Abstract
Initiation of DNA replication from the Escherichia coli chromosomal origin is highly regulated, assuring that replication occurs precisely once per cell cycle. Three mechanisms for regulation of replication initiation have been proposed: titration of free DnaA initiator protein by the datA locus, sequestration of newly replicated origins by SeqA protein and regulatory inactivation of DnaA (RIDA), in which active ATP-DnaA is converted to the inactive ADP-bound form. DNA microarray analyses showed that the level of initiation in rapidly growing cells that lack datA was indistinguishable from that in wild-type cells, and that the absence of SeqA protein caused only a modest increase in initiation, in agreement with flow-cytometry data. In contrast, cells lacking Hda overinitiated replication twofold, implicating RIDA as the predominant mechanism preventing extra initiation events in a cell cycle.
Keywords: replication, DnaA, oriC, Hda, initiation
Introduction
All organisms depend on tightly regulated initiation of chromosomal DNA replication for proper cell-cycle control. In Escherichia coli, the activity of the initiator protein, DnaA, is the focal point of this regulation. DnaA that is bound to recognition sites in the origin of chromosomal replication (oriC), accompanied by HU protein or integration host factor (IHF), initiates unwinding of a neighbouring AT-rich region (Messer, 2002). This, in turn, leads to the loading of the replicative helicase, DnaB (Sutton et al, 1998), replisome assembly and DNA replication.
Several independent mechanisms to ensure that initiation at an origin occurs only once per cell cycle have been proposed (reviewed by Boye et al, 2000; Katayama, 2001; Camara & Crooke, 2005). One such mechanism, termed regulatory inactivation of DnaA (RIDA), is a process that accelerates the hydrolysis of ATP bound to DnaA, thereby generating replicatively inactive ADP-DnaA. Essential components of RIDA include the Hda protein (Kato & Katayama, 2001; Camara et al, 2003) and the βsubunit of DNA polymerase III loaded as a sliding clamp on DNA (Katayama et al, 1998). Therefore, following βsubunit loading, which serves as a hallmark of replisome formation, DnaA is inactivated for re-initiation.
A second mechanism involves the high-affinity binding of SeqA to hemimethylated GATC sites (Lu et al, 1994; von Freiesleben et al, 1994; Brendler et al, 1995; Slater et al, 1995; Brendler & Austin, 1999; Skarstad et al, 2000), 11 of which exist in a newly replicated origin. It has been proposed that the SeqA-bound hemimethylated origin is sequestered at the cell membrane, where it is rendered inaccessible to DnaA. Thus, SeqA should assist in preventing untimely reinitiations until oriC is fully methylated, which occurs about one-third of a cell cycle after initiation has taken place.
Titration of free DnaA by the datA chromosomal locus, which shows an unusually high affinity for DnaA (Kitagawa et al, 1996, 1998), is a third mechanism proposed to have a role in restraining initiation of DNA replication: doubling of the datA locus copy number by chromosomal replication might rapidly diminish the amount of free DnaA that could otherwise initiate another round of replication in the same cell cycle.
On the basis of the analyses of flow-cytometry data, it has been thought that the disruption of any one of these three mechanisms might result in overinitiations (Lu et al, 1994; Kitagawa et al, 1998; Camara et al, 2003). Here, genomic microarray analysis (Khodursky et al, 2000), a technique previously used to investigate initiation frequency and replication-fork movement during DnaA overproduction (Simmons et al, 2004), was used to investigate the effects of deleting key components of the proposed sequestration, RIDA and datA DnaA titration regulatory mechanisms. We observed that cells that lacked Hda showed a significant increase in the ratio of the oriC-to-terminus regions of the chromosome, which was reflective of the importance of RIDA in preventing hyperinitiation. In contrast, deletion of the datA locus resulted in an oriC-to-terminus ratio indistinguishable from that for wild-type cells, and the loss of SeqA protein had only a minimal effect, indicating that initiator titration by datA and oriC sequestration do not contribute significantly to the prevention of overinitiations.
