Abstract
DNA topoisomerase II (topo II) is involved in unlinking replicating DNA and organizing mitotic chromosomes. Topo II is the target of many antitumour drugs. Topo II inhibition results in extensive catenation of newly replicated DNA and may potentially perturb chromatin assembly. Here, we show that the topo II inhibitor ICRF-193 does not prevent the bulk of nucleosome deposition, but perturbs nucleosome spacing in Xenopus egg extracts. This is due to the trapping of topo II-closed clamps on the DNA rather than increased DNA catenation. Inhibition of replicative DNA decatenation has in itself little or no effect on nucleosome deposition and spacing, suggesting that DNA can easily accommodate the sharp bending constraints imposed by the co-habitation of nucleosomes and catenane nodes. Chromatin perturbation by topo II clamps may explain some dominant cellular effects of ICRF-193. Nucleosome-driven bending of precatenane nodes may facilitate their unlinking by topo II during unperturbed replication.
Keywords: bisdioxopiperazines, DNA catenanes, DNA replication, nucleosomes, topoisomerase II
Introduction
Topoisomerase II (topo II) is a ubiquitous enzyme that is able to transiently break a DNA duplex and transport another DNA duplex through the break in an ATP-dependent manner. Topo II thereby contributes to unlinking DNA during replication, and is required for chromosome condensation and segregation at mitosis (Wang, 2002).
Topo II is the target of numerous antitumour drugs. ‘Topo II poisons', such as etoposide (VP-16), act by stabilizing a covalent topo II–DNA catalytic intermediate named the cleavable complex because it produces a protein-linked DNA break after protein-denaturant treatment. It is thought that cleavable complexes must be processed into irreversible double-strand breaks by cellular machineries to exert their lethality (Li & Liu, 2001). ‘Catalytic inhibitors', such as the bisdioxopiperazine ICRF-193, inhibit topo II without stabilizing cleavable complexes. ICRF-193 traps the enzyme on the DNA in the form of a non-covalent intermediate named the closed clamp (Roca et al, 1994). It is not clear whether the cellular effects of bisdioxopiperazines are due to inhibition of topo II activity or interference of closed clamps with DNA metabolism (Larsen et al, 2003).
Previously we have studied the effect of topo II inhibition on plasmid DNA replication in Xenopus egg extracts. In the presence of ICRF-193, replication intermediates show an excess of parental strand linkage ((+)ΔLk) and highly catenated dimers accumulate (∼1 catenane link per 200 base pairs (bp)), showing that topo II contributes to unlinking DNA during replication (Lucas et al, 2001). There are two possible forms of (+)ΔLk on replication intermediates and, thus, two possible pathways for unlinking replicating DNA. The unreplicated portion may form (+) supercoils, or the two replicated regions may wind around each other to form right-handed intertwinings. The latter will be trapped as (+) catenanes if not removed before termination and are therefore called (+) precatenanes (Schvartzman & Stasiak, 2004). To determine whether topo II unlinks supercoils in front of the forks or precatenanes behind the forks, we trapped topo-II-cleavable complexes with VP-16 and mapped them with respect to replication forks by two-dimensional (2D) gel electrophoresis. Cleavable complexes were detected only behind the replication forks, showing that topo II can unlink replicating DNA by precatenane removal (Lucas et al, 2001). Nevertheless, the universality of the precatenane model is still open to discussion (supplementary information online).
In this study, we have analysed the effect of ICRF-193 on replication-coupled and replication-independent chromatin assembly in Xenopus egg extracts. The results show that ICRF-193 does not prevent the bulk of nucleosome deposition but perturbs nucleosome spacing, because topo II clamps sterically hinder the assembly of a maximal density of nucleosomes. This specific chromatin perturbation may explain some previously reported cellular effects of ICRF-193. Surprisingly, DNA catenation per se has no detectable effect on replicative chromatin assembly. Nucleosomes may thus force precatenanes into a sharply bent conformation, helping their unlinking by topo II.
