Abstract
Production of large quantities of recombinant adeno-associated virus (AAV) is difficult and not cost-effective. To overcome this problem, we have explored the feasibility of creating a recombinant AAV encoding a 6×His tag on the VP3 capsid protein. We generated a plasmid vector containing a six-His (6×His)-tagged AAV VP3. A second plasmid vector was generated that contained the full-length AAV capsid capable of producing VP1 and VP2, but not VP3 due to a mutation at position 2809 that encodes the start codon for VP3. These plasmids, necessary for production of AAV, were transfected into 293 cells to generate a 6×His-tagged VP3mutant recombinant AAV. The 6×His-tagged VP3 did not affect the formation of AAV virus, and the physical properties of the 6×His-modified AAV were equivalent to those of wild-type particles. The 6×His-tagged AAV did not affect the production titer of recombinant AAV and could be used to purify the recombinant AAV using an Ni-nitrilotriacetic acid column. Addition of the 6×His tag did not alter the viral tropism compared to wild-type AAV. These observations demonstrate the feasibility of producing high-titer AAV containing a 6×His-tagged AAV VP3 capsid protein and to utilize the 6×His-tagged VP3 capsid to achieve high-affinity purification of this recombinant AAV.
Adeno-associated virus (AAV) capsids are composed of three proteins, VP1, VP2, and VP3 (4, 13-15, 32). Packaged within the capsid is a single-stranded DNA genome of 4,679 bases that contains two large open reading frames (ORFs), rep and cap (27). The three structural proteins, VP1 (87-kDa), VP2 (73-kDa), and VP3 (62-kDa), are encoded by a single ORF. Each of them are produced by alternative splicing of the transcript generated from the p40 promoter by use of alternative start codons at nucleotide position 2203 for VP1, 2614 for VP2, and 2809 for VP3 (15, 31, 32). The C-terminal region sequences that are common to all three capsid proteins promote folding of the C-terminal region of the polypeptide into a β-barrel structure, which is present in several viruses, including parvovirus B19 and porcine parvovirus (1, 5, 7, 18, 19, 26). The relative abundance of VP3 within the capsid is considerably higher (90%), than that of VP1 (5%) and VP2 (5%) (24, 29, 32).
Recently, AAV has attracted a significant amount of interest as a vector for gene therapy (28). AAV has a number of unique advantages that are potentially useful for gene therapy applications, including the ability to infect nondividing cells, a lack of pathogenicity, and the ability to establish long-term gene expression (16, 17). Attempts to alter the AAV capsid have been made in order to expand the tropism of AAV. Yang et al. (34) showed improved infectivity of hematopoietic progenitor cells by generating a chimeric recombinant AAV (rAAV) containing a single-chain antibody with specificity for human CD34. Girod et al. (11) showed that insertion of the L14 epitope into the capsid coding region can expand the tropism to mouse melanoma cell B16 cells that are nonpermissive for AAV infection.
Systematic site-directed mutagenesis of the entire capsid ORF has been carried out (12), with more than 40 substitution and insertion mutations being made in a search for regions that could tolerate substitutions and insertions (12). This study identified the critical regions within the capsid that are potentially responsible for receptor binding, DNA packaging, capsid formation, and infectivity. However, it remains difficult to achieve modification of AAV capsid proteins without affecting high titer of production rAAV.
A recent insertional mutation study of the AAV capsid proteins revealed that mutations in the capsid gene could affect AAV capsid assembly and infection (22). Wu et al. have analyzed the phenotypes of 93 AAV2 capsid mutants at 59 different positions within the capsid ORF (33). Several classes of mutants were analyzed, including epitope tag or peptide ligand insertion mutants, alanine-scanning mutants, and epitope substitution mutants. Notably, of the positions identified as being on the surface of the capsid, six were found potentially capable of accepting foreign epitope or ligand insertions for retargeting the viral capsid to alternative receptors. These are the N-terminal region of VP1 (near amino acid [aa] 34), the N terminus of VP2 (aa 138), the loop I region (aa 266), the loop IV region (near aa 447 and 591), and the loop V region (aa 664). All of these locations were capable of tolerating a hemagglutinin or serpin insertion but have the disadvantage that the recombinant virus titers are 1 to 2 logs lower than that achieved with wild-type AAV. Moreover, such modifications within the AAV capsid loop do not always lead to the desired novel tropism, possibly due to peptide constraints after insertion into the loop. The tumor-targeting sequence, NGRAHA, and a Myc epitope control were incorporated either as insertions or as replacements of the original capsid sequence. Viruses were assessed for packaging, accessibility of incorporated peptides, heparin binding, and transduction in a range of cell lines. Whereas recombinant viruses containing mutant capsid proteins were produced efficiently, transduction of several cell lines was impaired significantly for most modifications. Certain mutants containing the peptide motif NGR, which binds CD31 (a receptor expressed in angiogenic vasculature and in many tumor cell lines), displayed an altered tropism toward cells expressing this receptor. Notably, the carboxyl terminus of the VP3 loop, which was modified in this study, is not a constrained loop region of VP3 that has been identified previously as permissive of mutation. Ruffing et al. (23) have characterized deletions of the C terminus of the capsid ORF, and these deletions also were noninfectious.
To further address the issue as to whether VP3 can tolerate addition of a peptide, we used a novel approach to introduce a six-His (6×His) tag at the 5′ carboxyl terminus of VP3, without modifying VP1 or VP2. We found that this modification does not adversely affect the titer of AAV produced or its tropism. Addition of the 6×His tag, which binds to nickel-nitrilotriacetic acid (Ni-NTA) resin, facilitates purification of the AAV using this resin and subsequent elution with a high-salt buffer. The 6×His tag also facilitates detection using biotinylated anti-6×His antibody. Thus, the 6×His-modified AAV VP3 enables purification and detection of AAV without affecting tropism or production.
MATERIALS AND METHODS
Cell culture and reagents.
Human embryonic kidney 293, HeLa, human skin fibroblast, human B-lymphoma Raji, and Jurkat cells were purchased from the American Type Culture Collection (Manassas, Va.). 293 cells, HeLa cells, and human skin fibroblasts were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg of penicillin/ml, and 100 U of streptomycin/ml, and Raji and Jurkat cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 50 μM 2-mercaptoethanol (Invitrogen, Carlsbad, Calif.). The 6×His and RGD peptides were purchased (Invitrogen) and used for competition assays.
AAV vectors.
