the intracellular localization of pUL83 as a function of time after its synthesis. Infected cells were maintained for 1 h in medium lacking methionine and cysteine, pulse labeled for 10 min with [35S]methionine and [35S]cysteine (1 mCi/5 × 106 cells), washed, and refed with medium containing unlabeled methionine and cysteine for the chase period. After various periods of chase, cells were harvested, suspended in 10 volumes of hypotonic buffer containing protease inhibitors (5 mM sodium phosphate [pH 7.2], 2 mM MgCl2, 0.5 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin/ml, and 2 μg of aprotinin/ml), held on ice for 10 min, pelleted, and resuspended in the same buffer, and then nuclei were released by seven rapid passages of the cell suspension through a 25-gauge needle. Sucrose (0.1 M) was added to the broken cells, and nuclei were pelleted; the supernatant was the cytosolic fraction. The pelleted nuclei were centrifuged twice through a sucrose (0.54 M) cushion in hypotonic buffer and lysed in buffer containing 0.1% NP-40, 250 mM NaCl, and 50 mM HEPES, pH 7.0. Nuclear and cytosolic fractions were cleared of debris by centrifugation, and their pUL83 contents were analyzed by immunoprecipitation. To confirm that nuclear and cytoplasmic fractions were effectively separated, Western blot assays were performed to determine the distribution of nuclear (pUL123) and cytoplasmic (pUL99) viral proteins.
RESULTS
Construction and growth properties of mutant viruses lacking IRS1 or TRS1.
Recombination within infected primary human fibroblasts was employed to substitute a marker cassette for the central portions of either the IRS1 or TRS1 open reading frame (Fig. 1A). The marker cassette contains EGFP and puromycin-N-acetyltransferase coding regions (17). Expression of the markers is controlled by the SV40 early promoter, and translation of the downstream puromycin resistance-coding region is mediated by an internal ribosomal entry site. The markers replaced 863 and 890 bp of the IRS1 and TRS1 coding regions, respectively.
After three rounds of plaque purification, the IRS1-deficient mutant, ADsubIRS1, was judged to have the correct genomic structure and to be free of wild-type virus contamination by Southern blot analysis using probes corresponding to viral sequences flanking either side of the deleted sequences or to the marker cassette and by PCR assay using primers spanning the junctions of the predicted recombination regions (data not shown). A Western blot assay also was performed; it demonstrated that the mutant virus did not produce pIRS1 within infected cells but continued to express pTRS1 (Fig. 1B).
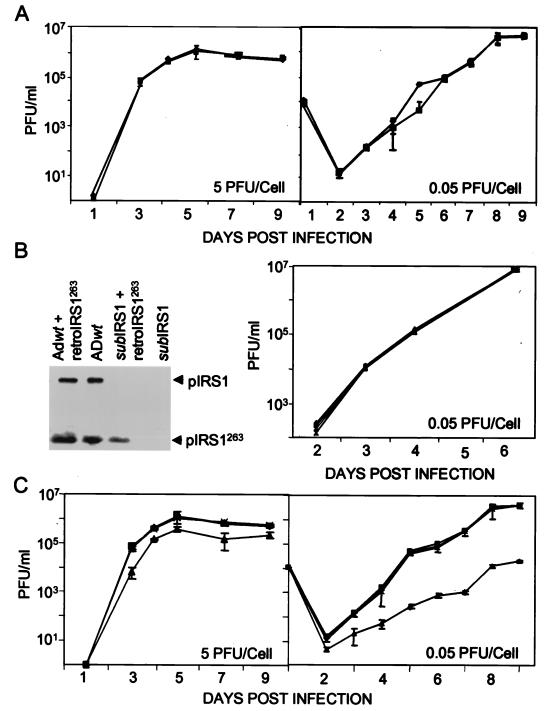
It was found that ADsubIRS1 grew with the same kinetics as its parental wild-type virus, ADwt, when growth was assayed after infection at relatively high (5 PFU/cell) and low (0.05 PFU/cell) multiplicities of infection (Fig. 2A). ADsubIRS1 fails to produce pIRS1263 in addition to pIRS1 (Fig. 2B, left). pIRS263, which is encoded by the 3′ domain of the IRS1 coding region in the same reading frame as that which encodes pIRS1 (Fig. 1A), has been shown to antagonize the transcriptional-activation function of pIRS1 and pTRS1 (19). Therefore, it was conceivable that the simultaneous loss of pIRS1 and pIRS1263 in ADsubIRS1 could mask a defect that would be observed if pIRS1 was lost while pIRS1263 was retained. This possibility was tested by infecting cells with a recombinant retrovirus expressing pIRS1263, retroIRS1263, prior to infection with ADsubIRS1. The retrovirus was used to infect cells at a multiplicity of infection of 3 transducing units/cell. It produced ∼50% of the amount of pIRS1263 that is found at 24 h after infection with wild-type HCMV (Fig. 2B, left), and immunofluorescence demonstrated that >95% of fibroblasts contained pIRS1263 (data not shown). Expression of pIRS1263, however, had no effect on the kinetics with which ADsubIRS1 produced progeny virus (Fig. 2B, right) or viral DNA (data not shown). ADsubIRS1 grew normally in the presence or absence of pIRS1263.