Results
Effects of ICRF-193 on replicative chromatin
The involvement of topo II in chromatin assembly on plasmid DNA has been studied previously in high-speed supernatants (HSS) of Xenopus eggs (Almouzni & Mechali, 1988a). Supercoiled DNA incubated in HSS is rapidly relaxed by endogenous topoisomerases. Then, the re-isolated DNA appears progressively more (−) supercoiled, due to the relaxation of the (+) supercoils introduced by nucleosome assembly. Camptothecin, a topo I inhibitor, inhibited (−) supercoiling, whereas topo II inhibitors did not. However, a small effect of topo II inhibition might have escaped detection because the most supercoiled topoisomers were not resolved. Furthermore, HSS do not replicate doublestranded DNA and therefore do not mimic constraints at the replication fork. We therefore studied the effects of ICRF-193 on chromatin assembly during replication of sperm nuclei in low-speed supernatants (LSS) of Xenopus eggs.
When demembranated sperm nuclei are incubated in LSS, there is an initial lag period of ∼25 min during which sperm chromatin decondenses and a new nuclear envelope reforms. After this lag, S phase starts abruptly and nuclei complete a rapid cycle of semiconservative DNA replication. We added ICRF-193 at t=20 min to avoid perturbation of the prereplicative period not relevant to replication-coupled chromatin assembly.
Pulse-chase labelling of replicating DNA and micrococcal nuclease digestion was used to monitor the structure of newly replicated chromatin at different durations after passage of the replication fork (Fig 1). Nuclease digestion of newly replicated DNA close to the fork (1 min pulse, no chase) produced two principal resistant fragments of ∼150 and ∼80 bp. The ∼150 bp fragment corresponds to the length of DNA protected by one nucleosome. We assume that the ∼80 bp fragment arises from the digestion of an assembly intermediate consisting of ∼80 bp of DNA wrapped around an (H3/H4)2 histone tetramer (Gruss et al, 1993). We do not think it represents a secondary product of nucleosome digestion, as even after extensive digestion, the relative intensity of the ∼150 and ∼80 bp bands did not vary (data not shown). After a 4 min chase, the ∼80 bp fragment was chased out, as expected for an assembly intermediate, and a ladder of mono-, di- and trinucleosomes was detected. The extent and regularity of this ladder continued to increase after a 10 min and a 90 min chase. In the presence of ICRF-193, however, the degree of regularity that was reached at 10 and 90 min was reproducibly lower than in the control reaction (in nine out of nine experiments), as shown by the smoother peaks and valleys in the scan profiles (Fig 1). Neither the mean spacing nor the amount of nuclease-resistant material was detectably altered. These results show that ICRF-193 does not prevent the bulk of nucleosome deposition, but perturbs the regular spacing of nucleosomes behind the replication fork.
Figure 1.
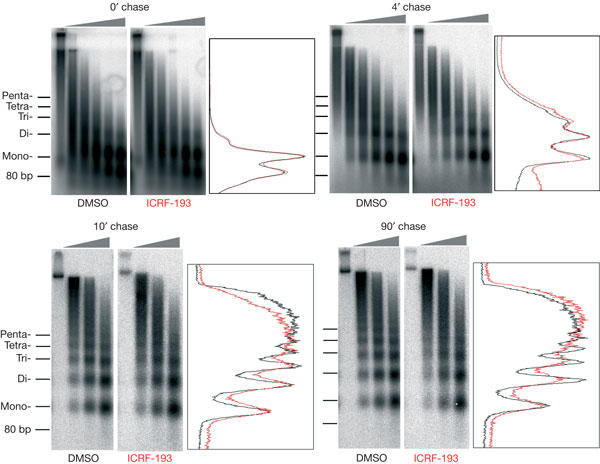
Effect of ICRF-193 on chromatin assembly during replication of sperm nuclei in egg extracts. Sperm nuclei were incubated in lowspeed supernatants at 1,000 nuclei/μl in the presence of 150 μM ICRF-193, or drug solvent alone (dimethylsulphoxide (DMSO)). Replicating DNA was pulse labelled for 1 min, then the label was chased for the indicated time and nucleosome assembly on newly replicated DNA was monitored by micrococcal nuclease digestion and gel electrophoresis. Consecutive lanes correspond to increasing digestion times (grey triangles). Density scans of representative lanes are shown in red (ICRF-193) and black (DMSO).
