Abstract
A single phosphorylation event at T-antigen residue Thr124 regulates initiation of simian virus 40 DNA replication. To explore this regulatory process, a series of peptides were synthesized, centered on Thr124. These peptides contain a nuclear localization signal (NLS) and a recognition site for cyclin/Cdk kinases. When unphosphorylated, the “CDK/NLS” peptides inhibit T-antigen assembly and bind non-sequence specifically to DNA. However, these activities are greatly reduced upon phosphorylation of Thr124. Similar results were obtained by using peptides derived from the CDK/NLS region of bovine papillomavirus E1. Related studies indicate that residues in the NLS bind to DNA, whereas those in the CDK motif regulate binding. These findings are discussed in terms of the control of T-antigen double hexamer assembly and initiation of viral replication.
Cell cycle-dependent phosphorylation events regulate the initiation of DNA synthesis in eukaryotes (18, 33, 50). At the molecular level, however, relatively little is known about these processes. Therefore, model systems, such as that based on simian virus 40 (SV40), are being used to gain insights into phosphorylation-dependent regulation of DNA replication. Initiation of SV40 DNA replication depends upon the binding of the virally encoded T-antigen (T-ag) to the SV40 origin (reviewed in references 3, 17, 18, and 58). Upon binding to the origin, T-ag monomers assemble sequentially into hexamers and then double hexamers (13, 15, 42, 66-68). When assembled into a double hexamer, T-ag becomes a DNA helicase that is able to unwind the SV40 origin (4, 14, 25, 59, 62, 71).
In the regulation of SV40 replication, phosphorylation of T-ag on Thr124 is the sole posttranslational modification required for initiation of replication (44, 57). Formation of the first hexamer is independent of the phosphorylation status of Thr124; therefore, the phosphorylation of Thr124 is required at a point subsequent to initial hexamer formation (1, 45, 47, 55). Consistent with these studies, on subfragments of the core origin that support double-hexamer formation (35, 61), phosphorylation of Thr124 selectively promotes the formation of the second hexamer (1). The region of T-ag flanking Thr124 contains additional residues that play regulatory roles during SV40 DNA replication. For instance, T-ag residues 126 to 132 contain the nuclear localization signal (NLS) used for nuclear translocation (reviewed in references 22 and 27). Furthermore, the T-ag NLS is flanked by residues that serve as the recognition site for cyclin/Cdk kinases (16, 48), an arrangement found in many other proteins (reviewed in references 26 and 28)
To further explore the regulation of T-ag assembly and initiation of viral replication, a series of peptides derived from the CDK/NLS region of T-ag, centered on Thr124, were synthesized. Initial studies revealed that certain of these peptides bind to DNA in a phosphate-dependent manner and interfered with the formation of T-ag hexamers and double hexamers. To establish whether these observations were unique to T-ag-based peptides, a set of peptides containing the CDK/NLS region of bovine papillomavirus (BPV) E1 were also synthesized. Experiments described in the present study demonstrate that peptides based on the BPV E1 CDK/NLS also bind to DNA and inhibit T-ag oligomerization events, in a phosphate-dependent manner. Finally, to characterize more fully peptide binding to DNA and its regulation, we have conducted experiments with T-ag derived mutant peptides containing alanine substitutions at critical residues (e.g., Thr124). The results from these studies are presented here.
MATERIALS AND METHODS
Commercial supplies of enzymes, DNA, reagents, oligonucleotides, and peptides.
T4 polynucleotide kinase was purchased from Gibco-BRL. Plasmid pBR322 DNA, used as competitor DNA, was purified according to standard procedures (56) and digested with HaeIII purchased from New England Biolabs.
Oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer, purified by electrophoresis through 10% urea-polyacrylamide gels, and isolated as described previously (56, 60). Double-stranded oligonucleotides, 32P labeled at their 5′ termini, were prepared by using standard procedures (56, 60).
Synthesis and purification of peptides.
Peptides were synthesized at the Tufts Core Facility on an Applied Biosystems 431A Peptide Synthesizer by using solid-phase methodologies. After cleavage and deprotection, the samples were ether precipitated, resuspended in distilled H2O, lyopholyzed, and purified by reversed-phase high-pressure liquid chromatography. The peptides were resuspended to 10 mM in equal volumes of 0.5% NH4HCO3 and 2× T-ag storage buffer (1) and then frozen at −20°C until use. Upon resuspension, the pH of all peptides was between 7.75 and 8.0.
EMSA.
T-ag-based electrophoretic mobility shift assays (EMSAs) (49) were conducted by using SV40 in vitro replication conditions (72); T-ag was isolated and stored as previously described (72). The reaction mixtures (20 μl) contained 7 mM MgCl2, 0.5 mM dithiothreitol (DTT), 4 mM AMP-PNP, 40 mM creatine phosphate (di-Tris salt; pH 7.6), 0.48 μg of creatine phosphate kinase, 5 μg of bovine serum albumin, 0.8 μg of HaeIII-digested pBR322 (∼6 pmol; used as a nonspecific competitor), ∼25 fmol of double-stranded oligonucleotide (∼10 6 cpm/pmol), 0.5 μg of T-ag (∼6 pmol), and the indicated amounts of peptide. After a 20-min incubation at 37°C, glutaraldehyde was added (0.1% final concentration), and the reactions were further incubated for 5 min. The reactions were stopped by the addition of 5 μl of 6× loading dye II (15% Ficoll, 0.25% bromophenol blue, and 0.25% xylene cyanol) (56) to the samples. The reaction products were applied to 3.5 to 12% gradient polyacrylamide gels and electrophoresed in 0.5% Tris-borate-EDTA (pH 8.4) for ∼1.5 h (10 W). Peptide-based band shift reactions were exactly as described above except that T-ag was omitted from the reactions and the final glutaraldehyde concentration was reduced to 0.05%. Moreover, the samples were loaded on 8% polyacryamide gels and electrophoresed in 0.5% Tris-borate-EDTA (pH 8.4) for ∼3 h (∼200 V). The gels were dried on Whatman 3MM paper, subjected to autoradiography, and quantitated by using a Molecular Dynamics PhosphorImager.