Results
Increased origin-to-terminus DNA in Δhda cells
Whole-genome microarrays were used to determine the effects that deletions of hda, datA or seqA had on the initiation of DNA replication. Genomic DNA was prepared from rapidly growing cells of each mutant strain, as well as from isogenic wild-type cells. The samples prepared from mutant cells were fluorescently labelled and hybridized, along with the wild-type genomic DNA labelled with a different fluorophore, to microarrays. The abundance of DNA hybridized to the arrays was plotted on the y-axis, and chromosomal position, relative to oriC, was plotted on the x-axis (Fig 1). The abundance of terminus DNA was normalized to lie on the x-axis. A mutation that increases initiation frequency would increase the copy number of chromosomal loci near oriC relative to the terminus, resulting in a peak at oriC and a minimum at the terminus.
Figure 1.
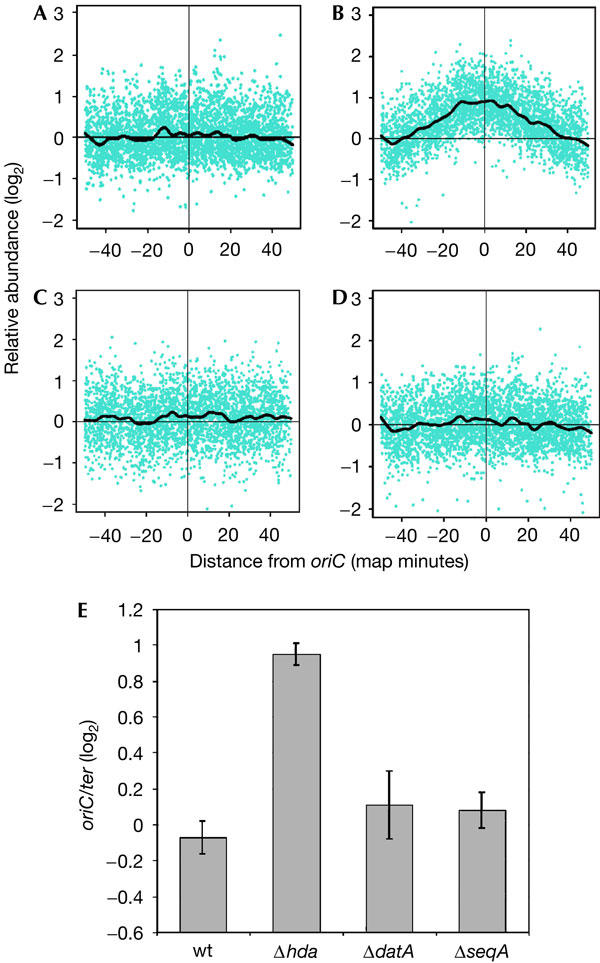
Lack of Hda leads to an increased oriC-to-terminus ratio in exponentially growing cells. (A–D) Genomic DNA prepared from exponentially growing cells of strains (A) MG1655, (B) JC125 (MG1655Δhda:tetR), (C) JC326 (MG1655ΔseqA:tetR) and (D) JC126 (MG1655ΔdatA:kanR) was analysed using microarrays containing 95% of Escherichia coli open reading frames. DNA from deletion-mutant cells was compared directly with log-phase DNA from the wild-type (wt) cells (MG1655). The ratios are represented on the y-axis as relative abundance on a base 2 logarithmic scale (log2). The x-axis represents the chromosomal location of each ORF in map minutes, with oriC located in the centre of each plot at 0 min. The black lines are plots of smoothed values from a moving median. (E). The bar graph shows the log2 ratio of oriC-to-terminus for wt and mutant cells, where each region was defined as a four-map-minute zone surrounding or diametrically opposed to oriC, respectively. Results are shown as the average±standard deviation (n=3).
As genomic DNA from exponentially growing wild-type cells serves as the internal control in these studies, it was compared with genomic DNA isolated from wild-type cells that had reached stationary phase. When genomic DNAs prepared from both exponentially growing and stationary-phase wild-type cells were hybridized to the same array, the oriC-to-terminus ratio for the exponentially growing cells was found to be around 2:1 (data not shown), similar to previous observations (Simmons et al, 2004).