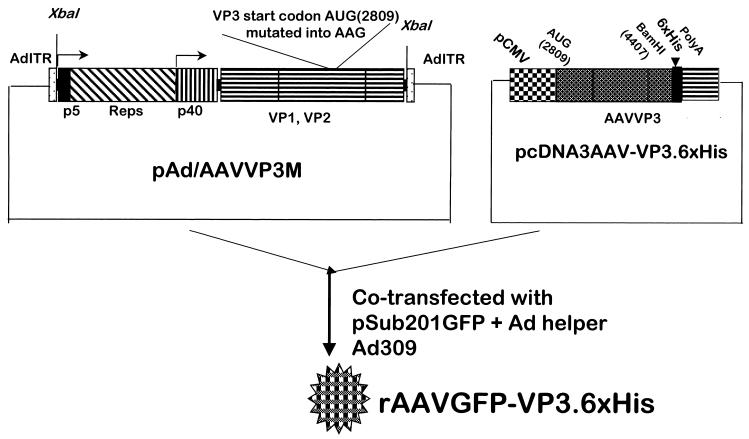
The AAV2-based vector pSub201GFP was used as described previously (35). pAd/AAV, provided by R. J. Samulski (University of North Carolina, Chapel Hill), was used for packaging pSub201GFP followed by infection with adenovirus type 309 (Ad309) as described previously (25). To generate a VP3 with the carboxyl-terminus addition of a 6×His-tagged peptide, the endogenous wild-type AUG translation start codon at position 2809 of AAV for VP3 was mutated to AAG using a site-directed mutagenesis kit (Stratagene, Palo Alto, Calif.), resulting in a mutated (M) pAd/AAV-VP3M (Fig. 1). The mutation was confirmed by partial sequence analysis of the mutated region of pAd/AAVVP3M. The 6×His-tagged VP3 vector was constructed by PCR amplification of AAV2 VP3 using pSub201 as a template. The 5′ oligonucleotide contained a HindIII linker and DNA sequences from positions 2809 to 2829 of wild-type AAV2 (5′-GGAAGCTTATGGCTACAGGCAGCGGCGCA-3′). The 3′ oligonucleotide contained a BamHI linker and DNA sequences from positions 4587 to 4607 of wild-type AAV2 contained in pSub201 (5′-CCGGATCCGAGGCCGGGCGACCAAAGGT-3′). The PCR product was digested with HindIII and BamHI and ligated to pcDNA3 (Invitrogen) as previously digested with the same enzymes to allow directionally insertion. The accuracy of the inserted VP3 was confirmed by direct sequencing. A linker sequence containing a 6×His tag and a flexible linker were synthesized (Invitrogen) and directionally inserted downstream of pcDNA3VP3 at the BamHI and XbaI sites, respectively, resulting in pcDNA3AAV-VP3.6×His.
FIG. 1.
Construction and production of rAAVGFP-VP3.6×His virus. The AAV2-based vector pSub201GFP was used as described previously (36). pAd/AAV provided by R. J. Samulski (University of North Carolina, Chapel Hill) was used for packaging pSub201GFP, and this was followed by infection with Ad309 as described elsewhere (36). To generate a VP3 with the carboxyl-terminal addition of a 6×His-tagged peptide, the endogenous wild-type AUG start codon at position 2809 of AAV for VP3 was mutated to AAG using a site-directed mutagenesis kit (Stratagene), resulting in mutated (M) pAd/AAVVP3M. The 6×His-tagged VP3 vector was constructed by PCR amplification of AAV2 VP3 using pSub201 as a template. A linker sequence containing a 6×His tag and a flexible linker was synthesized and directionally inserted downstream of pcDNA3VP3 at the BamHI and XbaI sites, respectively, to generate pcDNA3AAV-VP3.6×His.
Production and characterization of the 6×His-modified AAV vector.
rAAVGFP-VP3.6×His was generated by cotransfection of pSub201GFP vector (35) with packaging plasmids pAd/AAVVP3M and pcDNA3AAV-VP3.6×His into 293 cells (4 × 106 cells per 10-cm-diameter dish) followed by Ad309 infection (1 PFU/cell). The virus was harvested 60 h after Ad309 infection. Concentration and purification of AAV vectors were carried out as previously described (36). Briefly, clarified crude cell lysates obtained 3 days after transfection were concentrated by centrifugation through a sucrose cushion, followed by density banding on a CsCl gradient and dialysis in phosphate-buffered saline (PBS). Titers of rAAVGFP-VP3.6×His and rAAVGFP vector stocks were determined by counting the number of green fluorescence protein (GFP)-positive cells as previously described (36). Briefly, CsCl gradient-purified rAAVGFP-VP3.6×His virus was diluted 10-fold in PBS buffer and heat-inactivated at 56°C for 30 min to eliminate Ad309 contamination. The diluted and heat-inactivated rAAVGFP-VP3.6×His virus was added to 293 cells that were grown to 80% confluence. After 2 h of incubation in a 5% CO2 at 37°C in a humidified chamber, the cells were washed with PBS, followed by incubation for an additional 48 h at 37°C. The GFP-positive cells were then counted using a fluorescent light microscope. The titer, expressed in transduction units (TU) per cell, was determined as follows: (number of GFP-positive cells × dilution factor × volume of initial viral inoculation)/(total number of initially seeded 293 cells).
The level of contaminated wild-type AAV was determined using a virus replication assay as described previously (35) and was found to be less than one functional particle per 1011 rAAVGFP-VP3.6×His particles. Contamination with Ad309 helper adenovirus was evaluated by incubation of 1% of the purified vector stock with 293 cells and scoring for the adenovirus cytopathic effect after 7 days. Adenovirus contamination was consistently less than one infectious Ad309 particle per 1.8 × 1011 rAAVGFP-VP3.6×His particles.
Purification of rAAVGFP-VP3.6×His and rAAVGFP vectors on Ni-NTA resin columns.