Nitrocellulose filter-binding reactions.
The ability of NLS-based peptides to bind to DNA was also assayed by using previously described filter-binding assays (2, 41, 43, 60). The reaction mixtures (20 μl) contained the same components described in the EMSA section above and the indicated peptides. After incubation for 20 min at 37°C and addition of glutaraldehyde to 0.1%, the mixtures were filtered under suction through alkali-treated nitrocellulose filters (Millipore type HAWP [pore size, 0.45 μm]; stored in 100 mM Tris-HCl [pH 7.5]). The filters were then washed with 5 ml of 100 mM Tris-HCl (pH 7.5), dried, and counted in a Beckman LS 3801 scintillation counter.
RESULTS
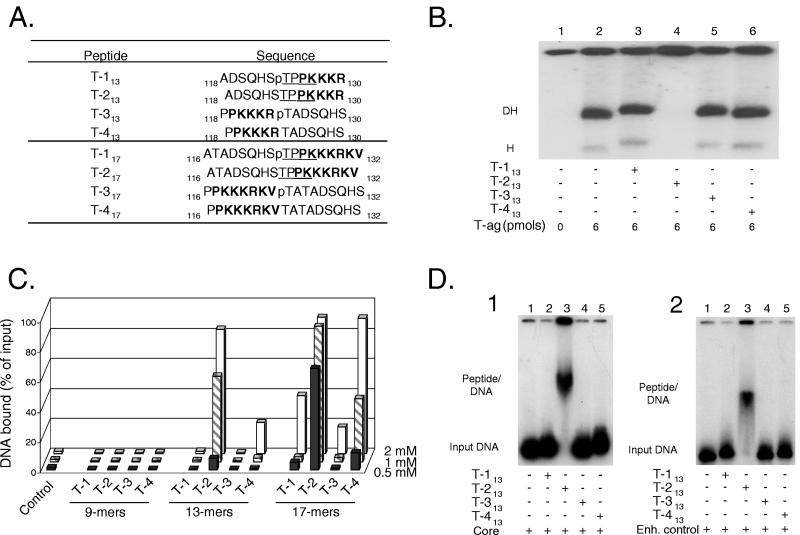
Two sets of T-ag (T)-derived peptides, centered on Thr124, are presented in Fig. 1A (the 13-mer and 17-mer sets). In both sets, peptides T-1 and T-2 differ in that Thr124 is phosphorylated on peptide T-1 but not on peptide T-2. Peptides T-3 and T-4 served as controls; they were formed by switching the amino acids normally positioned on either side of Thr 124 on peptides T-1 and T-2. In Fig. 1A, residues derived from the SV40 NLS are shown in boldface; those forming the consensus cyclin/Cdk recognition site (48) are underlined. In view of these features, these molecules are collectively referred to as CDK/NLS peptides. A third set of peptides centered on Thr124 was also synthesized (the 9-mer set), although it is not presented in Fig. 1A.
FIG. 1.
Studies conducted with the T-ag-based CDK/NLS peptides. (A) Peptides, centered on Thr124, derived from SV40 T-ag (T); the subscripts denote the size of the peptide. The peptide numbering system is based on the corresponding residues in full-length T-ag. Residues known to be part of the T-ag NLS are indicated in boldface; underlined amino acid residues define the recognition motif for the cyclin/Cdk kinase. Phosphorylated Thr residues are denoted by a lowercase “p”. (B) T-ag band shift reactions conducted in the presence of the “13-mer set” of peptides and the 64-bp core oligonucleotide. The reactions in lane 1 were conducted in the absence of either T-ag or peptide (input DNA is not shown). The control reaction displayed in lane 2 was conducted in the presence of T-ag (6 pmol). The reactions in lanes 3 to 6 were conducted in the presence of T-ag (6 pmol) and 20 nmol of the indicated peptides. The positions of T-ag hexamers (H) and double hexamers (DH) are indicated. (C) Detection of peptide-DNA interactions by nitrocellulose filter-binding assays. Reactions were conducted under replication conditions, employing a 64-bp double-stranded oligonucleotide containing the SV40 core origin, in the presence of three different concentrations of peptide (0.5, 1, and 2 mM). The percentage of input DNA bound to a given filter was determined by scintillation counting. As a control, the percentage of input DNA bound to a nitrocellulose filter, in the absence of peptide, was also determined (control). (D) Peptide band shift reactions conducted with the 13-mer set of peptides. The reactions in panel D1 were conducted under replication conditions in the presence of the 64-bp double-stranded oligonucleotide containing the SV40 core origin (core). The position of the input DNA is indicated in lane 1. The reaction products formed in the presence of peptides T-113, T-213, T-313, and T-413 are presented in lanes 2 to 5, respectively. The reactions in panel D2 were conducted in the presence of a 64-bp non-origin-containing oligonucleotide termed the enhancer control (29). The mobility of the input DNA is shown in lane 1; the reaction products formed in the presence of peptides T-113, T-213, T-313, and T-413 are presented in lanes 2 to 5, respectively. The positions of the peptide-DNA complexes are indicated in both figures.
Initially, we tested whether the 13-mer set of peptides (Fig. 1A) would interfere with T-ag assembly on a 64-bp oligonucleotide containing the SV40 core origin (Fig. 1B). In the absence of peptide, T-ag forms hexamers and double hexamers on this DNA substrate (lane 2). Inspection of lanes 3, 5, and 6 reveals that peptides T-113, T-313, and T-413 (20 nmol of each; final concentration of 1 mM) have little or no effect on T-ag assembly. However, in the presence of peptide T-213 (lane 4), hexamers and double hexamers were not detected. Identical experiments were conducted with the 9-mer and 17-mer sets of peptides. The 9-mer set of peptides did not inhibit T-ag assembly at any concentration tested (data not shown). Furthermore, as with the 13-mer set of peptides, there was no inhibition of T-ag assembly by peptides T-117 or T-317 when the concentration of peptide in the reactions was 1 mM. However, T-ag hexamers and double hexamers were not detected in the presence of peptide T-217 and to a lesser extent by peptide T-417. When these experiments were repeated at 0.5 mM, peptide T-217 continued to inhibit hexamer and double-hexamer formation but peptide T-417 did not (data not shown).