Genomic DNA from both ΔdatA and ΔseqA cells (Fig 1C and D, respectively) yielded oriC-to-terminus ratios and chromosome profiles comparable with those of wild-type cells (Fig 1A). Only the Δhda cells (Fig 1B) produced a significantly increased oriC-to-terminus ratio, around twofold over the ratio for wild-type cells, as indicated by the mean and standard deviation of three separate experiments of cell growth, genomic DNA isolation and microarray hybridization (Fig 1E).
The amounts of markers for origin DNA relative to markers for the terminus region of the chromosome were also compared by Southern hybridization analysis. In agreement with the microarray data, around twofold increases in oriC and dnaA (0.4 min) relative to terC and xasA (50 min), respectively, were detected in Δhda cells when compared with wild-type cells (Table 1), indicating significant overinitiation in Δhda cells. The ratios of oriC/ter and dnaA/xasA for ΔdatA and ΔseqA cells were similar to those for wild-type cells, which supported the suggestion from the microarray data that any overinitiation in ΔdatA and ΔseqA cells is modest. Marker frequency analysis, performed elsewhere (Morigen et al, 2005), also found that no or very little overinitiation occurs in ΔdatA cells.
Table 1.
Growth phenotypes, DnaA content and marker frequencies of strains cultured in LB medium
| Strain | Relevant genotype | τa (min) | OD600 nm in stationary phase |
Marker frequenciesb |
DnaA/total proteinc | |
|---|---|---|---|---|---|---|
| oriC/ter | dnaA/xasA | |||||
| MG1655 |
Wild type |
26 |
7.74 |
1.9 |
2.0 |
1.00±0.25 |
| JC125 |
Δhda |
30 |
5.70 |
4.1 |
3.9 |
1.15±0.22 |
| JC126 |
ΔdatA |
26 |
6.18 |
2.0 |
1.9 |
1.08±0.08 |
| JC326 | ΔseqA | 36 | 4.02 | 2.1 | 2.3 | 1.79±0.33 |
aLog-phase growth doubling time.
bAbundance of oriC relative to ter, and dnaA relative to xasA, as determined by Southern blot analysis.
cBased on immunoblots in Fig 3. Only experimental values (n=3–5) that fell within the range of the corresponding DnaA standard curves were used for further calculations.
DnaA content was calculated as DnaA/total protein and all values were normalized to the value for wild-type cells (MG1655), which was given an arbitrary value of 1.00.
The semi-log plot of the relative abundance of each gene versus its position on the chromosome descends linearly in both directions from oriC (Fig 1B), showing that DNA replication forks in Δhda cells showed no evidence of replication fork stalling. This contrasts with overinitiation induced through overexpression of DnaA, which resulted in stalling throughout the chromosome, but particularly near the origin, as indicated by a sharp apex at oriC (Simmons et al, 2004).
Effects of mutations on cell growth
When cell growth was monitored in rich medium, mimicking the culturing conditions for the cells that served as the source of genomic DNA, ΔdatA cells grew almost identically to wild-type cells; both showed a log-phase doubling time of 26 min in LB medium (Miller, 1972) at 37°C, with ΔdatA cells entering stationary phase at a slightly lower culture density (Table 1). The Δhda cells showed a slight increase in doubling time (30 min), similar to that previously reported (Camara et al, 2003), and seemed to enter stationary phase at a slightly lower cell density. The ΔseqA cells showed a larger difference in growth rate, doubling every 36 min, and these cells entered stationary phase at a culture density lower than that of the other mutant strains.
The growth rate of cells in a culture affects the frequency of initiation, and therefore the oriC-to-terminus ratio measured by microarrays. Compensating for the slower growth rate of the ΔseqA cells (Table 1) suggests that the oriC-to-terminus DNA ratio may be about 33% higher than the ratio for wild-type cells, assuming that the rates of fork progression are similar in wild-type and ΔseqA cells cultured in rich medium.