FIG. 2.
Production of infectious progeny in the medium of cultures infected with wild-type, mutant, or revertant viruses. HCMV was used to infect human fibroblasts at the indicated multiplicities of infection, and retroviruses were used at a multiplicity of infection of 3 transducing units/cell. Virus that accumulated in the medium of infected cultures was determined by plaque assays on fibroblasts (ADwt and ADsubIRS1) and retroTRS1-infected fibroblasts (ADsubTRS1 and ADrevTRS1) in duplicate. (A) Growth of ADwt (♦) and ADsubIRS1 (▪). (B) (Left) Western blot assay of pIRS1 and pIRS1263 expression after infection with ADwt or ADsubIRS1 alone or in combination with retroIRS263; (right) growth of ADwt (♦), ADsubIRS1 (▪), ADwt plus retroIRS1263 (▴), and ADsubTRS1 plus retroIRS1263 (×). (C) Growth of ADwt (♦), ADrevTRS1 (▪), ADsubTRS1 (▴), and ADsubTRS1 plus retroTRS1 (×).
After three rounds of plaque purification, the TRS1-deficient mutant, ADsubTRS1, was still contaminated with wild-type virus, suggesting that it was defective and required coinfecting wild-type virus to provide pTRS1 function. Three additional plaque purifications were performed on fibroblasts that were first infected at a multiplicity of infection of 3 transducing units/cell with a recombinant retrovirus expressing pTRS1, retroTRS1. At 24 h after infection of retroTRS1-containing cultures with ADsubTRS1, a Western blot experiment demonstrated that cells contained ∼18% as much pTRS1 as did ADwt-infected cultures (Fig. 1C, left). After growth in cells expressing pTRS1, ADsubTRS1 was judged to be free of wild-type virus contamination and to have the correct genomic structure by Southern blot, PCR, and DNA sequence analyses (data not shown), and a Western blot experiment confirmed that ADsubTRS1 failed to accumulate pTRS1 while producing pIRS1 (Fig. 1C, right).
Revertant virus ADrevTRS1 was generated by replacement of the ADsubTRS1 marker cassette with a TRS1 coding region that contained a 1-bp substitution (C to A at sequence position 228356; Fig. 1A), allowing it to be distinguished from wild-type virus. The structure of the revertant was confirmed by PCR and sequence analyses (data not shown), and it again produced pTRS1 (Fig. 1C, right).
The growth of the mutant, revertant, and wild-type viruses was evaluated by plaque assay of virus released into the medium of infected cells. The yield of ADsubTRS1 was reduced by a factor of ∼5 on days 5 to 9 after infection of human fibroblasts at a multiplicity of infection of 5 PFU/cell (Fig. 2C, left). When cells were infected at a multiplicity of infection of 0.05 PFU/cell, the mutant's growth defect was greater. Cell-free virus was reduced by a factor of ∼200 on days 6 to 9 after infection (Fig. 2C, right). ADrevTRS1 grew like ADwt at both multiplicities of infection, arguing that the ADsubTRS1 defect results from the substitution mutation and not from a spurious mutation at another location. In contrast to its defective growth on normal fibroblasts, ADsubTRS1 grew indistinguishably from ADwt or ADrevTRS1 in fibroblasts infected with retroTRS1 (Fig. 2C, right), confirming the expectation that its defect is due solely to the loss of pTRS1.
ADsubTRS1 accumulates DNA normally but fails to assemble infectious virus.