Effect of template concentration and replication
ICRF-193 inhibits topo II by trapping it in a closed clamped form onto DNA (Roca et al, 1994). The perturbation of nucleosome spacing by ICRF-193 may be due to topo II clamps, topo II catalytic inhibition, or both. To address this question, we varied the concentration of nuclei ([nuclei]) in the extract. As shown by the study of Luke & Bogenhagen (1989), there are ∼1010 topo II dimers per microlitre of LSS. Increasing the amount of added sperm nuclei (3 × 109 bp each) from 1,000 to 5,000 per microlitre would decrease the ratio of topo II dimers to nucleosomes from ∼0.66 to ∼0.13. This should decrease the density of topo II clamps along the DNA and attenuate the effect of ICRF-193 if it is due to topo II clamps but not if it is due solely to topo II catalytic inhibition. We found that increasing [nuclei] reproducibly (in four out of four experiments) diminished the effect of ICRF-193 on regular nucleosome spacing (Fig 2A), suggesting that this effect is due, at least in part, to topo II clamps. The lack of significant chromatin perturbation by ICRF-193 at high [nuclei] shows that topo II activity per se is dispensable for both the bulk of nucleosome deposition and their correct spacing on replicated DNA. Thus, unresolved DNA catenanes have surprisingly little, if any, effect on these processes (see the Discussion).
Figure 2.
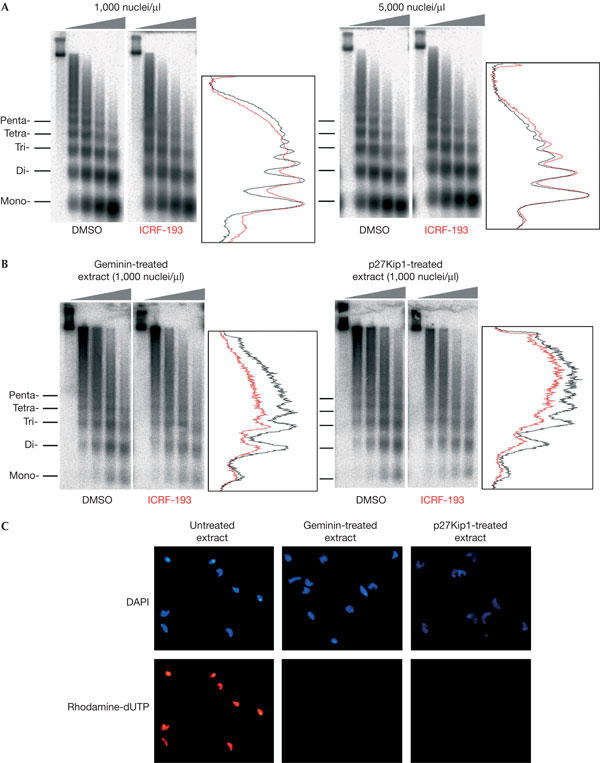
Effect of the concentration and replication of nuclei on the perturbation of chromatin structure by ICRF-193. (A) Effect of nuclei concentration on replicative chromatin assembly. Sperm nuclei were incubated in lowspeed supernatants (LSS) at 1,000 or 5,000 nuclei/μl and replicative chromatin was analysed as in Fig 1. The 90 min chase time points are shown. (B) Prereplicative chromatin assembly in geminin- and p27Kip1-treated LSS. Sperm nuclei were incubated in LSS for 90 min and nucleosome assembly was monitored as in Fig 1, except that DNA was hybridized with a total sperm DNA probe. (C) Microscopic visualization of DNA (4,6-diamidino-2-phenylindole) and rhodamine-dUTP incorporation in sperm nuclei incubated in control or geminin- or p27Kip1-treated extracts.