To avoid contamination by soluble VP3-6×His in the cultured supernatants, 60 h after transfection, the rAAV-transfected cells were harvested and washed three times with PBS. The virus particles were released from the 293 cells by three freeze-thaw cycles, and the free plasmid DNA were removed by first treatment with DNase I and deoxycholate followed by centrifugation at 21,000 × g for 15 min at 4°C as described previously (35). The soluble fraction containing rAAV was then mixed at a ratio of 50 to 1 (vol/vol) with Ni-NTA-agarose beads equilibrated in binding buffer (Qiagen, Stanford, Calif.) and incubated with agitation at 4°C overnight. The beads were then packed in a plastic disposable column (Bio-Rad, Hercules, Calif.) and washed with 100 bed volumes of wash buffer (10 mM Tris [pH 8.0], 300 mM NaCl, 20 mM imidazole [pH 7.0]) to remove nonspecifically bound material. Residual wash buffer was removed by centrifugation (2 min at 2,000 rpm). Ni-NTA-agarose-bound AAV was eluted by incubation in 1 bed volume of 100 or 500 mM imidazole (pH 7.0) elution buffers. The virus was then dialyzed against 4 liters of dialysis buffer containing 3% sucrose, 150 mM NaCl, 10 mM Tris (pH 7.4), and 1 mM MgCl2 for 4 h at 4°C and stored at −70°C.
The eluted fractions were collected and dot blot hybridization was carried out using a [32P]dCTP-labeled GFP probe as described in the published protocol (Amersham, Piscataway, N.J.) to quantify the eluted rAAVGFP-VP3.6×His or rAAVGFP.
6×His competition assay for rAAV-VP3.6×His virus.
Ni-NTA-agarose bead columns were prepared as described above, and then elevated doses (dose ranges from 0 to 100 mM) of 6×His or a control peptide RGD were mixed with 109 viral DNA particles of rAAVGFP-VP3.6×His or rAAVGFP. The viral suspensions were subsequently incubated with Ni-NTA-agarose beads as described above. The bound virus was eluted, and dot blot hybridization was carried out using a method identical to that described above. The intensity of each dot was scanned and quantified by the Quality One software program (Bio-Rad). The data were expressed as counts per minute.
Tropism of the virus.
To evaluate whether 6×His modification of AAV VP3 alters viral tropism, rAAVGFP-VP3.6×His and rAAVGFP (500 TU/cell) were incubated with a range of cells, including 293 cells, HeLa cells, normal human skin fibroblasts, Raji human B cells, and Jurkat human T cells (American Type Culture Collection). The cells were transfected by incubation with the virus for 2 h in medium supplements with 1% bovine serum albumin. The cells were washed and cultured for an additional 48 h. The cells underwent cytospinning and then were probed with a GFP probe to quantify viral DNA particles per cell. Briefly, the transfected cells were incubated with proteinase K (Boehringer, Petersburg, Va.) in a solution containing 10 mM Tris HCl (pH 8.0), 10 mM EDTA, and 1% sodium dodecyl sulfate for 1 h at 37°C and then blotted toa GeneScreen Plus (Stratagene, La Jolla, Calif.) membrane by using a dot blot manifold, followed by UV cross-linking at 60 mJ (UV Stratalinker 1800; Stratagene). Finally, the blot was hybridized with a [32P]dCTP-labeled GFP probe. The signal was scanned with a Cyclone phosphorimaging screen system (Packard Instrument, Meriden, Conn.) and quantified by the Quality One software program (Bio-Rad). To convert the dot blot DNA intensity to viral DNA particles, 1 ng of AAV viral genomic DNA was considered to be 3.8 × 108 viral DNA particles. A standard dot blot hybridization curve was generated using different known amounts of wild-type rAAVGFP as a standard compared with unknown amounts of rAAVGFP-VP3.6×His. The transduction efficiency of rAAVGFP-VP3.6×His was calculated and expressed as viral DNA particles per cell.
Western blot analysis.
The expression of capsid proteins produced by rAAVGFP or rAAVGFP-VP3.6×His was analyzed by Western blot analysis. In brief, total proteins from double-CsCl-purified rAAVGFP or rAAVGFP-VP3.6×His virus were extracted and separated on a sodium dodecyl sulfate-10% polyacrylamide gel and electrophoretically transferred onto a nitrocellulose membrane (35). The membrane was incubated with the AAV capsid-specific antibody B1 (Research Diagnostics Inc., Flanders, N.J.) or a 6×His antibody (Qiagen). The capsid proteins were visualized with a horseradish peroxidase (HRP)-coupled goat anti-mouse secondary antibody (Southern Research Institutes, Birmingham, Ala.) and a chemiluminescent detection system (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) (35). To quantify the amount of each of the AAV capsids, the signal intensities of VP1, VP2, and VP3 were determined using a Cyclone phosphorimaging screen system (Packard Instrument) and quantified by the Quality One software program (Bio-Rad). The ratios of VP1 to VP2 to VP3 were calculated.
Enzyme-linked immunosorbent assay (ELISA) to determine levels of 6×His-modified AAV.
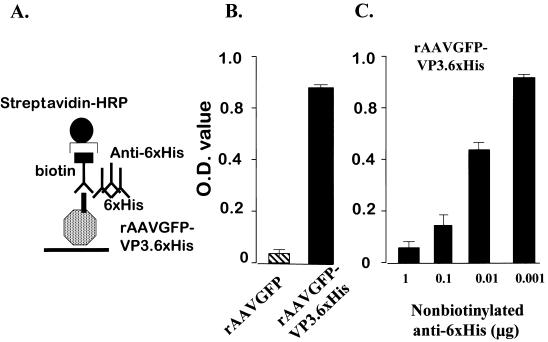
An ELISA was developed to determine the concentration of the 6×His-modified AAV capsid protein. First, the CsCl-purified rAAVGFP-VP3.6×His or rAAVGFP viruses was used to prevent potential contamination of soluble VP3-6×His protein. The plates were then coated with the 109 viral DNA particles of 6×His-modified rAAVGFP-VP3.6×His or rAAVGFP in carbonate coating buffer (pH 9.6), in 96-well plates. The virus was incubated in the 96-well plate overnight. After gently washing to remove unbound rAAVGFP-VP3.6×His or rAAVGFP, the rAAVGFP-VP3.6×His capsid protein was detected by incubation with biotinylated anti-6×His followed by washing. The third step was the addition of 1:3,000 diluted streptavidin-HRP (R&D, Minneapolis, Minn.) to detect biotinylated anti-6×His. To show specificity of binding, different levels of nonbiotinylated anti-6×His were added prior to the addition of the 6×His-biotin reagent, and the procedure was then carried out as above. The optical density at 409 nm was determined using an E-Max ELISA plate reader (Molecular Devices Corporation, Sunnyvale, Calif.).
RESULTS
Construction of rAAV-VP3.6×His-tagged rAAV.