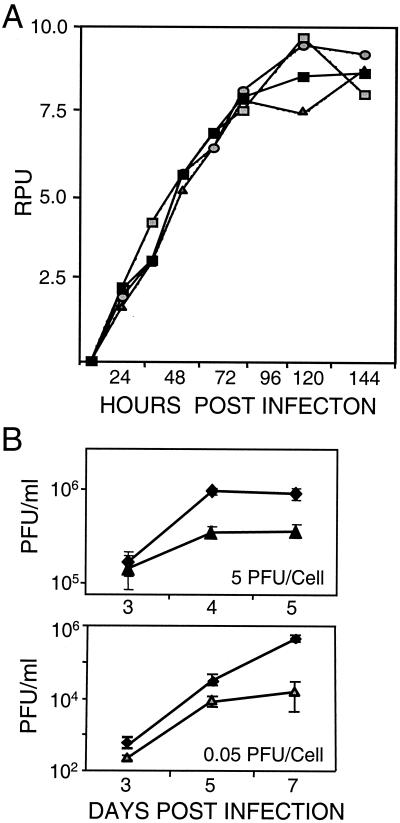
Two experiments were performed to roughly identify the point in the HCMV replication cycle at which pTRS1 is needed. Viral DNA accumulation was monitored by Southern blot assay after infection of human fibroblasts with ADsubTRS1 at a multiplicity of infection of 0.5 PFU/cell (Fig. 3A). Controls included ADsubIRS1-, ADwt-, and ADsubTRS1-infected fibroblasts containing pTRS1 supplied by retroTRS1. ADsubTRS1 DNA accumulation was indistinguishable from that of the controls, arguing that its defect occurs during the late phase of infection. Next, we examined the assembly of infectious virus. The growth experiments presented in Fig. 2 were performed by assaying cell-free virus. To determine whether ADsubTRS1 produced infectious particles that failed to escape from the cell, a growth experiment in which cells were lysed by two freeze-thaw cycles in their growth medium was performed. Then the total amount of infectivity produced in infected cultures (intracellular plus extracellular) was assayed (Fig. 3B). ADsubTRS1 failed to produce as much infectious intracellular virus as did ADwt. Consequently, the ADsubTRS1 defect must have its effect after DNA replication and before the assembly of infectious progeny.
FIG. 3.
Accumulation of DNA and intracellular virus after infection with wild-type or mutant virus. (A) DNA accumulation. Human fibroblasts were infected at a multiplicity of infection of 0.5 PFU/cell with ADwt (), ADsubIRS1 (igwidth>), ADsubTRS1 (▪), or ADsubTRS1 plus retroTRS1 (igwidth>). Total-infected-cell DNA was prepared at the indicated times, and viral DNA was assayed by Southern blotting with a 32P-labeled HCMV genomic probe DNA. Radioactivity was quantified with a phosphorimager, and results are presented as relative phosphorimager units (RPU). (B) Intracellular virus accumulation. Cells were infected at the indicated multiplicities of infection with ADwt (♦) or ADsubTRS1 (▴ or igwidth>). Cells were harvested at the indicated times and freeze-thawed twice, and infectious virus was quantified by plaque assay on retroTRS1-infected fibroblasts. Samples were assayed in duplicate.
ADsubTRS1 late mRNAs and proteins accumulate to nearly normal levels, but some proteins fail to localize properly.
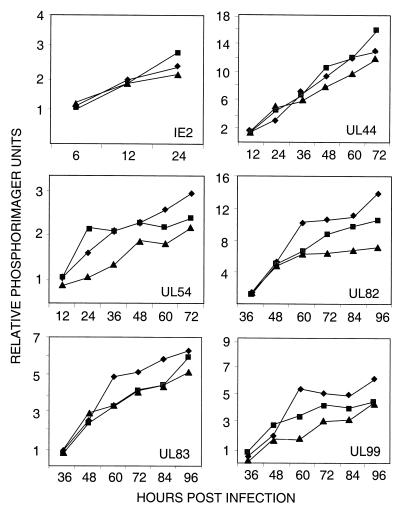
Earlier work employing transient assays showed that pIRS1 and pTRS1 could activate early and late HCMV promoters (8, 10, 19, 23). To test the possibility that ADsubTRS1 failed to express normal amounts of late mRNAs, fibroblasts were infected at a multiplicity of infection of 0.1 PFU/cell and the accumulation of a representative set of mRNAs (immediate early, IE2; early, UL44 and UL54; late, UL82, UL83, and UL99) was analyzed by Northern blot assay. The resulting RNA-specific bands were then quantified with a phosphorimager (Fig. 4). Although several mRNAs were produced in slightly reduced quantities after infection with ADsubTRS1 compared to quantities produced after infection with ADwt or ADsubIRS1, the reduction was never more than twofold.
FIG. 4.