However, flow cytometric data of ΔseqA cells (Fig 2) cultured under the same conditions as those used for the microarray studies show that the DNA/cell mass ratio is only 1.07-fold higher than that for wild-type cells. This suggests that initiation events are not highly elevated in cells that are unable to sequester oriC, and that the hypothetical 33% increase in oriC-to-terminus DNA may be an overestimation. The ΔseqA cells, although not significantly overinitiating, showed the expected high degree of asynchronous initiations (data not shown).
Figure 2.
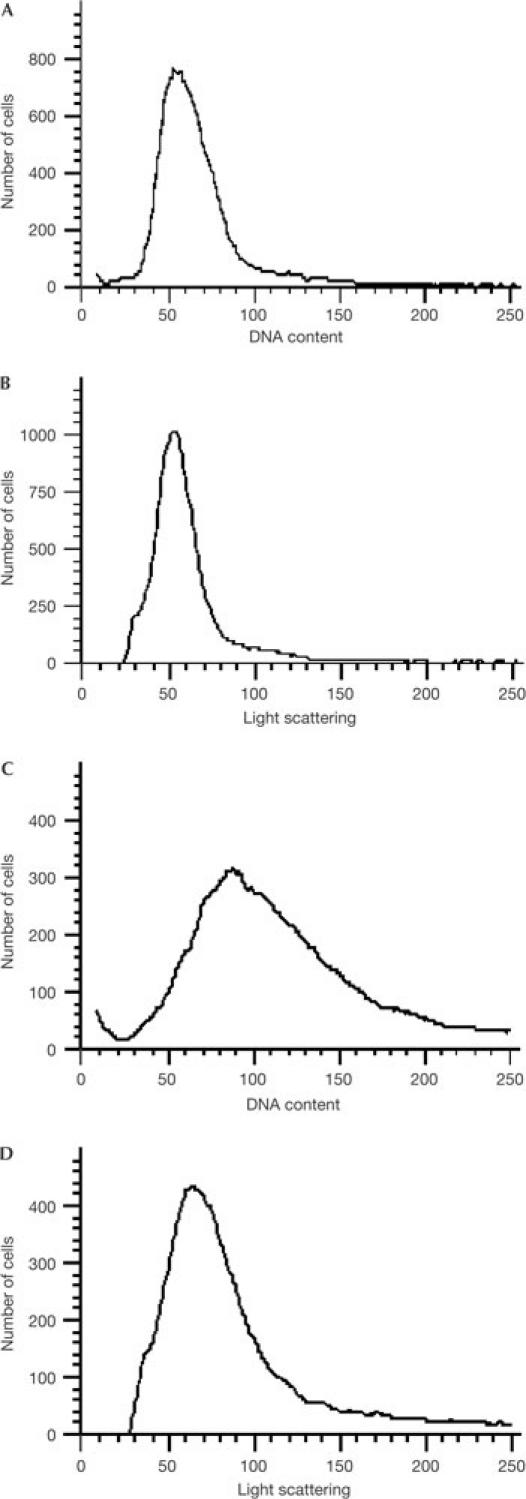
Flow cytometry of wild-type and ΔseqA cells. (A,B) Wild-type MG1655 and (C,D) JC326 (MG1655ΔseqA:tetR) cells were grown in M63+glucose medium (37°C, overnight), diluted into LB medium to an initial culture optical density (A600 nm) of 0.02. The cells were allowed to grow for 3 h at 37°C and maintained at or below an optical density of 0.1 with appropriate dilutions, and then immediately fixed in 77% ethanol. DNA content (A,C) and light scattering (B,D) distributions were determined by flow cytometry, as described previously (Li et al, 2002). Relative DNA/mass ratios (see text) were obtained using Winbrite 2.03 software (Bio-rad, Hercules, CA, USA) with the ratios of population mean values for DNA fluorescence and light scattering calculated. The ratio for the seqA mutant was normalized to that of the wild type to give the relative DNA/mass ratio of the mutant.
DnaA levels are affected only in the ΔseqA strain
Immunoblotting for DnaA was used to monitor whether deletion of hda, seqA or datA affected the levels of DnaA present in rapidly growing log-phase cells (Fig 3; Table 1). No significant difference was detected in the cellular content of DnaA in Δhda or ΔdatA cells when compared with wild-type cells. However, a roughly twofold increase in DnaA content was observed in ΔseqA cells, consistent with previous observations (von Freiesleben et al, 1994, 2000a).