To test whether ICRF-193 can perturb replication-independent chromatin assembly, sperm nuclei (1,000 nuclei/μl) were incubated in LSS in the presence of two different replication inhibitors. Geminin inhibits the assembly of the prereplication complexes (pre-RCs) at replication origins by blocking the function of Cdt1, an essential component of pre-RCs (McGarry & Kirschner, 1998; Tada et al, 2001). p27Kip1 is a Cdk2 inhibitor that does not block pre-RC formation but prevents origin firing (Toyoshima & Hunter, 1994). In the presence of either inhibitor, sperm chromatin decondensed and a new nuclear envelope reformed, but replication was efficiently blocked (Fig 2C). In both cases, chromatin assembled in the presence of ICRF-193 was less regularly spaced than that assembled in its absence (Fig 2B). This effect was weaker and less reproducible than for replication-dependent assembly, which may be due in part to the use of Southern blotting rather than metabolic labelling for DNA detection. Nevertheless, we have also found that ICRF-193 affects the spacing of nucleosomes assembled on sperm nuclei (see below, and data not shown) or on plasmid DNA (supplementary information online) incubated in HSS, which cannot replicate double-stranded DNA due to their inability to assemble a nuclear membrane. These results show that ICRF-193 can perturb nucleosome spacing independently of DNA replication and catenation.
Topo II clamps perturb chromatin structure
To examine further whether topo II clamps alone explain chromatin perturbation by ICRF-193, we studied chromatin assembly coupled to complementary strand synthesis of M13 single-stranded DNA in HSS (Almouzni & Mechali, 1988b). This process mimics replication-coupled chromatin assembly except that precatenane formation is eliminated because the DNA template is single stranded. The final nucleosome density reached after completion of complementary strand synthesis at different [DNA] in the presence or absence of ICRF-193 was measured by 2D chloroquine gel electrophoresis (Fig 3A). At 5 ng/μl DNA (∼7 topo II dimers per M13 DNA molecule), a significant decrease in nucleosome density due to ICRF-193 was reproducibly observed (in seven out of seven experiments). The shift in the topoisomer distribution peak between the control and the drug-treated sample was ΔLk∼4. Despite the ∼10% reduction in nucleosome density, the distance between the mono- and dinucleosome bands on micrococcal nuclease ladders was not detectably altered (Fig 3B), suggesting that the nucleosome deficit in the ICRF-193 sample was due to a minor fraction of abnormally long linker DNAs rather than an even increase of nucleosome spacing. At 15 ng/μl DNA (∼2 topo II dimers per M13 DNA molecule), this deficit was reproducibly (in five out of five experiments) much reduced (ΔLk∼1). These results confirm that ICRF-193 perturbs nucleosome spacing independently of DNA catenation, due to steric hindrance by topo II clamps. Some internucleosomal DNA linkers become abnormally long when they contain topo II clamps.
Figure 3.
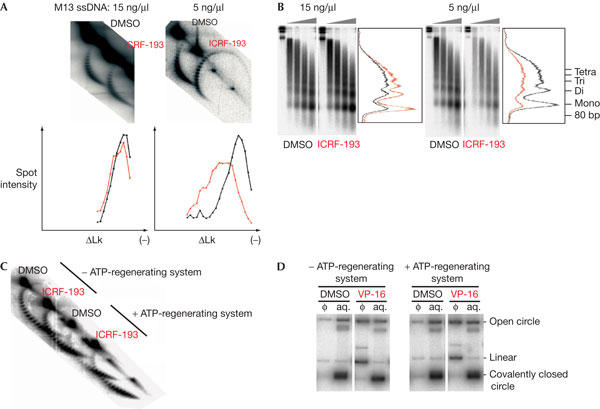
The effect of ICRF-193 on chromatin assembly coupled to the synthesis of a complementary DNA strand of M13 singlestranded DNA (ssDNA). (A) ICRF-193 reduces nucleosome density in a [DNA]-dependent manner. M13 ssDNA was incubated in high-speed supernatants (HSS) at 5 or 15 ng/μl with 150 μM ICRF-193 or drug solvent alone (dimethylsulphoxide (DMSO)). Replicating DNA was labelled during the 90 min incubation, then extracted and analysed by 2-dimensional chloroquine gel electrophoresis. Graphs represent spot intensity (percentage of total) for each topoisomer (DMSO in black and ICRF-193 in red). The difference in mean nucleosome density did not change on longer incubation (not shown). (B) Aliquots of the same reactions as in (A) analysed by micrococcal nuclease digestion and gel electrophoresis. (C) Same experiment as in (A) at 5 ng/μl ssDNA, with or without omission of an ATP-regenerating system. (D) Omission of an ATP-regenerating system does not affect VP-16-induced cleavage of plasmid DNA. pBR322 was incubated at 5 ng/μl for 90 min in the presence or absence of 50 μM VP-16 in HSS, with or without an ATP-regenerating system. DNA purified from cleavable complexes trapped by SDS addition and extracted with phenol (Φ), or DNA remaining in the aqueous phase (aq.), was electrophoresed in the presence of ethidium bromide, then blotted and hybridized with a pBR322 DNA probe.