One explanation for the failure to detect hexamers and double hexamers in the presence of the unphosphorylated peptides is that the peptides bind to DNA, thereby blocking subsequent T-ag assembly events. Alternatively, T-ag may assemble on the 64-bp core oligonucleotide but, owing to additional binding of the unphosphorylated peptides to the DNA, the complexes become trapped in the wells. While we have yet to distinguish between these hypotheses, both predict that the unphosphorylated CDK/NLS peptides bind to DNA.
To determine whether peptides derived from the CDK/NLS region of T-ag can bind to DNA.
To test this theory, a series of filter-binding assays were conducted with the T-ag-based CDK/NLS peptides. Results from these studies are presented in Fig. 1C. Inspection of this figure reveals that, over the range of peptide concentrations tested (0.5 to 2 mM), the 9-mer peptides bound at background levels to the 64-bp SV40 core origin containing oligonucleotide. In contrast, experiments conducted with the 13-mer set of peptides demonstrated that peptide T-213 is able to bind to DNA. Other peptides, such as peptide T-113, did not bind to the oligonucleotide at significant levels. However, low levels of binding were observed with peptide T-413 at high concentrations of peptide (e.g., 2 mM). DNA binding was also detected with the 17-mer set of peptides. At 0.5 mM, significant binding was observed only with peptide T-217. Given that peptide T-213 bound relatively poorly at 0.5 mM, whereas peptide T-217 bound significant levels of DNA at the same concentration, it is concluded that peptide T-217 has a higher affinity for DNA than peptide T-213. Moreover, when present at either 1 or 2 mM, peptide T-217 bound nearly all of the substrate DNA; thus, these reactions were saturated. Furthermore, at concentrations in which peptide T-217 binding was saturated, peptide-DNA interactions could be detected with additional members of the 17-mer set of peptides (e.g., peptide T-117 and peptide T-417).
To further characterize the peptide-DNA interactions, a series of “peptide band shift” experiments (Materials and Methods) were conducted (Fig. 1D). The initial set of reactions (Fig. 1) was performed in the presence of the 64-bp oligonucleotide containing the SV40 core origin; the position of the DNA substrate, in the absence of the peptide, is indicated (lane 1). The reaction products formed when the 13-mer set of peptides were added to the reactions are indicated in lanes 2 to 5 (20 nmol of peptide; 1 mM final concentration). It is apparent that peptide T-213 is unique in that it alone binds to DNA (lane 3). To determine whether peptide binding depends upon a high degree of sequence specificity, the experiment in Fig. 1D.1 was repeated with a control oligonucleotide (the 64-bp enhancer control [29]) (Fig. 1D.2). Inspection of lane 3 demonstrates that peptide T-213 is also able to bind the 64-bp enhancer control. Therefore, the binding of peptide T-213 is not sequence specific (an observation confirmed by additional filter-binding assays [data not shown]). As with the reactions conducted in the presence of the core origin, the other members of the 13-mer set of peptides (lanes 2, 4, to 5) did not bind to the 64-bp enhancer control. It is noted that the lane containing peptide T-213 has a relatively large amount of material in the well. This finding suggests that, in addition to the complex labeled “Peptide/DNA,” a larger aggregate formed upon peptide binding to DNA. The experiments presented in Fig. 1D were repeated with 20 nmol of the 17-mer set of peptides (1 mM final concentration), and nearly identical results were obtained. However, binding to DNA was not detected with the 9-mer set of peptides, even at a final concentration of 2 mM (data not shown). Collectively, the experiments in Fig. 1 demonstrate that peptides derived from the CDK/NLS region of T-ag preferentially bind to DNA when they lack a phosphate at Thr 124, they are at least 13 amino acids long and the residues on either side of Thr 124 have not been rearranged.
Testing whether a peptide containing the CDK/NLS motif from BPV E1 binds to DNA.
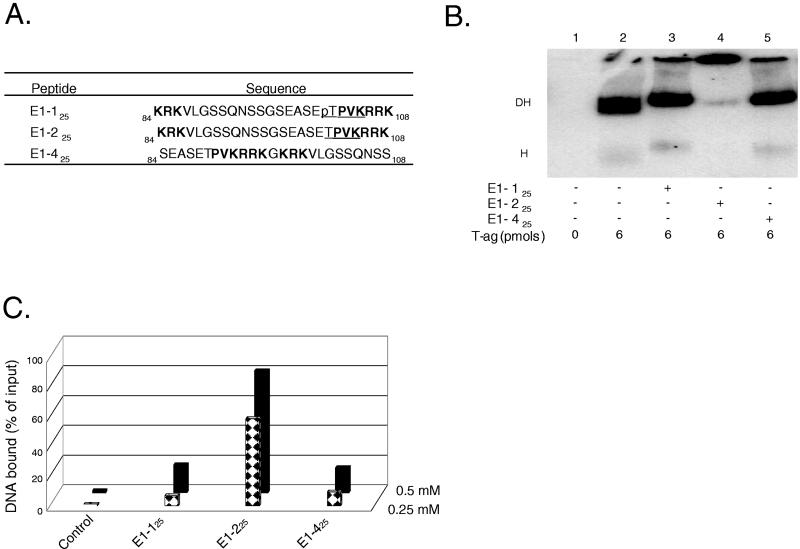
In view of the experiments described in the previous section, it was of interest to establish whether non-T-ag-based CDK/NLS peptides can also bind to DNA. The BPV E1 NLS, extending between residues 84 to 108, has been extensively characterized (38). It is a representative member of the bipartite NLS elements (28), and Thr102 is a putative site for phosphorylation by a cyclin/Cdk kinase (38). Moreover, BPV E1 protein also forms a double hexamer (20, 39). Therefore, two 25-residue-long peptides containing this region were synthesized. Peptide E1-125 contains a phosphate at Thr102, whereas peptide E1-225 does not (Fig. 2A). Furthermore, as a control for peptide E1-225, peptide E1-425 was synthesized by inverting the residues on either side of Gly96 (Fig. 2A).