VP3.6×His was constructed using the full-length of AAVVP3 ligated in frame to the His tag at the C terminus. This VP3.6×His was placed under the control of a cytomegalovirus promoter followed by a poly(A) tail and cloned into the pcDNA3 expression vector (Fig. 1). To prevent endogenous expression of VP3, the start codon of AAV VP3 was mutated from AUG to AAG (Fig. 1) and renamed pAd/AAVVP3M. The rAAVGFP-VP3.6×His virus was generated by cotransfection of three plasmids—pAd/AAVVP3M, pcDNA3-VP3.6×His, and pSub201GFP—as described previously, using the calcium phosphate precipitation method (36). The cells were then infected with Ad309 required for packaging of AAV.
6×His-tagged VP3 protein integrated into packaged rAAVGFP-6×HisVP3.
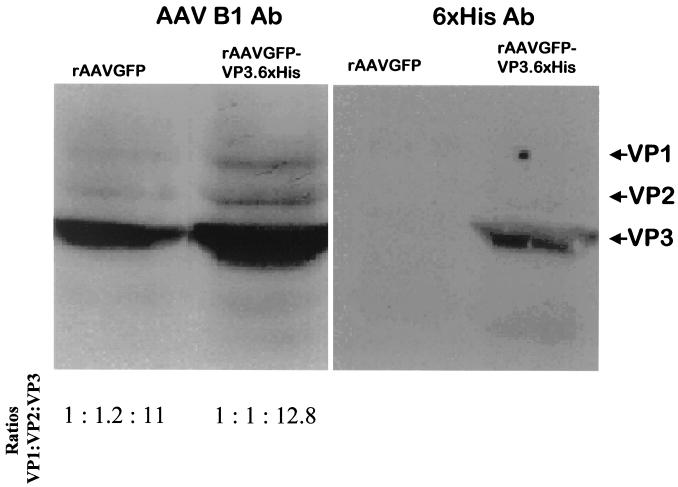
To determine if the 6×His-modified VP3 capsid was present in the intact rAAVGFP-6×HisVP3, Western blot analysis was carried out using this protein extracted from the purified non-capsid-modified rAAVGFP or capsid-modified rAAVGFP-VP3.6×His. The blots were probed with an anti-AAV B1 antibody that recognizes all three capsid proteins (VP1, VP2, and VP3) (Fig. 2). Compared to the rAAVGFP control, equivalent amounts of VP1, VP2, and VP3 were detected at the appropriate ratios in the rAAVGFP-VP3.6×His modified virus. Thus, the rAAVGFP-VP3.6×His containing the modified VP3 capsid protein does not result in an altered ratio of capsid proteins produced by rAAVGFP-VP3.6×His compared to non-capsid-modified rAAVGFP. When an identical blot was probed with a 6×His-specific antibody, a protein with a molecular weight equivalent to that of VP3 was detected in the rAAVGFP-VP3.6×His-modified virus, but not in the wild-type virus rAAVGFP (Fig. 2). Thus, the 6×His tag was detected in the AAV capsid VP3 protein such that it retained its reactivity with an anti-6×His antibody.
FIG. 2.
6×His-tagged VP3 is incorporated into the rAAVGFP-VP3.6×His virion. Protein from approximately 107 viral particles as determined by DNA dot blot hybridization were loaded and probed either with an anti-AAV capsid antibody (clone B1) that recognizes all three viral particle capsid proteins (VP1, VP2, and VP3) or with an anti-6×His antibody, followed by detection using HRP-conjugated goat anti-mouse antibody. The membrane was developed as described in Materials and Methods.
The 6×His modification of VP3 does not affect the rAAVGFP-VP3.6×His viral titer.
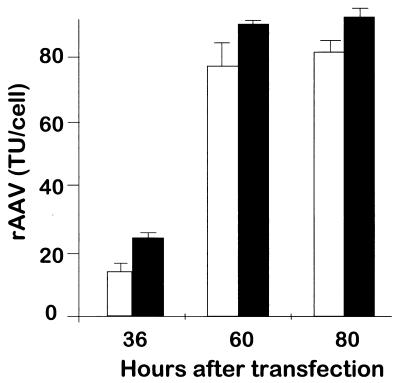
To determine if modification of the VP3 protein by 6×His in rAAVGFP-VP3.6×His affects the growth and final titer of the virus, wild-type rAAVGFP and rAAVGFP-VP3.6×His were titrated by counting the number of GFP-positive transfected 293 cells. There was no delay in the initial production of rAAVGFP-VP3.6×His compared to wild-type rAAVGFP as determined by the number of TU per cell at 36 h (Fig. 3). There also was no decrease in the maximal yield of rAAVGFP-VP3.6×His compared to wild-type rAAVGFP as determined by the number of TU per cell at 60 and 80 h (Fig. 3). Thus, the modification of the VP3 capsid protein with the 6×His did not inhibit the production of virus.
FIG. 3.
Addition of the 6×His tag to AAV VP3 does not affect viral titer. Both wild-type rAAVGFP and rAAVGFP-VP3.6×His were produced in the 293 cells and harvested at different time points as described in the text. Titers of rAAVGFP-VP3.6×His and rAAVGFP vector stocks were determined by counting the number of GFP-positive cells at 48 h after transfection. The GFP-positive cells were counted using a fluorescent light microscope. The titer, expressed as TU per cell, was determined as follows: (number of GFP positive cells × dilution factor × volume of initial viral inoculation)/(total number of initially seeded 293 cells). Each bar represents the mean of three independent experiments (error bars, standard errors of the means). ▪, rAAVGFP; □, rAAVGFP-VP3.6×His.
The 6×His modification does not affect the AAV viral tropism.
Modification of the capsids can alter the native tropism of the virus, making it difficult to both transfect 293 cells that might affect production capacity and to utilize the virus in gene therapy. To determine the tropism of the rAAVGFP-VP3.6×His with the 6×His-modified VP3, members of a panel of five different human cell lines were transfected with equivalent amounts of either rAAVGFP or rAAVGFP-VP3.6×His. The cells were harvested 48 h after transfection, and the viral particles per cell were determined by DNA dot blot analysis. As previously described, unmodified AAV exhibits high tropism for 293 cells and low tropism for lymphoma cells lines, such as Raji and Jurkat (Table 1). The rAAVGFP-VP3.6×His exhibited a similar cellular tropism compared to wild-type rAAVGFP-transfected cells, indicating that the 6×His modification of VP3 does not alter endogenous viral tropism.
TABLE 1.