Figure 3.
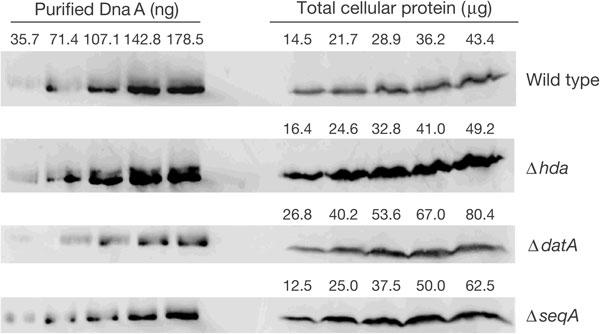
Immunoblot of DnaA protein in exponentially growing cells. DnaA standards and total cellular protein from MG1655, JC125 (MG1655Δhda:tetR), JC126 (MG1655ΔdatA:kanR) and JC326 (MG1655ΔseqA:tetR) cells were probed with anti-DnaA antiserum.
Discussion
We have used genomic microarrays to investigate the frequency at which chromosomal replication is initiated and the characteristics of subsequent replication fork progression in cells that lack Hda, datA and SeqA. Strains deficient in each of these elements have previously been reported to overinitiate DNA replication, on the basis of flow cytometric analysis after treatment with rifampicin and cephalexin (Lu et al, 1994; von Freiesleben et al, 1994; Kitagawa et al, 1998; Ogawa et al, 2002; Camara et al, 2003). However, the genomic DNA microarray analysis presented here, which is non-invasive, non-perturbing and avoids the use of powerful drugs, shows that only Δhda cells have a significantly increased oriC-to-terminus ratio. No increase in overall cellular DnaA content was associated with this overinitiation (Fig 3; Table 1), which suggests, at least for rapidly growing cells, that the nucleotide-bound state has a central role in suppressing reinitiation and that cells do not seem to downregulate expression of the initiator to compensate for increased initiation potential.
The Δhda cells seem to cope with overinitiation of DNA replication, with minimal effect on their overall growth rate, providing an interesting contrast with earlier experiments in which DnaA was overexpressed (Simmons et al, 2004). Although both overexpression of DnaA and deletion of hda resulted in overinitiation, replication fork stalling occurred only in the former case. This uncoupling of effects suggests that whereas the level of ATP-DnaA controls initiation, increases in overall DnaA concentration (combined ADP-DnaA and ATP-DnaA) have negative effects on replication fork elongation. This concept is supported by experiments in which cells harbouring multicopy datA-bearing plasmids showed an increased rate of chromosomal replication (Morigen et al, 2003).
In agreement with our finding that the loss of Hda leads to overinitiation, cells with a temperature-sensitive allele of hda, when cultured at a non-permissive temperature, have been reported to have an oriC-to-terminus ratio three- to fourfold higher than that of wild-type cells (Kato & Katayama, 2001). The heightened overinitiation seen by these authors, relative to the twofold increase presented here, may be attributed to differences in strains, growth conditions and the examination of a temperaturesensitive versus deleted hda allele.