FIG. 2.
Studies conducted with CDK/NLS peptides derived from BPV E1. (A) Peptides derived from BPV E1 in the vicinity of the putative CDK/NLS element. Peptide numbering is based on the system used to designate residues in BPV E1. Peptides E1-125 and E1-225 both contain the bipartite NLS found in BPV E1; they differ in that peptide E1-125 contains a phosphate (“p”) on Thr102. The control peptide E1-425 was formed by swapping the residues normally found on either side of Gly96. Residues in boldface define the bipartite BPV E1 NLS, and underlined residues represent the putative recognition motif for the cyclin/Cdk kinase (38). (B) T-ag band shift reactions conducted in the presence of the BPV E1 CDK/NLS set of peptides. The reactions in lane 1 were conducted in the absence of either T-ag or peptide (input DNA is not shown). The reaction in lane 2 was conducted in the presence of T-ag (6 pmol). The reactions in lanes 3 to 5 were conducted in the presence of T-ag and 5 nmol of the indicated peptides. (C) Determination of whether peptides derived from the CDK/NLS region of BPV E1 bind to DNA. Nitrocellulose filter-binding assays, conducted under replication conditions, were performed with the indicated peptides and the 64-bp SV40 core origin-containing oligonucleotide. In this series of experiments, two different concentrations of peptide (0.25 and 0.5 mM) were used. The percentage of input DNA bound to a given filter was determined by scintillation counting. As a control, the percentage of input DNA bound to a nitrocellulose filter in the absence of peptide was also determined.
As with the T-ag-based peptides, the E1-based peptides (5 nmol of peptide; final concentration of 0.25 mM) blocked T-ag assembly when unphosphorylated but not when phosphorylated (Fig. 2B, lanes 4 and 3, respectively). Moreover, peptide E1-425, the control peptide for E1-225, did not block T-ag assembly (lane 5). In view of these observations and those made with the T-ag-based peptides (Fig. 1), it was of interest to establish whether peptide E1-225 not only blocked T-ag assembly but also bound to DNA. Therefore, an additional series of filter-binding experiments were conducted (Fig. 2C). Inspection of this figure demonstrates that of the three E1-based peptides that were synthesized, peptide E1-225 preferentially bound to DNA. Indeed, it bound at a relatively low concentration of peptide (0.25 mM); thus, it is a better DNA binder than the T-ag-derived peptide T-217. That peptide E1-225 is a relatively strong DNA binder is also indicated by the observation that EMSA experiments can be performed with peptide E1-225 in the absence of cross-linking with glutaraldehyde (data not shown). In contrast, peptide E1-125, the peptide containing a phosphate on Thr102, binds to DNA at levels similar to the control peptide E1-425. Thus, peptides derived from the NLS regions of both E1 and T-ag bind to DNA in a manner that is regulated by phosphorylation and dependent on an intact CDK/NLS region.
Further insights into the regulation and sequence requirements of the peptide-DNA interactions.
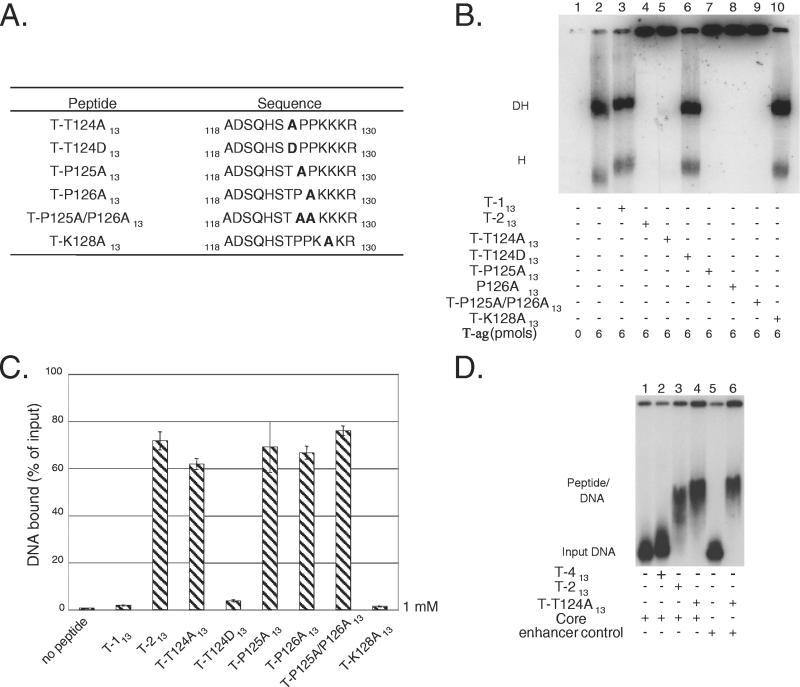
Previous studies demonstrated that T-ag residue Thr124 is involved in the regulation of SV40 DNA replication (1, 44-47, 57), a conclusion supported by the finding that peptide-DNA interactions are regulated by phosphorylation of this residue. In view of these observations, we sought to determine whether Thr124 is essential for DNA binding and whether a Thr-to-Asp substitution could functionally simulate phosphorylation of Thr124. In addition, we wanted to establish whether flanking residues of interest, such as Pro125 or -126, are also required for DNA binding. Interest in these residues stems from the fact that Pro125 is part of the CDK/cyclin recognition site (48), that both residues are required for viral DNA replication (31), and that short proline-rich segments are binding targets of proteins containing WW domains (63). Furthermore, it was of interest to determine whether a mutation in the NLS region, a Lys-to-Ala substitution at position 128, disrupted DNA binding.