Addition of 6×His tag to AAV VP3 does not change AAV capsid viral tropisma
| Cell line | Mean no. of viral DNA particles/cell (103) after transfection with:
|
|
|---|---|---|
| rAAGFP-VP3.6×His | rAAVGFP | |
| 293 | 448 | 392 |
| HeLa | 392 | 312 |
| Skin fibroblast | 168 | 152 |
| Raji (B cell) | 3.2 | 0.8 |
| Jurkat (T cell) | 4.4 | 3.6 |
Five different human cell lines were transfected with equivalent numbers (500 TU/cell) of either rAAVGFP or rAAVGFP-VP3.6×His. The cells were harvested after 48 h, and the number of viral particles per cell was determined by DNA dot blot analysis as described elsewhere (33). Data are expressed as means of the results of three independent experiments.
Ni-NTA resin column purification of rAAV-VP3.6×His.
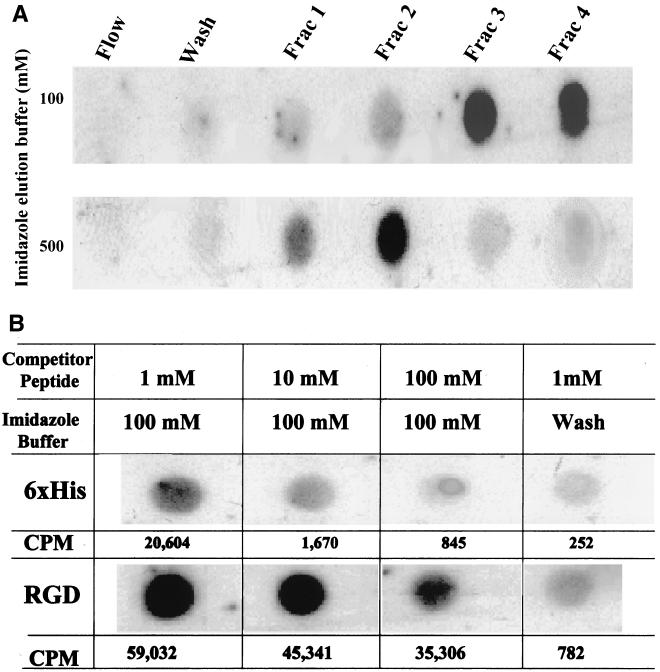
The 6×His modification of VP3 was made with the intent of enabling enrichment and purification of the AAV using the Ni-NTA resin that has been used for purification of proteins tagged with six consecutive histidine residues. The binding of the AAVGFP-VP3.6×His virus particles to the column was confirmed by loading rAAV-VP3.6×His virus particles in low-salt binding buffer onto a 2-ml Ni-NTA resin column. The column was first washed three times with the low-salt binding buffer and then the bound virus particles were eluted in either 100 mM imidazole (pH 7.0) or 500 mM imidazole (pH 7.0) (Fig. 4A). No detectable AAV virus was eluted in the low-salt binding buffer used to load the column (flow). Also, no AAV was detectable by addition of a 0.5-ml fraction of low-salt buffer (wash), indicating that the 6×His-tag VP3 was bound to the column. Optimal elution of the rAAFGFP-VP3.6×His occurred either in the third or fourth elution fractions using 100 mM elution buffer (fraction 3 and fraction 4). The most optimal elution of the rAAVGFP-VP3.6×His occurred in the second fraction after elution with 500 mM imidazole (pH 7.0), and most of the virus was eluted in this single 0.5-ml fraction (Fig. 4A).
FIG. 4.
Affinity purification of rAAVGFP-VP3.6×His by an Ni-NTA column. An Ni-NTA column was constructed using a 2-ml pipette as described in the text. (A) rAAVGFP-VP3.6×His virus particles released from 293 cells were loaded on the column in 50 μl of 20 mM imidazole salt binding buffer. The last fraction of loading sample is referred to as flow. The column was washed three times with this low-salt buffer, and the last fraction is referred to as wash. rAAVGFP-VP3.6×His was eluted in 0.5-ml fractions (Frac) of elution buffer consisting of either 100 mM imidazole or 500 mM imidazole (pH 7.0) buffer. Viral elution was determined by dot blot analysis. (B) The binding specificity of rAAVGFP-VP3.6His virus to the Ni-NTA column was demonstrated by addition of different concentrations of competitor 6×His peptide or a control peptide RGD (concentrations ranged from 1 to 100 mM) to the rAAVGFP-VP3.6×His. The virus was eluted in fraction 3 of 100 mM imidazole (pH 7.0) as predetermined in Fig. 5A. The intensity of each dot was quantified using a phosphorimaging system as described in the text. The data are presented as counts per minute of [32P]dCTP radioisotope.
It is possible that the rAAVGFP-VP3.6×His could be nonspecifically bound to the Ni-NTA resin or specifically bound by interaction with the 6×His incorporated in the rAAVGFP-VP3.6×His virus. To distinguish between these possibilities, a column was constructed as described above and loaded with rAAVGFP-VP3.6×His virus (106 TU) in a low-salt buffer containing different concentrations of competitor peptide 6×His or a control peptide RGD at 1, 10, or 100 mM. The column was washed with 20 mM imidazole (pH 7.0) buffer. Most of the virus was eluted in a third fraction of 0.5 ml of 100 mM imidazole (pH 7.0) buffer (Fig. 4A). Viral binding was greatly reduced by addition of 1 mM 6×His to the binding buffer, which resulted in a threefold decrease in the amount of virus eluted in this fraction (Fig. 4B, column 1). The ability of the 6×His tag to inhibit binding of the virus was even more apparent using binding buffer that contained a 10 or 100 mM concentration of the 6×His peptide. This result indicates that free 6×His effectively competes with 6×His expressed on the VP3 component of the virus capsid and is significantly more efficient at competition of binding compared to the control RGD peptide. A high concentration of imidazole (100 mM, pH 7.0) buffer is required for elution of bound virus, since elution with low-salt buffer (Fig. 4B, wash) does not elute significant viral particles from the 6×His-pretreated column or the RGD-pretreated column.
ELISA for detection of rAAV-VP3.6×His.