As stated earlier, cells deleted for datA have been thought to overinitiate, on the basis of flow-cytometry data of cephalexin- and rifampicin-treated cells. However, an alternative possibility is that datA cells carry out rifampicin-resistant initiations, but do not overinitiate in the absence of rifampicin. Rifampicin, an inhibitor of RNA synthesis, prevents further initiations from occurring, but allows continuing rounds of replication to persist; cephalexin inhibits cell division, and therefore, replicated daughter chromosomes will reside within the same cell (Skarstad et al, 1986). Thus, the number of fully replicated chromosomes in a cell is indicative of the number of origins present at the time of rifampicin and cephalexin treatment. However, rifampicin-resistant initiations can occur, such as in E. coli overexpressing DnaA (Pierucci et al, 1987). Rifampicin-resistant, DnaA-dependent initiation of replication has also been observed in other genetic backgrounds such as ihf mutants (von Freiesleben et al, 2000b). Without being properly accounted for, rifampicin-resistant initiations can lead to misinterpretation of flow-cytometry data, resulting in the overestimation of initiation events. Recent studies by Skarstad and co-workers (Morigen et al, 2005) showed that ΔdatA cells indeed carry out rifampicin-resistant initiations. These results support the conclusion from our microarray and Southern blot analyses, and the Southern blot analysis performed by Morigen and colleagues (2005), that any hyperinitiation that may occur in ΔdatA cells is slight. Interestingly, Ogawa and co-workers (Kitagawa et al, 1998) observed a modest decrease in the apparent oriC-to-terminus ratio when ΔdatA cells were cultured in rich medium, as used here, versus a minimal medium. Our conclusion that datA does not significantly contribute to prevention of hyperinitiation does not rule out the possibility that titration of DnaA by other DnaA-binding sites on the chromosome helps regulate the initiation of chromosomal replication (Hansen et al, 1991).
Our microarray analysis also shows that overinitiation occurs to a significantly lesser extent in ΔseqA cells grown in rich medium than in Δhda cells cultured under comparable conditions. The oriC-to-terminus ratio was twofold higher in cells that lacked Hda than in wild-type cells. For ΔseqA cells, the microarray profile and oriC-to-terminus ratio were indistinguishable from those of wild-type cells (Fig 1). Adjusting for the slower growth rate of ΔseqA cells (Table 1) suggests that the oriC-to-terminus ratio may be at most 1.3-fold over the ratio for wild-type cells. That ΔseqA cells have limited levels of hyperinitiation is supported by our flow cytometry analysis and by previous studies. Here, a 1.07-fold increase of the DNA/cell mass ratio was observed for ΔseqA cells (Fig 2), similar to the 1.19-fold increase seen in seqA2 mutants (von Freiesleben et al, 1994). Boye et al (1996) found that initiation occurred, on average, at a cell mass about 10–20% lower for seqA mutants than for wild-type cells, consistent with a small increase in DNA/cell mass, assuming that the initiation events were followed by completed rounds of replication. Under growth conditions different from those used here, an increase in DNA/cell mass of as much as 1.5-fold in ΔseqA cells has been reported (Bach & Skarstad, 2004). However, the authors also present compelling data that the loss of SeqA affects functions other than chromosomal replication.
SeqA has been implicated in chromosomal segregation and organization (Hiraga et al, 1998; Brendler et al, 2000; Bach et al, 2003), cellular events that, therefore, would be affected by the lack of SeqA. When origin sequestration is disrupted instead by mutating the GATC sites within oriC, but wild-type SeqA protein levels are still maintained, the degree of overinitiation observed by flow cytometry is significantly less, with only a 1.1-fold increase in DNA/mass when compared with wild-type cells (Bach & Skarstad, 2004), in close agreement with the 1.07-fold increase that we observed. This provides further support for the proposed roles of SeqA beyond its function as a negative regulator of initiation events, and supports our conclusion that oriC sequestration by SeqA is not a principal mechanism in preventing overinitiation.
Interestingly, in the absence of Hda, overinitiation is only twofold, suggesting that other mechanisms are in place to prevent even higher levels of hyperinitiation. SeqA and datA, although minimally involved in preventing reinitiation, probably have important roles in assuring the proper timing of initiation during the cell cycle. Answers to how various mechanisms work in concert to promote initiation and prevent reinitiation should be of great interest.
Methods
Bacterial strains. Strains JC125, JC126 and JC326 were constructed by transducing the appropriate allele from strains JE202 (Δhda:tetR; Camara et al, 2003), RSD448 (ΔdatA:kanR; Kitagawa et al, 1998) and CC1064 (ΔseqA:tetR; Lu et al, 1994), respectively, into MG1655 by P1-mediated transduction, as described previously (Camara et al, 2003). Transductants that showed appropriate antibiotic resistance were screened by PCR to confirm that the antibiotic resistance gene was inserted into the respective hda, datA or seqA gene.