To test whether particular residues are essential for DNA binding, derivatives of peptide T-213 were synthesized that contained, in most instances, alanine substitutions at the indicated locations (Fig. 3A). The mutant peptides were termed T-T124A13, T-T124D13, T-P125A13, T-P126A13, T-P125A/P126A13, and T-K128A13 (Fig. 3A). The T-P125A/P126A13 double mutant was synthesized to eliminate the possibility that a single proline would suffice for DNA binding. In an initial series of reactions, we examined whether the mutant set of peptides had any effect on the formation of T-ag hexamers and double hexamers (Fig. 3B). As controls, reactions were conducted in the absence of protein (lane 1) or in the presence of T-ag (6 pmol; lane 2). Additional controls were conducted in the presence of T-ag (6 pmol) and 20 nmol of either peptide T-113 (lane 3) or peptide T-213 (lane 4). As expected, peptide T-213 disrupted standard-hexamer and double-hexamer formation, whereas peptide T-113 did not. (The increased size of the double hexamers formed in the presence of peptide T-113 indicates that this peptide binds to T-ag [see also Fig. 1B]). The reaction products formed in the presence of T-ag (6 pmol) and 20 nmol of the “mutant set” of peptides are shown in lanes 5 to 10. As with peptide T-213, mutant peptides T-T124A13 (lane 5), T-P125A13 (lane 7), T-P126A13 (lane 8), and T-P125A/P126A13 (lane 9) disrupted standard T-ag assembly events. In contrast, mutant peptides T-T124D13 (lane 6) and T-K128A13 (lane 10) had little or no effect on the assembly of T-ag oligomers on DNA.
FIG. 3.
Use of mutant versions of T-ag-derived peptide T-213 to explore peptide-DNA interactions and the regulation of this process. (A) The mutant set of peptides are presented on the left, whereas the sequences of the corresponding peptides are shown on the right. Mutant peptides were formed, in general, by substituting the indicated wild-type residues with alanines (shown in boldface). In the T-P125A/P126A13 double mutant, two alanines replaced the prolines at positions 125 and 126. The T-T124D mutation was formed as a model for phosphorylation of Thr124. (B) Determination of whether the mutant set of peptides disrupted T-ag assembly on a 64-bp oligonucleotide containing the SV40 core origin. The reaction in lane 1 was conducted in the absence of protein (input DNA not shown), whereas the reaction in lane 2 was performed in the presence of T-ag (6 pmol). The locations of T-ag hexamers (H) and double hexamers (DH) are indicated. The reactions in lanes 3 to 10 were conducted in the presence of T-ag and 20 nmol of peptide T-113, T-213, T-T124A13, T-T124D13, T-P125A13, T-P126A13, the T-P125A/P126A13 double mutant, and T-K128A, respectively. (The experiments described below in panel C suggest that materials in the wells in lanes 4, 5, 7, 8, and 9 are due to peptide-DNA complexes.) (C) Detection of interactions between the mutant set of peptides with DNA by nitrocellulose filter-binding assays (the average of three experiments). The reactions contained the 64-bp SV40 core origin oligonucleotide and 20 nmol of the indicated peptides. The percentage of input DNA bound to a given filter was determined by scintillation counting. (D) EMSAs performed with peptide T-T124A13. The products formed in reactions containing the SV40 core origin but lacking peptide are shown in lane 1. As additional controls, the products of reactions conducted in the presence of peptides T-413 and T-213 (20 nmol of each) are shown in lanes 2 and 3, respectively. The reaction products formed in the presence of 20 nmol of peptide T-T124A13 are shown in lane 4. An identical reaction, performed in the presence of the 64-bp enhancer control, is shown in lane 6; the mobility of the 64-bp enhancer control, in the absence of peptide, is shown in lane 5.
Given the results shown in Fig. 3B, additional filter-binding assays were performed to determine which of the mutant set of peptides bind to DNA (Fig. 3C). It is apparent from Fig. 3C, conducted with 20 nmol of the indicated peptides (1 mM final concentration), that peptides T-T124A13, T-P125A13, T-P126A13, and T-P125A/P126A13 bind to DNA, as well as the peptide T-213 control. In contrast, peptides T-T124D13 and T-K128A13 bind to DNA at significantly lower levels, a result consistent with the data in Fig. 3B. To determine whether the mutant peptides that bind to DNA form similar peptide-DNA complexes, as does peptide T-213, additional peptide-gel shift assays were performed. The results obtained with peptide T-T124A13 are shown in Fig. 3D. The reactions in lanes 1 to 4 were conducted with the 64-bp SV40 core origin oligonucleotide. The reaction in lane 1 was performed in the absence of peptide, whereas the reactions displayed in lanes 2 and 3 were conducted in the presence of peptides T-413 and T-213, which served as negative and positive controls, respectively. The reaction in lane 4 was conducted with 20 nmol of peptide T-T124A13; it is apparent that, as with peptide T-213, peptide T-T124A13 binds to DNA and forms a similar peptide-DNA complex. The reactions in lanes 5 and 6 were conducted with the 64-bp enhancer control oligonucleotide. The reaction in lane 6 demonstrates that, as with peptide T-213, the binding of peptide T-T124A13 to DNA requires little or no sequence specificity.
In summary, the experimental results shown in Fig. 3 demonstrate that T-ag residues Thr124, Pro125, and Pro126 are not needed for DNA binding. In contrast, Lys128 is among the residues necessary for this interaction. Furthermore, like peptide T-113 (phosphorylated), peptide T-T124D failed to bind to DNA. Collectively, these observations indicate that the basic residues in the T-ag NLS are important for DNA binding, whereas the residues comprising the cyclin/Cdk element are involved in the regulation of this process. Residues with similar functions may govern the peptide E1-225-DNA interactions.