Accumulation of viral mRNAs after infection with wild-type or mutant virus. Fibroblasts were infected at a multiplicity of infection of 0.1 PFU/cell with ADwt (♦), ADsubIRS1 (▪), or ADsubTRS1 (▴). Total cellular RNA was prepared at the indicated times, viral mRNAs were assayed by Western blotting with 32P-labeled probe DNAs, and radioactivity relative to that for a cellular mRNA (cPLA2) was quantified with a phosphorimager. Similar results were obtained in two independent experiments.
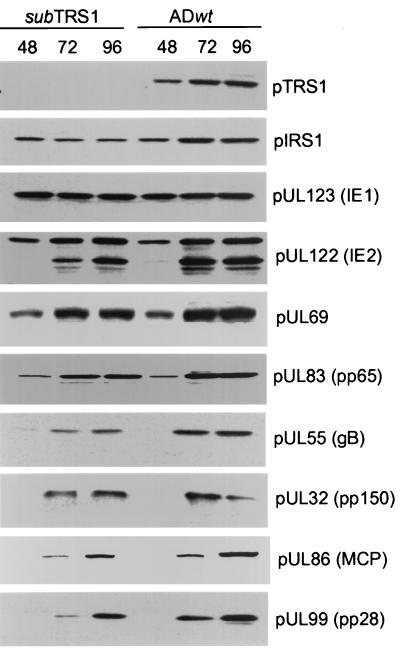
Protein accumulation was monitored by Western blotting, again after infection at a multiplicity of infection of 0.1 PFU/cell (Fig. 5). As expected, pIRS1 was made, but no pTRS1 was detected in ADsubTRS1-infected cells. Immediate-early (IE1 and IE2), early (pUL69), and late proteins (pUL32, pUL55, pUL82, pUL83, pUL86, and pUL99) accumulated to approximately wild-type levels. Although the levels of pUL55 and pUL99 were somewhat reduced after infection with ADsubTRS1 in the experiment displayed, this reduction was not reproduced in repeated experiments.
FIG. 5.
Accumulation of viral proteins after infection with wild-type or mutant virus. Fibroblasts were infected at a multiplicity of infection of 0.5 PFU/cell with ADwt or ADsubTRS1. Cell extracts were prepared at the indicated times (hours postinfection), and viral proteins were assayed by Western blotting with antibodies specific for the indicated proteins. gB, glycoprotein B; MCP, major capsid protein.
Late proteins pUL82 and pUL83 are abnormally localized in ADsubTRS1-infected cells.
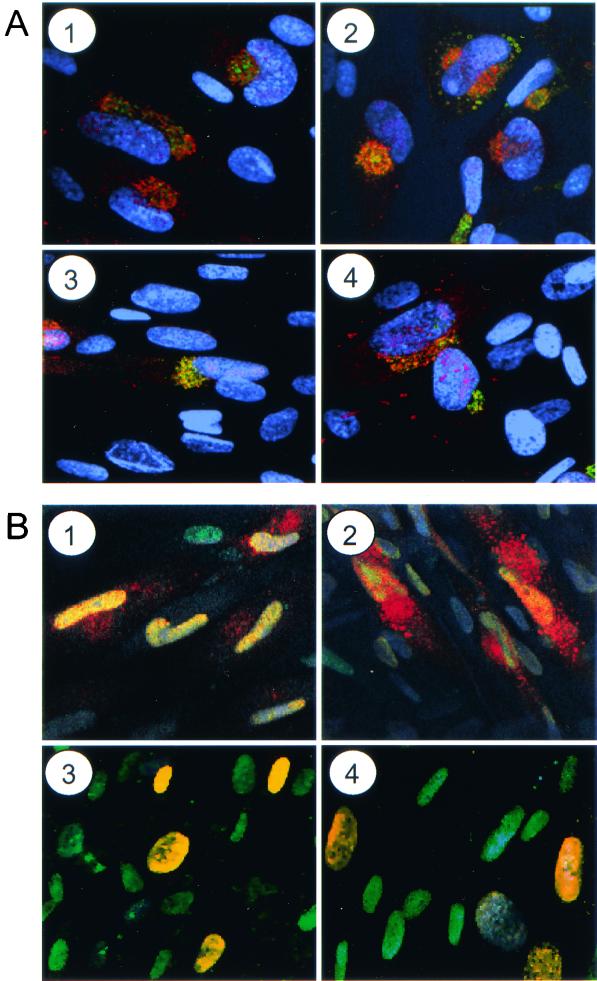
Since the late mRNAs and proteins that were tested accumulated to nearly normal levels in the absence of pTRS1 (Fig. 4 and 5) but less infectious virus was made in ADsubTRS1-infected cells than in wild-type-virus-infected cells (Fig. 3B), we considered the possibility that late proteins might be improperly localized and examined the distribution of several late proteins within ADsubTRS1-infected cells. The tegument protein, pUL99 (also termed pp28), and envelope protein, pUL55 (also termed glycoprotein B), normally accumulate in the cytoplasm of HCMV-infected cells (22). Their locations after infection with ADsubTRS1 could not be distinguished by immunofluorescence analysis from their positions in ADwt-infected cells (Fig. 6A and data not shown). Both proteins resided in the cytoplasm near a Golgi marker. Two other tegument proteins, pUL82 (also termed pp71) and pUL83 (also termed pp65), normally accumulate predominantly in the nucleus in association with the nuclear matrix (6, 21). The location of pUL83 in ADsubTRS1-infected cells was altered in compared to that in ADwt-infected cells. Immunofluorescence analysis at 60 h after infection with the wild-type virus revealed that, although pUL83 resided predominantly in the nucleus, it also was detected in the cytoplasm (Fig. 6B, 1). In contrast, pUL83 appeared to be located exclusively in the nucleus at this time after infection with ADsubTRS1 (Fig. 6B, 3). The protein was also present in both the nucleus and cytoplasm at 72 h after infection with ADwt (Fig. 6B, 2), and it was again predominantly nuclear in ADsubTRS1-infected cells at this time (Fig. 6B, 4). Similar results were obtained for pUL82 (data not shown).