The above results indicate that the 6×His epitope specifically binds to the Ni-NTA resin and can be inhibited by excess amounts of 6×His. Conversely, to show that the anti-6×His antibody specifically recognizes the 6×His epitope tag on the virus, an ELISA was developed to determine if anti-6×His can specifically block binding of a biotinylated anti-6×His to the virus (Fig. 5A). rAAVGFP-VP3.6×His and controlled rAAVGFP virus were purified over two CsCl gradients. Ninety-six-well plates were first coated with equivalent amounts (106 TU) of either rAAVGFP or rAAVGFP-VP3.6×His virus. A two-step ELISA was carried out by first washing free virus and then incubating the wells with biotinylated antibody specific for 6×His and carrying out detection using a secondary streptavidin-HRP as a reporter. There was high ELISA activity in wells coated with the rAAVGFP-6×His.VP3, but not in the control wells coated with rAAVGFP (Fig. 5B). To next determine if binding of biotinylated anti-6×His could be specifically inhibited by an unlabeled anti-6×His antibody, 96-well plates that had previously been coated with rAAVGFP-VP3.6×His and washed were first incubated with different concentrations of a nonbiotinylated anti-6×His, ranging from 1 to 0.001 μg/ml. After washing, the ELISA was carried out using the biotinylated anti-6×His followed by streptavidin-HRP as described above. There was a dose-dependent inhibition of the binding of the biotinylated anti-6×His after pretreatment with nonbiotinylated anti-6×His, indicating that the binding of biotinylated anti-6×His was specific for the 6×His epitope on rAAVGFP-VP3.6×His (Fig. 5C).
FIG. 5.
ELISA to detect 6×His-tagged VP3 integrated into AAV. (A) ELISA. After CsCl purification, 109 viral DNA particles of either 6×His-modified rAAVGFP-VP3.6×His or rAAVGFP viral particles was suspended in carbonate coating buffer (pH 9.6) and used to coat 96-well plates overnight. The VP3.6×His was detected using a biotin-labeled mouse anti-6×His antibody with and without different concentrations of nonbiotinylated 6×His antibody. The biotinylated 6×His antibody was detected using HRP-conjugated streptavidin. (B) Plates were coated with 109 6×His-modified rAAVGFP-VP3.6×His or rAAVGFP viral DNA particles, and the VP3.6×His antigen was detected using a biotin-labeled mouse anti-6×His antibody followed by HRP-conjugated streptavidin. The VP3.6×His antigen was then quantified by the ELISA. The data represent the means of three separate experiments (error bars, standard errors of the means). (C) Plates were coated with 109 6×His-modified rAAVGFP-VP3.6×His viral DNA particles. The wells were first incubated with different concentrations of nonbiotinylated 6×His antibody, and then the ELISA was carried out as described above. The results represent the means of three separate experiments (error bars, standard errors of the means).
DISCUSSION
Previous investigators have analyzed regions of the AAV capsid that can be mutated, or can tolerate insertion of a peptide, to identify modifiable sites compatible with production of AAV. Mutation of the carboxyl-terminal end has been analyzed by Ruffing et al. (23) and has been shown to be not permissive for production of stable virus. Capsid initiation codon mutagenesis studies suggested that both VP2 and VP3 were required for capsid formation and production of infectious particles, and either VP1 or VP2 was required for nuclear localization of VP3. A recent insertional mutation study on AAV capsid protein revealed that mutations in the capsid gene could affect AAV capsid assembly and infection. Addition of a 6×His tag has not previously been investigated. We, therefore, first modified the AAV by an in-frame addition of the 6×His coding sequence to the 3′ end of AAV. This would result in production of VP1, VP2, and VP3 with a 6×His modification of the carboxyl terminus. This construct was cotransfected with pSub201GFP plus the adenovirus helper Ad309 into 293 cells. We found that no AAV was produced under these circumstances (data not shown). This result demonstrates that addition of the 6×His tag to all three of the AAV capsid proteins at their carboxyl terminus is incompatible with formation of viral particles or production of a stable virus.
In the present experiments, we therefore have constructed the pAd/AAV-VP3M mutant vector, in which the VP3 starting translational codon has been mutated from AUG to AAG at position 2809. In this vector, VP1 and VP2 transcription was under the control of the endogenous p40 promoter,and the rep genes were produced using the endogenous p5 promoter. This resulted in production of rep and cap genes except with VP3 due to the mutation of the start codon of VP3 at position 2809. By itself, this construct does not result in production of AAV since VP3 is not produced. We therefore constructed a second vector, pcDNA3AAV-VP3.6×His, that contains the AAV VP3 ORF fused with 6×His tag at the C terminus. The resultant vector, pcDNA3AAV-VP3.6×His, is shown in Fig. 1 and enables the production of AAV VP3 fused with a 6×His tag. Cotransfection of pAd/AAV-VP3M plus pcDNA3AAV-VP3.6×His into 293 cells with the pSub201GFP plus adenovirus helper Ad309 resulted in equivalent and high-level production of the rAAV containing the VP3.6×His-modified capsid. This result shows that modification of all three capsids at the carboxyl terminus with 6×His is not permissive to production of AAV, but wild-type VP1 and VP2 along with modification of only VP3 with a 6×His tag at the carboxyl terminus does result in the production of equivalent titers of AAV that incorporates this VP3.6×His tag. We propose that wild-type VP1 and VP2 along with mutated VP3 result in a stable capsid formation. This is important for future capsid development with AAV, since VP1, VP2, and VP3 are expressed in the capsid at a 1:1:10 ratio. However, an intact VP1 and VP2 are sufficient to enable production of AAVs that have mutations of VP3 alone. This suggests the possibility of analyzing the retargeting and mutations of each capsid protein individually, such as demonstrated here with VP3, which might permit a broader range of mutations or modifications that are not possible if all of the capsid proteins are modified.
To determine if the modification of VP3 resulted in altered tropism of rAAVGFP-6×His.VP3, we analyzed the transfection efficiency of AAV in four cell lines by two methods. The first method was the number of GFP-positive cells at different dilutions of rAAVGFP-6×His compared to rAAVGFP. There was no difference in the efficiency of transfection of 293 cells when rAAVGFP and rAAVGFP-6×His.VP3 were compared. We also compared the expression of rAAVGFP and rAAVGFP-6×His.VP3 in other cell lines, including foreskin fibroblasts, Raji B cells, and Jurkat T cells. The peak expression of GFP was delayed for up to 3 days in foreskin fibroblasts and for more than 3 days in Raji B cells and Jurkat T cells (data not shown). It was therefore difficult to accurately quantify the transduction efficiency in these relatively nonpermissive cells, using the GFP method. Therefore, to avoid underestimation of the transfection efficiency of rAAV due to this variable and delayed peak expression in different cell lines, we also used a dot blot method to verify the GFP results in 293 cells and to accurately quantify the transfection efficiency in relatively nonpermissive cell types. The dot blot method does not depend on expression of GFP and directly measures AAV genomic viral particle DNA. Using the dot blot method, we confirmed that there was no difference in the transfection efficiency of our AAVGFP compared to rAAVGFP-6×His.VP3 in either permissive 293 cells and HeLa cells and also relatively nonpermissive foreskin fibroblasts, Raji B cells, and Jurkat T cells. Therefore, together these results indicate that the VP3.6×His modification does not effect transfection efficiency of rAAVGFP-6×His.VP3.