DISCUSSION
The present studies demonstrate that peptides derived from the CDK/NLS regions of SV40 T-ag and BPV E1 bind to DNA when unphosphorylated. Other classes of peptides that bind to DNA or RNA were previously described. For instance, zinc finger (10, 34, 36, 40), bZip basic region (see, for example, reference 65), α-helix-containing (9), and small acidic (8) peptides all bind to duplex DNA. Peptides that bind to the minor groove of DNA, some preferentially at A/T-rich and GC sequences, have also been described (11, 23, 24, 54, 64). Moreover, peptides that bind to single-stranded DNA have been reported (see, for example, references 7 and 69). In addition, a peptide mimetic of the human immunodeficiency virus type 1 Tat NLS is known to bind to RNA (21). Interest in these molecules reflects, in part, their potential use in gene therapy (37, 40). Regarding the sequence specificity of peptide binding to duplex DNA, the α-helix and zinc finger peptides recognized 5 and 3 bp, respectively (9, 51). Whether the CDK/NLS-based peptides have limited sequence specificity has yet to be determined. However, given that the CDK/NLS peptides are among the smallest DNA binders, this and related issues merit further investigation.
Upon phosphorylation of the T-ag-derived peptides on Thr124 or the BPV-E1-derived peptides on Thr102, the peptides bind to DNA at greatly reduced levels. Therefore, phosphorylation serves as part of a switch that downregulates DNA binding. The underlying mechanism(s) responsible for the phosphorylation-dependent switch is currently not understood. One possibility is that upon phosphorylation, the CDK/NLS peptides no longer bind to DNA owing to electrostatic repulsion. This conclusion is supported by the observation that an aspartic acid at position 124 substitutes for a phosphorylated threonine and by previous studies of the SV124E T-ag mutation (57). A related possibility is that phosphorylation or the presence of a negative charge induces structural changes that disrupt DNA binding. This hypothesis is consistent with studies indicating that phosphorylation of Ser/Thr residues regulates the cis/trans isomerization rate of Ser/Thr-Pro bonds (reviewed in references 19 and 73). Indeed, peptide bond isomerization has been compared to a mechanical on-off switch (reviewed in reference 19).
Regarding the function(s) of CDK/NLS motifs in full-length proteins, it has been reported that most NLS elements do not bind to DNA (12). Nevertheless, certain viral proteins have NLS elements that bind to nucleic acids (reviewed in reference 32). Furthermore, nonviral proteins are also known to contain NLS motifs that bind to DNA. For example, the NLS in c-fos is used for DNA binding (5, 12). Whether the CDK/NLS motifs in full-length T-ag and BPV-E1 are DNA-binding elements has yet to be determined. However, Prives et al. (53) previously speculated that T-ag residues 125 through 131 might be involved in DNA binding. This possibility is also consistent with the observation that T-ag residues 121 to 135, a region spanning the T-ag NLS, are required for untwisting the A/T-rich region in the SV40 core origin (6). In addition, previous studies of mutant T-ag molecules have demonstrated that residues in the CDK/NLS motif are required for both nuclear entry and some aspect of viral replication. For instance, T-ag molecules containing the P125S mutation (52), the P126S and P126L T-ag mutations (6, 31, 52), the K127T or K127I mutations (30), and the K129T and R130I mutations (31) enter the nucleus but fail to support SV40 replication. Thus, a number of studies, including the present one, are consistent with the hypothesis that the CDK/NLS region in T-ag (and perhaps other proteins) plays two important roles: namely, nuclear entry and an additional function in DNA replication that may include cell cycle-dependent binding to DNA.
Finally, it is of interest to consider how cell cycle-dependent binding of the T-ag CDK/NLS region to DNA might regulate viral replication. It was previously noted that phosphorylation of Thr124 is not required for assembly of the initial hexamer (1, 45, 47, 57). In contrast, on origin subfragments containing single assembly units (61), phosphorylation of Thr124 promotes the assembly of the second hexamer (1). Furthermore, on the full-length core origin, “proper” interactions between hexamers are required for origin unwinding (70). In view of the observations presented here, it is proposed that in the initial hexamer, region(s) of T-ag encompassing the CDK/NLS motif(s) bind to DNA and thereby block the proper assembly of the double hexamer. However, upon cell cycle-dependent phosphorylation of Thr124, residues comprising the CDK/NLS motif are released from DNA. As a result, the second hexamer is free to complete its assembly on the core origin. A related possibility is that binding of the CDK/NLS region to DNA may help to inhibit viral replication during non-S phases of the cell cycle. Subsequent studies will be required to test these models and to determine whether the assembly of other viral initiators is regulated in a similar manner.
Acknowledgments
R.J.K. and S.M. contributed equally to this study.
We thank Anita Hong and James Sudmeier for advice on resuspending the peptides, Brett Barbaro and Andrea Prack for help with preliminary studies, Bruce Malcom for helpful conversations, and A. J. Bullock for comments on the manuscript.
This study was funded by a grant from the National Institutes of Health (9R01GM55397).