FIG. 6.
Localization of late viral proteins in infected cells. Fibroblasts were infected at a multiplicity of infection of 0.5 PFU/cell with ADwt (1 and 2) or ADsubTRS1 (3 and 4). Cells were fixed 60 (1 and 3) or 72 h (2 and 4) later and treated with RNase A. pUL99 (A) was visualized by using a specific monoclonal antibody plus a fluorescein-conjugated secondary antibody (green); Golgi bodies were stained with an antibody to a Golgi-specific marker (mannosidase II) plus a Cy5-conjugated secondary antibody (red), and DNA was stained with Hoechst 33342 dye (blue). Limited colocalization of pUL99 and mannosidase II produces a yellow signal. pUL83 (B) was detected with a specific antibody plus a Cy5-conjugated secondary antibody (red), and DNA was stained with YOYO-1 (green). Colocalization of pUL83 and nuclear DNA yields a yellow signal.
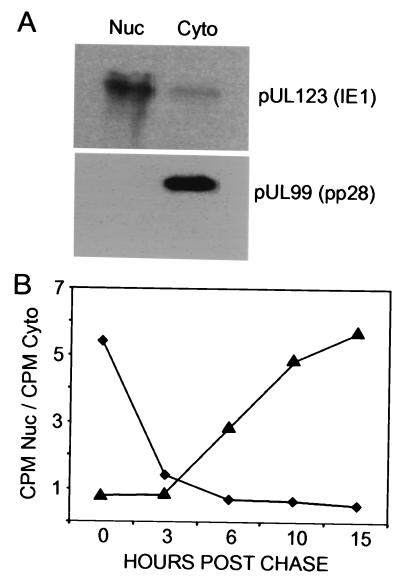
To monitor the kinetics of pUL83 localization after its synthesis, cells were pulse-labeled with [35S]methionine plus [35S]cysteine for 10 min and then chased for several hours after infection with the mutant or wild-type virus at a multiplicity of infection of 0.5 PFU/cell. Cells were then fractionated into nuclear and cytoplasmic compartments, and, to control for successful separation, the locations of nuclear IE1 protein and cytoplasmic pUL99 were assayed. Although about 5% of the IE1 proteins were found in the cytoplasmic fraction, the bulk of the two proteins partitioned into the proper fractions (Fig. 7A). The pUL83 in the two fractions was then isolated by immunoprecipitation, and the band corresponding to pUL83 was quantified with a phosphorimager. The trafficking of newly synthesized pUL83 in the mutant virus-infected cells was markedly different from that in wild-type-virus-infected cells (Fig. 7B). In ADwt-infected cells, the ratio of nuclear to cytoplasmic pUL83 was 5.5 at the end of the 10-min pulse, indicating that most protein synthesized during the brief labeling period had moved from its site of synthesis in the cytoplasm to the nucleus. After a 3-h chase, the ratio was 1.4, and after longer periods it was reduced to 0.7, showing that somewhat more pUL83 was in the cytoplasm than in the nucleus after an extended chase period. In ADsubTRS1-infected cells, the ratio of nuclear to cytoplasmic pUL83 was 0.7 after the pulse period, indicating that much of the newly synthesized protein remained in the cytoplasm. Then the ratio slowly increased until it reached 5.8 after 15 h of chase. Thus, there are two differences between ADsubTRS1 and its wild-type parent. Newly synthesized pUL83 rapidly localizes to the nucleus and then moves back to the cytoplasm in ADwt-infected cells, whereas most of it translocates slowly to the nucleus and then remains there in ADsubTRS1-infected cells.