We elected to use a strong cytomegalovirus promoter to express AAV VP3 since we want to assure efficient production of the carboxyl terminus 6×His mutated form of AAV VP3 capsid encoding transcript and protein. Use of this promoter did not interfere with production of AAV, and there was equivalent or higher production of rAAVGFP-VP3.6×His under these conditions. Furthermore, several arguments can be made that the capsid-modified VP3 is incorporated into AAV capsids. (i) It is unlikely that wild-type VP3 would be produced by the second plasmid vector that produced VP1 and VP2, since this vector has a mutation in the start codon for VP3. Therefore, in the capsid modified AAV, all VP3 should contain the 6×His modification. (ii) The ratio of VP1, VP2, and VP3 capsid proteins of both wild-type AAV and 6×His-modified AAV origin are equivalent (Fig. 2A). A dramatic increase in VP1 and VP2, such as would be seen with formation of pseudocapsids containing high ratios of VP1 or VP2, would be expected to alter this ratio. (iii). Unincorporated soluble 6×His was not a major component of the preparation, since it was eliminated by the AAV purification procedures. If this were present, the ratio of VP3, relative to VP1 and VP2, would be expected to be increased. (iv). Although the intensity of the VP3 was different in the Western blot using the B1 antibody compared to that using the 6×His antibody, this may be due to differences in the affinity of the antibody for 6×His-modified VP3 compared to the affinity of the antibody for the unmodified capsid. In addition, the secondary biotinylated antibody may contribute to the intensity difference. (v) Altered levels of VP3 would be expected to lead to capsid instability and difference in production titer in wild-type and capsid-modified AAV. However, capsid-modified AAV VP3 leads to the formation of a stable capsid and there is no difference in titer (Fig. 3). These results show that the heterologous and high expressive promoter pCMV can be used to efficiently drive expression of the rAAVGFP-VP3.6×His modified capsid protein and that this may result in stable utilization of this modified VP3 capsid protein.
One major limitation of AAV includes the lack of a high-affinity method for purification of AAV (2, 3, 8, 10, 20) or the necessity for HPLC purification (6, 9). AAV can also be purified over a column of heparin sulfate column, which is one of the target binding molecules for the AAV capsid. Other target binding molecules and therefore columns that have been produced include the heparin sulfate proteoglycan and αvβ5 integrin adhesion molecule column (6, 30). These columns improve purification, but the affinity of AAV for these columns is variable and the elution procedures can sometimes be too harsh, therefore neutralizing some of the virus or limiting the concentration of the virus. A third limitation of AAV is the difficulty of producing high-titer virus. This is related, in part, to isolation of pure virus and difficulty in production of infectious virus made with capsid modification. In this study, we demonstrated that the 6×His-modified AAV was further utilized for purification of the AAV that enables high binding to an Ni-NTA column. This modification results in production of a virus that exhibits normal AAV tropism and does not inhibit production of AAV. The 6×His tag binds to an Ni-NTA resin, and therefore AAV exhibits tight binding to this resin and can be easily eluted with a salt buffer. The binding is specific, since pretreatment of the column with 6×His peptide prevents binding of the AAV to the resin. The 6×His tag exhibits high-affinity binding to biotinylated anti-6×His antibody, which can be detected by the second reagent (streptavidin-HRP). This activity can be inhibited by nonbiotinylated 6×His antibody.
The 6×His modification and the modification of VP3 in the presence of wild-type VP1 and VP2 could therefore offer a versatile ligand which can be used for retargeting of the AAV. AAV type 2 infects a broad range of cells by binding to its primary receptor, heparin sulfate proteoglycan (30). Two types of coreceptors, αvβ5 integrin and fibroblast growth factor receptor-1, have been implicated in the subsequent internalization process (21, 30). The present results demonstrate that a biotin anti-6×His antibody combined with the 6×His-modified AAV VP3 with high affinity can be specifically inhibited with an anti-6×His antibody. This could be used to create a biotin bridge to enable altered tropism for cells that exhibit low transduction by unmodified AAV. Modification of VP3 in the presence of wild-type VP1 and VP2 will enable development of stable AAV with additional novel mutations of VP3. These mutations may confer desirable properties, such as novel tissue tropism, to AAV.
Acknowledgments
We thank E. Hunter for expert review of the manuscript and L. Flurry for expert secretarial assistance.
Hui-Chen Hsu is a recipient of a grant from the Center for Aging at the University of Alabama at Birmingham. Huang-Ge Zhang is a recipient of an Arthritis Foundation Investigator Award. This work is supported by NIH grants R01 AG 11653, N01 AR 6-2224, and RO1 AI 42900 and a Birmingham VAMC merit review grant to J.D.M. and H.-G.Z.