Before serving as a source of genomic DNA or having their cellular DnaA content measured, MG1655, JC125 and JC126 cells were cultured overnight in LB medium (Miller, 1972) that contained no antibiotic, tetracycline (10 μg/ml) or kanamycin (20 μg/ml), respectively. JC326 cells were cultured overnight in M9+glucose (0.2%) medium (Miller, 1972) that also contained tetracycline (25 μg/ml). Samples from overnight cultures were diluted in the ratio of 1:400 into LB medium (37°C) that contained antibiotic at the same concentration as the overnight cultures, except for JC326 cells, which were diluted into LB medium (37°C) that contained tetracycline at 10 μg/ml. All cultures were then incubated at 37°C with vigorous shaking.
Microarray analysis. Cells of each strain were grown, as stated above, to a final optical density (OD)600 nm=0.3–0.4. Sodium azide was added to a final concentration of 0.2% (w/v), and cells were collected by centrifugation (5,000g, 10 min at 4°C). Genomic DNA was extracted using the DNeasy Tissue Kit (Qiagen, Valencia, CA, USA), digested with AluI (New England Biolabs, Beverly, MA, USA) and purified by phenol:chloroform extraction followed by ethanol precipitation (Sambrook et al, 1989). DNA concentration was determined spectrophotometrically and samples were stored at −80°C.
Fluorescent labelling of DNA samples and microarray hybridization were performed, as described previously (Simmons et al, 2004). Microarray fluorescence intensity data were acquired using an Axon 4000B scanner and GenePix 3.0 software. Data were processed further using the R language and environment, version 1.3.0 (http://www.r-project.org) as follows. ‘Ratio of medians' values were extracted for each spot; spots appearing visually deficient or with fewer than 60% of pixels one standard deviation above background were rejected. The oriC-to-terminus ratios plotted in Fig 1E were calculated as the median of the four map minutes centred on oriC. The smoothed traces shown in Fig 1A–D were obtained using the lowess algorithm, with a value of 0.075 for the f parameter.
Marker frequency analysis. Cells were cultured and prepared as for the microarray analysis. Genomic DNA was digested with BamH1 for Southern hybridization analysis of oriC and ter with probes generated by PCR, as described previously (Kato & Katayama, 2001). Independently, genomic DNA was digested with NcoI and PstI for Southern blot analysis of dnaA (0.4 min) and xasA (50 min), with probes generated by PCR, as described previously (Zheng et al, 2001).
Quantitative immunoblot analysis. Cells of each strain were grown, as stated above, to a final OD600 nm=0.3–0.4. Samples of total cellular protein were separated on 15% SDS–polyacrylamide gel electrophoresis gels along with standards of purified DnaA. Resolved proteins were transferred to nitrocellulose and probed with anti-DnaA antiserum, which was visualized by enhanced chemifluorescence with a STORM 840 imaging system (Molecular Dynamics, Piscataway, NJ, USA). Intensities of DnaA bands in the samples were compared with the bands for the DnaA standards using ImageQuant (Molecular Dynamics) software.
Acknowledgments
This work was supported in part by the Cosmos Club Foundation's Grants-in-Aid to Scholars (to J.E.C.) and grants from National Institutes of Health (R01GM49700 and R01 GM031655, to E.C. and N.R.C., respectively).