FIG. 7.
Kinetic analysis of pUL83 location after synthesis in infected cells. (A) Fractionation of cells. Fibroblasts were infected with ADwt at a multiplicity of infection of 0.05 PFU/cell. At 72 h later, cells were subjected to mechanical fractionation in hypotonic buffer, separating nuclear (Nuc) and cytoplasmic (Cyto) fractions. The locations of the nuclear pUL123 (IE1) and cytoplasmic pUL99 viral proteins were ascertained by Western blot assay. (B) Localization of pUL83. Fibroblasts were infected at a multiplicity of infection of 0.05 PFU/cell with ADwt (♦) or ADsubTRS1 (▴). At 65 h later, cells were labeled with [35S]methionine plus [35S]cysteine for 10 min, washed, and then chased in the presence of unlabeled methionine plus cysteine. Lysates were prepared at the indicated times, and immunoprecipitated pUL83 from nuclear and cytoplasmic fractions was analyzed by electrophoresis. The radioactivity in pUL83-specific bands was quantified with a phosphorimager, and the ratio of 35S-labeled pUL83 in the nucleus to that in the cytoplasm was calculated.
ADsubTRS1 produces abnormal virus particles.
Some late viral proteins were aberrantly localized in the absence of newly synthesized pTRS1 (Fig. 6 and 7), so we considered the possibility that defective particles lacking some of the appropriate protein constituents were assembled. Initially, we determined the relative PFU/genome ratios for ADsubTRS1, ADsubIRS1, and ADwt. Extracellular virus produced in noncomplementing fibroblasts was partially purified by sedimentation through a sorbitol cushion. The relative genome numbers were estimated in a slot blot experiment, and infectivity was assayed by plaque assay on fibroblasts that were previously infected with retroTRS1. The PFU/genome ratios for ADwt and ADsubIRS1 were similar (12.3 and 11.4, respectively), whereas the ratio for ADsubTRS1 was decreased by a factor of ∼10 (1.2).
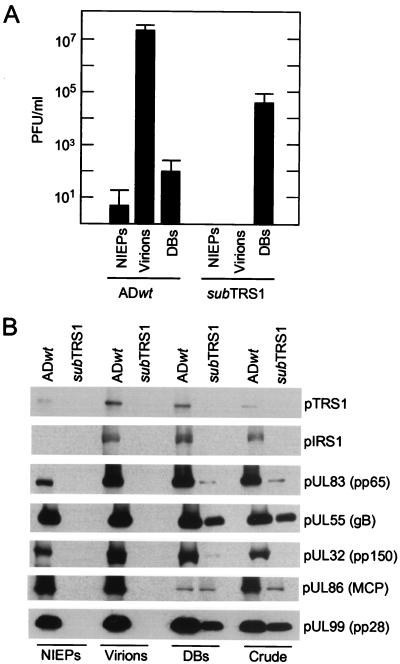
To further characterize the infectious particles produced in ADsubTRS1-infected fibroblasts, mutant and wild-type virus particles from the medium of infected cultures were pelleted through sorbitol cushions and then sedimented in glycerol-tartrate gradients. Three types of particles produced by wild-type virus have been described previously (2, 7, 25): infectious virions, NIEPs, and noninfectious DBs. NIEPs contain empty capsids, whereas DBs do not contain organized capsids. These three classes of particles were obtained from the ADwt virus preparation, but only material sedimenting at the position of DBs was evident in gradients that received ADsubTRS1 virus. All of the ADsubTRS1 infectivity was located in the DB fractions (Fig. 8A), and this infectivity accounted for all of the gradient's input infectivity.
FIG. 8.
Infectivity and protein constituents of wild-type and mutant virus particles. Virus particles from the medium of fibroblasts infected with ADwt or ADsubTRS1 were partially purified by sedimentation through a sorbitol cushion to produce partially purified particles (crude). The particles in the crude fraction were further purified by sedimentation in a glycerol-tartrate gradient. Virions, NIEPs, and (DBs) were separated on the gradient containing the ADwt preparation. No virions or NIEPs were evident in the ADsubTRS1 preparation, and the regions of the gradient that would normally contain these particles were collected in blind fashion and processed. (A) The infectivity present in fractions from glycerol-tartrate gradients was analyzed by plaque assay on retroTRS1-infected fibroblasts. Three independent experiments were performed, and the averages plus standard deviations are displayed. (B) Viral proteins in gradient fractions were analyzed by Western blotting with antibodies to the indicated proteins. gB and MCP are as defined for Fig. 5.