ICRF-193 interferes with close nucleosome packing
In the above 2D gel experiments, nucleosome assembly was monitored through a change in Lk. However, topo II clamps may increase Lk by constraining positive writhe ((+)Wr), for example, by stabilizing DNA crossings rather than interfering with nucleosome assembly. This seems unlikely because the binding to plasmid DNA of yeast topo II in either the open or the closed clamp form does not alter its Lk distribution on relaxation by topo I (Trigueros et al, 2004). To further exclude this hypothesis, we studied the effect of ICRF-193 at low [DNA] (5 ng/μl), under conditions where nucleosome assembly is decreased by omission of the ATP-regenerating system (Fig 3C). In such conditions, ICRF-193 did not change the Lk distribution of the DNA template (Fig 3C). It has been reported that ATP is not required for maintaining the closed clamp form of topo II (Roca et al, 1994). Furthermore, omission of the ATP-regenerating system did not alter the trapping of topo II–DNA complexes by etoposide (Fig 3D). Therefore, the lack of change in Lk argues against a stabilization of (+)Wr by ICRF-193.
Following addition of an ATP-regenerating system to the same extract, however, nucleosome assembly was more extensive, and the reduction of final nucleosome density by ICRF-193 was again observed (Fig 3C). This experiment was performed twice, with identical results. These results show that at suboptimal nucleosome density, topo II clamps and nucleosomes can coexist on the same DNA molecule without perturbing each other. Only when nucleosomes become closely packed do topo II clamps significantly limit nucleosome assembly, presumably due to steric hindrance.
Effects of topo II addition or depletion
Purified human topo II was added to LSS or HSS in sufficient quantity to increase topo II activity ∼3-fold, as assessed by decatenation of kinetoplast DNA (not shown). We found that topo II addition strongly increased the perturbation of chromatin structure by ICRF-193 during replication-independent chromatin assembly on sperm nuclei in HSS and replication-dependent assembly on M13 DNA in HSS (Fig 4). These results confirm that ICRF-193 acts, at least in part, by trapping topo II clamps.
Figure 4.
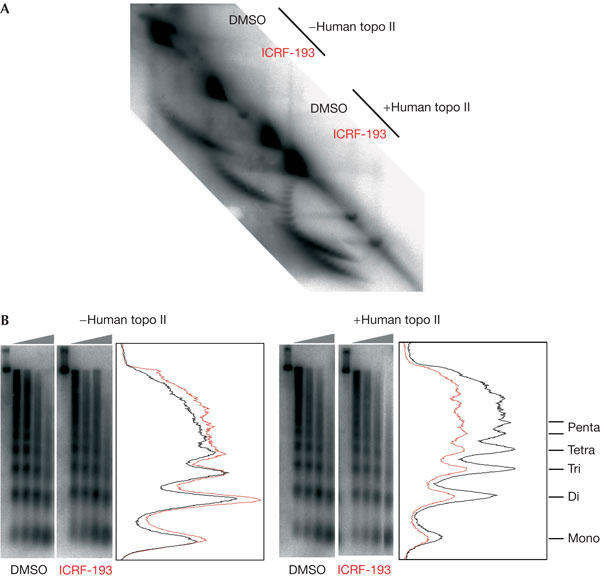
Addition of exogenous topoisomerase II (topo II) enhances the effect of ICRF-193 on chromatin assembly. Highspeed supernatants (HSS) were added with either buffer or human topo IIα. (A) Replication-dependent assembly on M13 DNA (15 ng/μl) and (B) replication-independent chromatin assembly on sperm nuclei (5,000 nuclei/μl) in HSS, added with 150 μM ICRF-193 or drug solvent alone (dimethylsulphoxide (DMSO)), were analysed as shown in Figs 2 and 3.