REFERENCES
- 1.Barbaro, B. A., K. R. Sreekumar, D. R. Winters, A. E. Prack, and P. A. Bullock. 2000. Phosphorylation of simian virus 40 T-antigen on Thr124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borowiec, J. A., and J. Hurwitz. 1988. ATP stimulates the binding of the simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc. Natl. Acad. Sci. USA 85:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 4.Bullock, P. A., Y. S. Seo, and J. Hurwitz. 1991. Initiation of simian virus 40 DNA synthesis in vitro. Mol. Cell. Biol. 11:2350-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L., J. N. M. Glover, P. G. Hogan, A. Rao, and S. C. Harrison. 1998. Structure of the DNA-binding domains form NFAT, Fos, and Jun bound specifically to DNA. Nature 392:42-48. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., W. S. Joo, P. A. Bullock, and D. T. Simmons. 1997. The N-terminal side of the origin-binding domain of simian virus 40 large T antigen is involved in A/T untwisting. J. Virol. 71:8743-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, X., B. K. Kay, and R. L. Juliano. 1996. Identification of a biologically significant DNA-binding peptide motif by use of a random phage display library. Gene 171:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Chillemi, F., G. Lugaro, D. Boari, E. Cardellini, M. Bramucci, A. Miano, D. Amici, G. L. Gianfranceschi, and E. Durban. 1991. Acidic pentapeptide phosphorylated in vitro by calf thymus protein kinase NII binds to DNA in the presence of Mg2+ cations. FEBS Lett. 291:67-70. [DOI] [PubMed] [Google Scholar]
- 9.Chin, J. W., and A. Schepartz. 2001. Concerted evolution of structure and function in a miniature protein. J. Am. Chem. Soc. 123:2929-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo, Y., I. Sánchez-Garcia, and A. Klug. 1994. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature 372:642-645. [DOI] [PubMed] [Google Scholar]
- 11.Churchill, M. E. A., and A. A. Travers. 1991. Protein motifs that recognize structural features of DNA. Trends Biochem. Sci. 16:92-97. [DOI] [PubMed] [Google Scholar]
- 12.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, F. B., J. A. Borowiec, T. Eki, and J. Hurwitz. 1992. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 267:14129-14137. [PubMed] [Google Scholar]
- 14.Dean, F. B., P. Bullock, Y. Murakami, C. R. Wobbe, L. Weissbach, and J. Hurwitz. 1987. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc. Natl. Acad. Sci. USA 84:16-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean, F. B., M. Dodson, H. Echols, and J. Hurwitz. 1987. ATP-dependent formation of a specialized nucleoprotein structue by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc. Natl. Acad. Sci. USA 84:8981-8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draettta, G. 1990. Cell cycle control in eukaryotes: molecular mechanism of cdc2 activation. Trends Biochem. Sci. 15:378-383. [DOI] [PubMed] [Google Scholar]
- 17.Fanning, E. 1992. Simian virus 40 large T antigen: the puzzle, the pieces, and the emerging picture. J. Virol. 66:1289-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, G. 2000. Chemical aspects of peptide bond isomerisation. Chem. Soc. Rev. 29:119-127. [Google Scholar]
- 20.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 21.Friedler, A., D. Friedler, N. W. Luedtke, Y. Tor, A. Loyter, and C. Gilon. 2000. Development of a functional backbone cyclic mimetic of the HIV-1 Tat arginine-rich motif. J. Biol. Chem. 275:23783-23789. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Bustos, J., J. Heitman, and M. N. Hall. 1991. Nuclear protein localization. Biochem. Biophys. Acta 1071:83-101. [DOI] [PubMed] [Google Scholar]
- 23.Geierstanger, B. H., M. Mrksich, P. B. Dervan, and D. E. Wemmer. 1994. Design of a GC-specific DNA minor groove-binding peptide. Science 266:646-650. [DOI] [PubMed] [Google Scholar]
- 24.Geierstanger, B. H., B. F. Volkman, W. Kremer, and D. E. Wemmer. 1994. Short peptide fragments derived from HMG-I/Y proteins bind specifically to the minor groove of DNA. Biochemistry 33:5347-5355. [DOI] [PubMed] [Google Scholar]
- 25.Goetz, G. S., F. B. Dean, J. Hurwitz, and S. W. Matson. 1988. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263:383-392. [PubMed] [Google Scholar]
- 26.Jans, D. A. 1995. The regulation of protein transport to the nucleus by phosphorylation. Biochem. J. 311:705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jans, D. A., C. K. Chan, and S. Huebner. 1998. Signals mediating nuclear targeting and their regulation: application in drug delivery. Med. Res. Rev. 18:189-223. [DOI] [PubMed] [Google Scholar]
- 28.Jans, D. A., and S. Hübner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 29.Joo, W. S., H. Y. Kim, J. D. Purviance, K. R. Sreekumar, and P. A. Bullock. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear locataion of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 31.Kalderon, D., and A. E. Smith. 1984. In vitro mutagenesis of a putative DNA binding domain of SV40 large-T. Virology 139:109-137. [DOI] [PubMed] [Google Scholar]
- 32.Kasamatsu, H., and A. Nakanishi. 1998. How do animal DNA viruses get to the nucleus? Annu. Rev. Microbiol. 52:627-686. [DOI] [PubMed] [Google Scholar]
- 33.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 34.Kim, C. A., and J. M. Berg. 1995. Serine at position 2 in the DNA recognition helix of a Cys2-His2 zinc finger peptide is not, in general, responsible for base recognition. J. Mol. Biol. 252:1-5. [DOI] [PubMed] [Google Scholar]
- 35.Kim, H. Y., B. A. Barbaro, W. S. Joo, A. Prack, K. R. Sreekumar, and P. A. Bullock. 1999. Sequence requirements for the assembly of simian virus 40 T-antigen and T-antigen origin binding domain on the viral core origin of replication. J. Virol. 73:7543-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, J.-S., and C. O. Pabo. 1998. Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc. Natl. Acad. Sci. USA 95:2812-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, J.-S., and C. O. Pabo. 1997. Transcriptional repression by zinc finger peptides. J. Biol. Chem. 272:29795-29800. [DOI] [PubMed] [Google Scholar]
- 38.Lentz, M. R., D. Pak, I. Mohr, and M. R. Botchan. 1993. The E1 replication protein of bovine papillomavirus type 1 contains an extended nuclear localization signal that includes a p34cdc2 phosphorylation site. J. Virol. 67:1414-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, J.-S., S.-R. Kuo, A. M. Makhov, D. M. Cyr, J. D. Griffith, T. R. Broker, and L. T. Chow. 1998. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J. Biol. Chem. 273:30704-30712. [DOI] [PubMed] [Google Scholar]