Several viral structural proteins in the material that was pelleted through the sorbitol cushion (crude fraction) and in the three fractions obtained from the glycerol-tartrate gradient were assayed (Fig. 8B). No viral proteins were detected in the NIEP and virion fractions derived from the ADsubTRS1 infection, consistent with our inability to observe particles in the glycerol-tartrate gradients. Substantial amounts of pUL99 and pUL55 were present in the crude and DB fractions from both mutant and wild-type virus preparations. The ADsubTRS1 crude fraction contained detectable pUL86 (also termed major capsid protein), but much less than was found in the crude fraction from ADwt. The two viruses contained similar, modest amounts of pUL86 in the DB fraction, consistent with earlier work showing that DBs do not contain organized capsid structures (17). pUL83 was found in both the crude and DB fractions from ADsubTRS1, but the amounts were less than those in the ADwt-derived fraction. Finally, although a substantial amount of pUL32 (also termed pp150) was present in wild-type DBs, none was detected in the equivalent fraction from the mutant.
DISCUSSION
The loss of pIRS1 had no discernible effect on the replication of ADsubIRS1 in human fibroblasts (Fig. 2A), consistent with earlier work showing that blocks of HCMV genes, including IRS1, could be deleted with little effect on virus growth (4, 9). ADsubIRS1 failed to produce not only pIRS1 but also pIRS1263 (Fig. 2B). pIRS1263 can antagonize the transcriptional activation function of pIRS1 and pTRS1 (19), but its addition to ADsubIRS1-infected cells did not detectably influence viral replication (Fig. 2B). Consequently, we do not detect a phenotype resulting from the loss of pIRS1 plus pIRS1263 or pIRS1 alone. It is likely that pTRS1 duplicates all pIRS1 functions needed for growth in fibroblasts. The N-terminal 549 amino acids of the two proteins are identical, and, although their C-terminal regions diverge, they nevertheless remain 55% identical in the divergent region, as well.
In contrast to pIRS1, pTRS1 is required for efficient growth of HCMV in fibroblasts (Fig. 2C). The growth defect of ADsubTRS1 is multiplicity of infection dependent, being greater at lower multiplicities of infection. ADsubTRS1 DNA accumulation is normal (Fig. 3A), arguing that most immediate-early and early viral functions are intact. All of the late viral mRNAs and proteins that were assayed accumulated to nearly normal levels in ADsubTRS1-infected fibroblasts (Fig. 4 and 5), but two virion tegument proteins, pUL82 and pUL83, were aberrantly localized (Fig. 6 and data not shown). Whereas the proteins were present in both the nuclei and cytoplasm of ADwt-infected cells, they were detected only in the nuclear compartment at 60 h, and primarily in the nucleus at 72 h, after infection with ADsubTRS1. A pulse-chase analysis of pUL83 confirmed its altered localization in mutant virus-infected cells (Fig. 7B). In fibroblasts infected with ADwt, newly synthesized pUL83 rapidly accumulated in the nucleus and then slowly moved to the cytoplasm. Presumably, pUL83 associates with newly assembled capsids as well as other tegument proteins that accumulate in the nucleus; then, after passing through the inner and outer nuclear envelope, it reenters the cytoplasm as part of a capsid-tegument protein complex. The complex subsequently buds through a virus-modified membrane to produce infectious virions (reviewed in reference 22). We assume that cytoplasmic pUL83 is detected by immunofluorescence in wild-type-virus-infected cells (Fig. 6) either because the capsid-tegument protein complex is not immediately packaged or not quantitatively packaged into virus particles or because newly assembled particles are disrupted by the fixation process. In contrast to what is found for the wild-type situation, much of the newly synthesized pUL83 in ADsubTRS1-infected cells translocated slowly to the nucleus and then stayed there (Fig. 7B). The failure of pUL83 to efficiently depart the nucleus likely reflects a breakdown in the assembly process. Conceivably, pUL83 and perhaps other viral structural proteins reach the nuclei of ADsubTRS1-infected cells in an abnormal state that is not competent for assembly into a capsid-tegument complex that is able to move out of the nucleus. The failure of the majority of pUL83 proteins to exit the nucleus in the pulse-chase experiment probably explains the absence of a cytoplasmic signal for the protein in the immunofluorescence assay (Fig. 6).