We assessed the effect of topo II immunodepletion on replication-coupled chromatin assembly on M13 DNA (5 ng/μl) in HSS (Fig 5). Addition of ICRF-193 to mock-depleted extracts perturbed chromatin assembly as with undepleted extracts. However, topo II depletion (>95%) almost completely abolished the effect of ICRF-193 on chromatin assembly (Fig 5). These results (reproduced twice) show that ICRF-193 has no detectable effect on chromatin assembly other than by topo II inhibition. Note that topo II depletion alone slightly altered the efficiency of chromatin assembly compared with mock depletion (Fig 5), and this effect was annihilated by re-addition of purified topo II (data not shown). This suggests that topo II contributes slightly to nucleosome assembly in these conditions. However, the effect of topo II depletion on chromatin assembly was always much weaker than ICRF-193 addition, confirming that most of the effect of ICRF-193 is attributable to topo II clamps.
Figure 5.
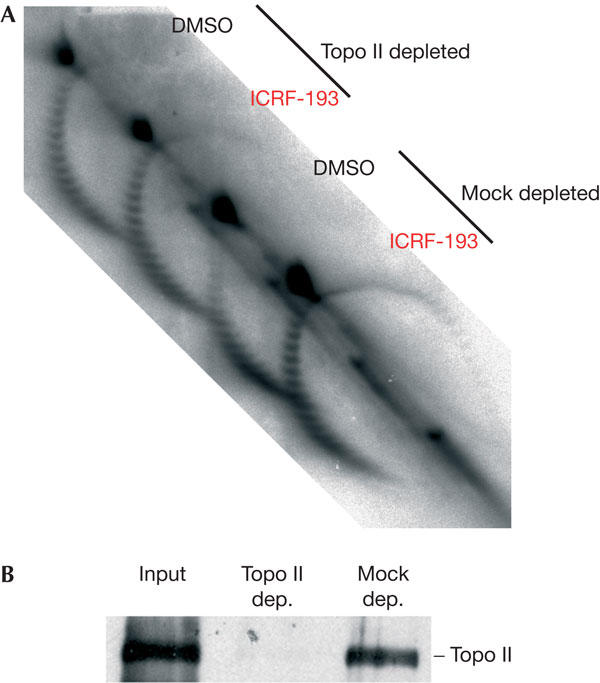
Immunodepletion of topoisomerase II (topo II) abolishes the effect of ICRF-193 on chromatin assembly. (A) Mock- or topo-II-depleted highspeed supernatants were added to M13 single-stranded DNA (5 ng/μl) and 150 μM ICRF-193 or drug solvent alone (dimethylsulphoxide (DMSO)) and the reactions were analysed as in Fig 3. (B) Control of the extent of topo II depletion by western blotting.
Discussion
Topo II clamps and cellular effects of ICRF-193
The finding that topo II-closed clamps trapped by ICRF-193 perturb the chromatin structure provides a new basis to understand the cellular effects of bisdioxopiperazines. Expression of wild-type human topo IIα in yeast cells sensitizes them to bisdioxopiperazines even when coexpressed with a drug-resistant form of topo II, consistent with a dominant, intrinsic toxicity of topo II clamps (Jensen et al, 2000). Given that chromatin is intimately involved in many DNA transactions, perturbation of the nucleosome array may compromise genome function in a variety of ways. For example, the reported effects of ICRF-193 on cellular differentiation (Tsutsui et al, 2001; Niitsu et al, 2002) may result from transcriptional reprogramming of key control genes after chromatin perturbation by topo II clamps.