- 40.Liu, Q., D. J. Segal, J. B. Ghiara, and C. F. Barbas. 1997. Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc. Natl. Acad. Sci. USA 94:5525-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorimer, H. E., E. H. Wang, and C. Prives. 1991. The DNA-binding properties of polyomavirus large T antigen are altered by ATP and other nucleotides. J. Virol. 65:687-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mastrangelo, I. A., P. V. C. Hough, J. S. Wall, M. Dodson, F. B. Dean, and J. Hurwitz. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658-662. [DOI] [PubMed] [Google Scholar]
- 43.McEntee, K., G. M. Weinstock, and I. R. Lehman. 1980. RecA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc. Natl. Acad. Sci. USA 77:857-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McVey, D., L. Brizuela, I. Mohr, D. R. Marshak, Y. Gluzman, and D. Beach. 1989. Phosphorylation of large tumor antigen by cdc2 stimulates SV40 DNA replication. Nature 341:503-507. [DOI] [PubMed] [Google Scholar]
- 45.McVey, D., S. Ray, Y. Gluzman, L. Berger, A. G. Wildeman, D. R. Marshak, and P. Tegtmeyer. 1993. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J. Virol. 67:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McVey, D., B. Woelker, and P. Tegtmeyer. 1996. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J. Virol. 70:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moarefi, I. F., D. Small, I. Gilbert, M. Hopfner, S. K. Randall, C. Schneider, A. A. R. Russo, U. Ramsperger, A. K. Arthur, H. Stahl, T. J. Kelly, and E. Fanning. 1993. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J. Virol. 67:4992-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moreno, S., and P. Nurse. 1990. Substrates for p34cdc2: in vivo veritas? Cell 61:549-551. [DOI] [PubMed] [Google Scholar]
- 49.Murakami, Y., and J. Hurwitz. 1993. DNA polymerase α stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J. Biol. Chem. 268:11018-11027. [PubMed] [Google Scholar]
- 50.Norbury, C., and P. Nurse. 1992. Animal cell cycles and their control. Annu. Rev. Biochem. 61:441-470. [DOI] [PubMed] [Google Scholar]
- 51.Pavletich, N. P., and C. O. Pabo. 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science 252:809-817. [DOI] [PubMed] [Google Scholar]
- 52.Peden, K. W. C., and J. M. Pipas. 1985. Site-directed mutagenesis of the simian virus 40 large T-antigen gene: replication-defective amino acid substitution mutants that retain the ability to induce morphological transformation. J. Virol. 55:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prives, C., B. Barnet, A. Scheller, G. Khoury, and G. Jay. 1982. Discrete regions of simian virus 40 large T antigen are required for nonspecific and viral origin-specific DNA binding. J. Virol. 43:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reeves, R., and M. S. Nissen. 1990. The AT-DNA-binding domain of mammalian high mobility group I chromosomal proteins. J. Biol. Chem. 265:8573-8582. [PubMed] [Google Scholar]
- 55.Reynisdottir, I., H. E. Lorimer, P. N. Friedman, E. H. Wang, and C. Prives. 1993. Phosphorylation and active ATP hydrolysis are not required for SV40 T antigen hexamer formation. J. Biol. Chem. 268:24647-24654. [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 57.Schneider, J., and E. Fanning. 1988. Mutations in the phosphorylation sites of simian virus 40 (SV40) T antigen alter its origin DNA-binding specificity for sites I or II and affect SV40 DNA replication activity. J. Virol. 62:1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 59.Smelkova, N. V., and J. A. Borowiec. 1997. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 71:8766-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sreekumar, K. R., B. A. Barbaro, A. Prack, and P. A. Bullock. 2000. Methods for studying interactions between simian virus 40 T-antigen and the viral origin of replication, p. 49-67. In L. Raptis (ed.), Methods in molecular biology: SV40 protocols. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 61.Sreekumar, K. R., A. E. Prack, D. R. Winters, B. A. Barbaro, and P. A. Bullock. 2000. The simian virus 40 core origin contains two separate sequence modules that support T-antigen double-hexamer assembly. J. Virol. 74:8589-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stahl, H., P. Droge, and R. Knippers. 1986. DNA helicase activity of SV40 large tumor antigen. EMBO J. 5:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Staub, O., and D. Rotin. 1996. WW domains. Structure 4:495-499. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki, M. 1989. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 8:797-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talanian, R. V., C. J. McKnight, R. Rutkowski, and P. S. Kim. 1992. Minimum length of a sequence-specific DNA binding peptide. Biochemistry 31:6871-6875. [DOI] [PubMed] [Google Scholar]
- 66.Valle, M., C. Gruss, L. Halmer, J. M. Carazo, and L. E. Donate. 2000. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 20:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.VanLoock, M. S., A. Alexandrov, X. Yu, N. R. Cozzarelli, and E. H. Egelman. 2002. SV40 large T antigen hexamer structure: domain organization and DNA-induced conformational changes. Curr. Biol. 12:472-476. [DOI] [PubMed] [Google Scholar]
- 68.Virshup, D. M., A. A. R. Russo, and T. J. Kelly. 1992. Mechanism of activation of simian virus 40 DNA replication by protein phosphatase 2A. Mol. Cell. Biol. 12:4883-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voloshin, O. N., L. Wang, and R. D. Camerini-Otero. 1996. Homologous DNA pairing promoted by a 20-amino-acid peptide derived from RecA. Science 272:868-872. [DOI] [PubMed] [Google Scholar]
- 70.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wessel, R., J. Schweizer, and H. Stahl. 1992. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 66:804-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wobbe, C. R., F. Dean, L. Weissbach, and J. Hurwitz. 1985. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc. Natl. Acad. Sci. USA 82:5710-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou, X. Z., P. J. Lu, G. Wulf, and K. P. Lu. 1999. Phosphorylation-dependent prolyl isomerization: a novel signaling regulatory mechanism. Cell. Mol. Life Sci. 56:788-806. [DOI] [PMC free article] [PubMed] [Google Scholar]