A defect in assembly is consistent with the failure of ADsubTRS1 to accumulate normal quantities of intracellular infectious particles (Fig. 3B). A defect in assembly also fits well with the observation that ADsubTRS1 produces particles of reduced infectivity that sediment abnormally in a glycerol-tartrate gradient (Fig. 8A).
What is the role of pTRS1 late after infection? It might act indirectly to influence assembly of virus particles. Both pIRS1 and pTRS1 have been shown to activate transcription in cooperation with other viral activators (8, 10, 19, 23). pTRS1 might be required to efficiently activate the expression of a late protein whose accumulation we did not monitor and that functions during assembly. Alternatively, pTRS1 might function directly in assembly. For example, it could serve as a chaperone, helping to properly fold pUL83, assemble it into a complex, or to deliver it to the nucleus in a functional state. A failure to properly fold pUL83 could explain our inability to detect newly synthesized pUL83 in the cytoplasm of ADsubTRS1-infected cells (Fig. 6), even though its movement to the nucleus is delayed (Fig. 7B).
Although transfection experiments with reporter genes have indicated that pIRS1 and pTRS1 can cooperate with other immediate-early proteins to activate early and late viral promoters (8, 10, 19, 23), we failed to detect a significant alteration in the accumulation of the set of viral mRNAs in ADsubTRS1-infected cells that were tested (Fig. 4). Since pTRS1 is packaged in the virion, ADsubTRS1 made in complementing cells should contain the protein, but we have not purified a sufficient quantity of infectious ADsubTRS1 particles to confirm this supposition. Nevertheless, if there are viral genes that depend for their normal activation on pTRS1 delivered in virions but not on newly synthesized pTRS1, then they might have been missed in our assays. We examined only a small subset of viral mRNAs, and we may have failed to assay one or more viral genes that are dependent on TRS1 for their full activation. Finally, since both pIRS1 and pTRS1 are able to elevate the expression of reporter constructs, it is possible that the retention of just one of the two proteins is sufficient for transcriptional activation. We are currently constructing a virus unable to express either pIRS1 or pTRS1 to test the hypothesis that a transcriptional activation function resides in the conserved N-terminal domains of pIRS1 and pTRS1.
Acknowledgments
We thank W. Gibson for a generous gift of antibodies to HCMV proteins and J. Adamo and W. Brune for critical reading of the manuscript.
This work was supported by a grant from the National Cancer Institute (CA85786).
REFERENCES
- 1.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (UL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chee, M. S., et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 4.Greaves, R. F., J. M. Brown, J. Vieira, and E. S. Mocarski. 1995. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J. Gen. Virol. 76:2151-2160. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for the efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hensel, G. M., H. H. Meyer, I. Buchmann, D. Pommerehne, S. Schmolke, B. Plachter, K. Radsak, and H. F. Kern. 1996. Intracellular localization and expression of the human cytomegalovirus matrix phosphoprotein pp71 (ppUL82): evidence for its translocation into the nucleus. J. Gen. Virol. 77:3087-3097. [DOI] [PubMed] [Google Scholar]
- 7.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 8.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of seven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, T. R., and V. P. Muzithras. 1992. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5 and the US6 family. J. Virol. 66:2541-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerry, J. A., M. A. Priddy, T. Y. Jervey, C. P. Kohler, T. L. Staley, C. D. Vanson, T. R. Jones, A. C. Iskenderian, D. G. Anders, and R. M. Stenberg. 1996. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J. Virol. 70:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 12.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 13.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132:325-338. [DOI] [PubMed] [Google Scholar]
- 14.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Patterson, C. E., and Shenk, T. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowe, W. P., J. W. Hartley, S. Waterman, H. C. Turner, and R. J. Huebner. 1956. Cytopathic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc. Soc. Exp. Biol. Med. 92:418-424. [PubMed] [Google Scholar]
- 21.Sanchez, V., P C. Angeletti, J. A. Engler, and W. J. Britt. 1998. Localization of human cytomegalovirus structural proteins to the nuclear matrix of infected human fibroblasts. J. Virol. 72:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter required the a gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stinski, M. F. 1976. Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J. Virol. 19:594-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talbot, P., and J. D. Almeida. 1977. Human cytomegalovirus: purification of enveloped virions and dense bodies. J. Gen. Virol. 36:345-349. [DOI] [PubMed] [Google Scholar]
- 26.Weston, K., and B. G. Barrell. 1986. Sequence of the short unique region, short repeats, and part of the long repeats of human cytomegalovirus. J. Mol. Biol. 192:177-208. [DOI] [PubMed] [Google Scholar]
- 27.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]