Our findings that ICRF-193—and, to a lesser extent, topo II immunodepletion—perturb chromatin assembly does not necessarily contradict the study of Almouzni & Mechali (1988a), who failed to observe an effect of topo II inhibitors during chromatin assembly on plasmid DNA in HSS. First, the simple one-dimensional gel electrophoretic assay they used may well have missed the subtle changes that our 2D chloroquine gels can detect. Second, a different drug (novobiocin) was used to inhibit topo II. Third, we have found that, at the DNA concentration used for most of their experiments (15 ng/μl), the effect of ICRF-193 is strongly attenuated.
ICRF-193 can arrest mammalian cells in G2 or in mitosis by a checkpoint that has been suggested to monitor chromatin decatenation (Downes et al, 1994; Skoufias et al, 2004). However, the idea that catenation triggers a checkpoint is not easily conciliated with the fact that cells normally enter anaphase when their sister chromatids are still catenated (Shamu & Murray, 1992). Also, it is not clear how the cell could measure precisely sister chromatid catenation—a global topological property that has no simple relationship to the local occurrence of DNA crossings. Interestingly, recent reports raise the possibility that chromatin-based defects may act as direct initiators of cell-cycle checkpoints (Bakkenist & Kastan, 2003; Mikhailov et al, 2004). Therefore, we suggest that chromatin perturbation induced by ICRF-193, rather than DNA catenation per se, may trigger the ‘decatenation' checkpoint. All our results are otherwise consistent with the results of Skoufias et al (2004), showing that the checkpoint triggered by ICRF-193 is distinct from the DNA damage checkpoint.
DNA catenation and chromatin assembly
Our results show that inhibition of decatenation per se has little or no effect on the deposition and correct spacing of nucleosomes in sperm nuclei replicating in LSS. This is surprising given the high specific catenation reached by plasmid DNA replicating in LSS in the presence of ICRF-193 (about one catenane per 200 bp; Lucas et al, 2001). As there is no practical way to measure the catenation of long linear DNA molecules, we cannot exclude the possibility that sister chromatid catenation after topo II inhibition is much lower for sperm chromosomes than for plasmid DNA. Conversely, we have found that nucleosome deposition does occur on plasmid DNA catenanes (data not shown), suggesting that DNA can accommodate the sharp bending constraints imposed by a tight co-habitation of nucleosomes and catenane nodes. It was recently suggested that DNA is much more prone to sharp bending than was previously thought (Cloutier & Widom, 2004; but see Du et al, 2005). This inherent flexibility may facilitate the sequestration of sharply bent, hooked juxtapositions of catenated DNA strands between nucleosomes (Fig 6). Buck & Zechiedrich (2004) have proposed that type-2 topoisomerases use the local information at helix–helix juxtapositions to distinguish entanglements from two helices that are juxtaposed but not entangled. We speculate that during the unperturbed S phase, replication-coupled nucleosome assembly may facilitate (pre)catenane unlinking by transiently forcing (pre)catenane nodes into a hooked conformation that is a preferred substrate for their removal by topo II.
Figure 6.
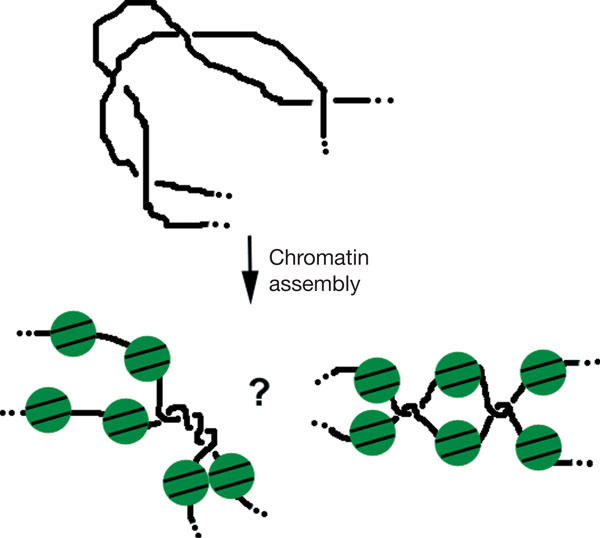
Nucleosome assembly may drive catenane nodes into a hooked conformation. Catenated DNAs unassembled or assembled into nucleosomes are schematically drawn. Catenane nodes may either alternate with or segregate from nucleosomes. In both cases, nucleosome assembly is proposed to drive a sharp bending of the juxtaposed linker DNA segments.