REFERENCES
- 1.Agbandje, M., S. Kajigaya, R. McKenna, N. S. Young, and M. G. Rossmann. 1994. The structure of human parvovirus B19 at 8 Å resolution. Virology 203:106-115. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R., I. Macdonald, T. Corbett, A. Whiteway, and H. G. Prentice. 2000. A method for the preparation of highly purified adeno-associated virus using affinity column chromatography, protease digestion and solvent extraction. J. Virol. Methods 85:23-34. [DOI] [PubMed] [Google Scholar]
- 3.Auricchio, A., G. Kobinger, V. Anand, M. Hildinger, E. O'Connor, A. M. Maguire, J. M. Wilson, and J. Bennett. 2001. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum. Mol. Genet. 10:3075-3081. [DOI] [PubMed] [Google Scholar]
- 4.Cassinotti, P., M. Weitz, and J. D. Tratschin. 1988. Organization of the adeno-associated virus (AAV) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein 1. Virology 167:176-184. [PubMed] [Google Scholar]
- 5.Chu, C. Y., M. J. Pan, and J. T. Cheng. 2001. Genetic variation of the nucleocapsid genes of waterfowl parvovirus. J. Vet. Med. Sci. 63:1165-1170. [DOI] [PubMed] [Google Scholar]
- 6.Clark, K. R., X. Liu, J. P. McGrath, and P. R. Johnson. 1999. Highly purified recombinant adeno-associated virus vectors are biologically active and free of detectable helper and wild-type viruses. Hum. Gene Ther. 10:1031-1039. [DOI] [PubMed] [Google Scholar]
- 7.Costello, F., N. Steenfos, K. T. Jensen, J. Christensen, E. Gottschalck, A. Holm, and B. Aasted. 1999. Epitope mapping of Aleutian mink disease parvovirus virion protein VP1 and 2. Scand. J. Immunol. 49:347-354. [DOI] [PubMed] [Google Scholar]
- 8.Debelak, D., J. Fisher, S. Iuliano, D. Sesholtz, D. L. Sloane, and E. M. Atkinson. 2000. Cation-exchange high-performance liquid chromatography of recombinant adeno-associated virus type 2. J. Chromatogr. B Biomed. Sci. Appl. 740:195-202. [DOI] [PubMed] [Google Scholar]
- 9.Drittanti, L., C. Jenny, K. Poulard, A. Samba, P. Manceau, N. Soria, N. Vincent, O. Danos, and M. Vega. 2001. Optimised helper virus-free production of high-quality adeno-associated virus vectors. J. Gene Med. 3:59-71. [DOI] [PubMed] [Google Scholar]
- 10.Gao, G., G. Qu, M. S. Burnham, J. Huang, N. Chirmule, B. Joshi, Q. C. Yu, J. A. Marsh, C. M. Conceicao, and J. M. Wilson. 2000. Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum. Gene Ther. 11:2079-2091. [DOI] [PubMed] [Google Scholar]
- 11.Girod, A., M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deleage, and M. Hallek. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5:1052-1056. [DOI] [PubMed] [Google Scholar]
- 12.Grifman, M., M. Trepel, P. Speece, L. B. Gilbert, W. Arap, R. Pasqualini, and M. D. Weitzman. 2001. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 3:964-975. [DOI] [PubMed] [Google Scholar]
- 13.Hermonat, P. L., M. A. Labow, R. Wright, K. I. Berns, and N. Muzyczka. 1984. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J. Virol. 51:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoque, M., N. Shimizu, K. Ishizu, H. Yajima, F. Arisaka, K. Suzuki, H. Watanabe, and H. Handa. 1999. Chimeric virus-like particle formation of adeno-associated virus. Biochem. Biophys. Res. Commun. 266:371-376. [DOI] [PubMed] [Google Scholar]
- 15.Jay, F. T., C. A. Laughlin, and B. J. Carter. 1981. Eukaryotic translational control: adeno-associated virus protein synthesis is affected by a mutation in the adenovirus DNA-binding protein. Proc. Natl. Acad. Sci. USA 78:2927-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotin, R. M., J. C. Menninger, D. C. Ward, and K. I. Berns. 1991. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics 10:831-834. [DOI] [PubMed] [Google Scholar]
- 17.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olijslagers, S., A. Y. Dege, C. Dinsart, M. Voorhoeve, J. Rommelaere, M. H. Noteborn, and J. J. Cornelis. 2001. Potentiation of a recombinant oncolytic parvovirus by expression of apoptin. Cancer Gene Ther. 8:958-965. [DOI] [PubMed] [Google Scholar]
- 20.O'Riordan, C. R., A. L. Lachapelle, K. A. Vincent, and S. C. Wadsworth. 2000. Scaleable chromatographic purification process for recombinant adeno-associated virus (rAAV). J. Gene Med. 2:444-454. [DOI] [PubMed] [Google Scholar]
- 21.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 22.Rabinowitz, J. E., W. Xiao, and R. J. Samulski. 1999. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology 265:274-285. [DOI] [PubMed] [Google Scholar]
- 23.Ruffing, M., H. Heid, and J. A. Kleinschmidt. 1994. Mutations in the carboxy terminus of adeno-associated virus 2 capsid proteins affect viral infectivity: lack of an RGD integrin-binding motif. J. Gen. Virol. 75:3385-3392. [DOI] [PubMed] [Google Scholar]
- 24.Ruffing, M., H. Zentgraf, and J. A. Kleinschmidt. 1992. Assembly of viruslike particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J. Virol. 66:6922-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samulski, R. J., L. S. Chang, and T. Shenk. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 63:3822-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson, A. A., B. Hebert, G. M. Sullivan, C. R. Parrish, Z. Zadori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189-1198. [DOI] [PubMed] [Google Scholar]
- 27.Smith, R. H., and R. M. Kotin. 2000. An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-stranded AAV ori DNA. J. Virol. 74:3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith-Arica, J. R., and J. S. Bartlett. 2001. Gene therapy: recombinant adeno-associated virus vectors. Curr. Cardiol. Rep. 3:43-49. [DOI] [PubMed] [Google Scholar]
- 29.Steinbach, S., A. Wistuba, T. Bock, and J. A. Kleinschmidt. 1997. Assembly of adeno-associated virus type 2 capsids in vitro. J. Gen. Virol. 78:1453-1462. [DOI] [PubMed] [Google Scholar]
- 30.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tratschin, J. D., I. L. Miller, and B. J. Carter. 1984. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J. Virol. 51:611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trempe, J. P., and B. J. Carter. 1988. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J. Virol. 62:3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Q., M. Mamounas, G. Yu, S. Kennedy, B. Leaker, J. Merson, F. Wong-Staal, M. Yu, and J. R. Barber. 1998. Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Hum. Gene Ther. 9:1929-1937. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, H. G., Y. M. Wang, J. F. Xie, X. Liang, H. C. Hsu, X. Zhang, J. Douglas, D. T. Curiel, and J. D. Mountz. 2001. Recombinant adenovirus expressing adeno-associated virus cap and rep proteins supports production of high-titer recombinant adeno-associated virus. Gene Ther. 8:704-712. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, H. G., J. Xie, P. Yang, Y. Wang, L. Xu, D. Liu, H. C. Hsu, T. Zhou, C. K. Edwards III, and J. D. Mountz. 2000. Adeno-associated virus production of soluble tumor necrosis factor receptor neutralizes tumor necrosis factor alpha and reduces arthritis. Hum. Gene Ther. 11:2431-2442. [DOI] [PubMed] [Google Scholar]