References
- Bach T, Skarstad K (2004) Re-replication from nonsequesterable origins generates three-nucleoid cells which divide asymmetrically. Mol Microbiol 51: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Bach T, Krekling MA, Skarstad K (2003) Excess SeqA prolongs sequestration of oriC and delays nucleoid segregation and cell division. EMBO J 22: 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E, Lobner-Olesen A, Skarstad K (2000) Limiting DNA replication to once and only once. EMBO Rep 1: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E, Stokke T, Kleckner N, Skarstad K (1996) Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc Natl Acad Sci USA 93: 12206–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T, Abeles A, Austin S (1995) A protein that binds to the P1 origin core and the oriC 13mer region in a methylationspecific fashion is the product of the host seqA gene. EMBO J 14: 4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T, Austin S (1999) Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J 18: 2304–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T, Sawitzke J, Sergueev K, Austin S (2000) A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J 19: 6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara JE, Crooke E (2005) Initiation of chromosomal replication. In The Bacterial Chromosome, Higgins NP (ed) pp 177–192. Washington, DC, USA: ASM Press [Google Scholar]
- Camara JE, Skarstad K, Crooke E (2003) Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein. J Bacteriol 185: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen FG, Christensen BB, Atlung T (1991) The initiator titration model: computer simulation of chromosome and minichromosome control. Res Microbiol 142: 161–167 [DOI] [PubMed] [Google Scholar]
- Hiraga S, Ichinose C, Niki H, Yamazoe M (1998) Cell cycle-dependent duplication and bi-directional migration of SeqA-associated DNA–protein complexes in E. coli. Mol Cell 1: 381–387 [DOI] [PubMed] [Google Scholar]
- Katayama T (2001) Feedback controls restrain the initiation of Escherichia coli chromosomal replication. Mol Microbiol 41: 9–17 [DOI] [PubMed] [Google Scholar]
- Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94: 61–71 [DOI] [PubMed] [Google Scholar]
- Kato J, Katayama T (2001) Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J 20: 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR (2000) Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc Natl Acad Sci USA 97: 9419–9424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Mitsuki H, Okazaki T, Ogawa T (1996) A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol Microbiol 19: 1137–1147 [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Ozaki T, Moriya S, Ogawa T (1998) Negative control of replication initiation by a novel chromosomal locus exhibiting affinity for Escherichia coli DnaA protein. Genes Dev 12: 3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sergueev K, Austin S (2002) The segregation of the Escherichia coli origin and terminus of replication. Mol Microbiol 46: 985–996 [DOI] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N (1994) SeqA: a negative modulator of replication initiation in E. coli. Cell 77: 413–426 [DOI] [PubMed] [Google Scholar]
- Messer W (2002) The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev 26: 355–374 [DOI] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics, pp 431–434. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory [Google Scholar]
- Morigen, Lobner-Olesen A, Skarstad K (2003) Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol Microbiol 50: 349–362 [DOI] [PubMed] [Google Scholar]
- Morigen, Molina F, Skarstad K (2005) Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J Bacteriol 187: 3913–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Yamada Y, Kuroda T, Kishi T, Moriya S (2002) The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol Microbiol 44: 1367–1375 [DOI] [PubMed] [Google Scholar]
- Pierucci O, Helmstetter CE, Rickert M, Weinberger M, Leonard AC (1987) Overexpression of the dnaA gene in Escherichia coli B/r: chromosome and minichromosome replication in the presence of rifampin. J Bacteriol 169: 1871–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989), In Molecular Cloning: A Laboratory Manual, Nolan C (ed) 2nd edn, pp E.3–E.10. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory [Google Scholar]
- Simmons LA, Breier AM, Cozzarelli NR, Kaguni JM (2004) Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol Microbiol 51: 349–358 [DOI] [PubMed] [Google Scholar]
- Skarstad K, Boye E, Steen HB (1986) Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J 5: 1711–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K, Lueder G, Lurz R, Speck C, Messer W (2000) The Escherichia coli SeqA protein binds specifically and co-operatively to two sites in hemimethylated and fully methylated oriC. Mol Microbiol 36: 1319–1326 [DOI] [PubMed] [Google Scholar]
- Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N (1995) E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82: 927–936 [DOI] [PubMed] [Google Scholar]
- Sutton MD, Carr KM, Vicente M, Kaguni JM (1998) Escherichia coli DnaA protein: the N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J Biol Chem 273: 34255–34262 [DOI] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV, Schaechter M (1994) SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol 14: 763–772 [DOI] [PubMed] [Google Scholar]
- von Freiesleben U, Krekling MA, Hansen FG, Lober-Olesen A (2000a) The eclipse period of Escherichia coli. EMBO J 19: 6240–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV, Atlung T, Hansen FG (2000b) Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol Microbiol 37: 1087–1093 [DOI] [PubMed] [Google Scholar]
- Zheng W, Li Z, Skarstad K, Crooke E (2001) Mutations in DnaA protein suppress the growth arrest of acidic phospholipid-deficient Escherichia coli cells. EMBO J 20: 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]