Methods
All methods are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Material
Acknowledgments
We thank D. Bogenhagen for the generous gift of anti-topo-II antiserum, J. Blow and J. Walter for plasmids, K. Marheineke for recombinant p27 and T. Strick, K. Marheineke and H. Labit for reading the paper. This work was supported by the Association pour la Recherche sur le Cancer and the Ligue Nationale Contre le Cancer (Comité de Paris). T.G. was supported by fellowships from the MENESR, the Ligue Nationale Contre le Cancer and the Asssociation pour la Recherche sur le Cancer.
References
- Almouzni G, Mechali M (1988a) Assembly of spaced chromatin involvement of ATP and DNA topoisomerase activity. EMBO J 7: 4355–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, Mechali M (1988b) Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. EMBO J 7: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 [DOI] [PubMed] [Google Scholar]
- Buck GR, Zechiedrich EL (2004) DNA disentangling by type-2 topoisomerases. J Mol Biol 340: 933–939 [DOI] [PubMed] [Google Scholar]
- Cloutier TE, Widom J (2004) Spontaneous sharp bending of doublestranded DNA. Mol Cell 14: 355–362 [DOI] [PubMed] [Google Scholar]
- Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT (1994) A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature 372: 467–470 [DOI] [PubMed] [Google Scholar]
- Du Q, Smith C, Shiffeldrim N, Vologodskaia M, Vologodskii A (2005) Cyclization of short DNA fragments and bending fluctuations of the double helix. Proc Natl Acad Sci USA 102: 5397–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss C, Wu J, Koller T, Sogo JM (1993) Disruption of the nucleosomes at the replication fork. EMBO J 12: 4533–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LH, Nitiss KC, Rose A, Dong J, Zhou J, Hu T, Osheroff N, Jensen PB, Sehested M, Nitiss JL (2000) A novel mechanism of cell killing by anti-topoisomerase II bisdioxopiperazines. J Biol Chem 275: 2137–2146 [DOI] [PubMed] [Google Scholar]
- Larsen AK, Escargueil AE, Skladanowski A (2003) Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol Ther 99: 167–181 [DOI] [PubMed] [Google Scholar]
- Li TK, Liu LF (2001) Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol 41: 53–77 [DOI] [PubMed] [Google Scholar]
- Lucas I, Germe T, Chevrier-Miller M, Hyrien O (2001) Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J 20: 6509–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke M, Bogenhagen DF (1989) Quantitation of type II topoisomerase in oocytes and eggs of Xenopus laevis. Dev Biol 136: 459–468 [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Mikhailov A, Shinohara M, Rieder CL (2004) Topoisomerase II and histone deacetylase inhibitors delay the G2/M transition by triggering the p38 MAPK checkpoint pathway. J Cell Biol 166: 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niitsu N, Higashihara M, Honma Y (2002) The catalytic DNA topoisomerase II inhibitor ICRF-193 and all-trans retinoic acid cooperatively induce granulocytic differentiation of acute promyelocytic leukemia cells: candidate drugs for chemo-differentiation therapy against acute promyelocytic leukemia. Exp Hematol 30: 1273–1282 [DOI] [PubMed] [Google Scholar]
- Roca J, Ishida R, Berger JM, Andoh T, Wang JC (1994) Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA 91: 1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JB, Stasiak A (2004) A topological view of the replicon. EMBO Rep 5: 256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Murray AW (1992) Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol 117: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL (2004) Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol Cell 15: 977–990 [DOI] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol 3: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima H, Hunter T (1994) p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78: 67–74 [DOI] [PubMed] [Google Scholar]
- Trigueros S, Salceda J, Bermudez I, Fernandez X, Roca J (2004) Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J Mol Biol 335: 723–731 [DOI] [PubMed] [Google Scholar]
- Tsutsui K, Sano K, Kikuchi A, Tokunaga A (2001) Involvement of DNA topoisomerase IIβ in neuronal differentiation. J Biol Chem 276: 5769–5778 [DOI] [PubMed] [Google Scholar]
- Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3: 430–440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


