Abstract
During asymmetric cell division in Drosophila sensory organ precursors (SOPs), the Numb protein segregates into one of the two daughter cells, in which it inhibits Notch signalling to specify pIIb cell fate. We show here that Numb acts in SOP cells by inducing the endocytosis of Sanpodo, a four-pass transmembrane protein that has previously been shown to regulate Notch signalling in the central nervous system. In sanpodo mutants, SOP cells divide symmetrically into two pIIb cells. We show that Sanpodo is cortical in pIIa, but colocalizes with Notch and Delta in Rab5- and Rab7-positive endocytic vesicles in pIIb. Sanpodo endocytosis requires α-Adaptin, a Numb-binding partner involved in clathrin-mediated endocytosis. In numb or α-adaptin mutants, Sanpodo is not endocytosed. Surprisingly, this defect is observed already before and during mitosis, which suggests that Numb not only acts in pIIb, but also regulates endocytosis throughout the cell cycle. Numb binds to Sanpodo by means of its phosphotyrosine-binding domain, a region that is essential for Numb function. Our results establish numb- and α-adaptin-dependent endocytosis of Sanpodo as the mechanism by which Notch is regulated during external sensory organ development.
Keywords: asymmetric cell division, cell polarity, endocytosis, nervous system development
Introduction
The four different cells that form Drosophila external sensory (ES) organs arise from a single sensory organ precursor (SOP) cell in a series of asymmetric cell divisions (Jan & Jan, 2001; Bardin et al, 2004). First, the SOP cell divides into a smaller anterior (pIIb) and a larger posterior (pIIa) cell. Next, the pIIb cell divides into the pIIIb cell and a small glial cell, which does not become part of the ES organ (Gho et al, 1999). Finally, the pIIa cell gives rise to the externally visible hair and the socket cell, whereas the pIIIb cell generates the neuron and the sheath cell, which are not visible from the outside. Correct cell fate specification in the SOP lineage requires Numb (Uemura et al, 1989). Numb is a membrane-associated protein that concentrates at the anterior cell cortex in prophase and segregates into the pIIb cell after cytokinesis (Rhyu et al, 1994). In the absence of Numb, SOP cells divide into two identical pIIa cells, whereas numb overexpression leads to the opposite cell fate transformation. Numb performs a similar function during pIIa and pIIIb division so that in numb-null mutants, abnormal ES organs with four socket cells are generated (Rhyu et al, 1994).
Numb specifies cell fate by repressing signal transduction by means of the Notch receptor. Notch is a large transmembrane receptor, which is proteolytically cleaved after binding of its ligand Delta (Schweisguth, 2004). After cleavage, the Notch intracellular domain translocates into the nucleus, where it can act as a transcriptional coactivator (Bailey & Posakony, 1995; Lecourtois & Schweisguth, 1998; Struhl & Adachi, 1998). Although both Notch and Delta are expressed in all cells of the SOP lineage, Notch is activated only in daughter cells that do not inherit Numb (Guo et al, 1996). Several experiments have suggested that this is because Numb might induce the endocytosis of Notch. First, Numb acts upstream of Notch (Guo et al, 1996; Spana & Doe, 1996). Second, Numb binds to the endocytic protein α-Adaptin and regulates its preferential segregation with Numb into pIIb (Berdnik et al, 2002). Third, Numb interacts with Notch in vitro and in the two-hybrid system (Guo et al, 1996). Although this is an attractive model, the situation might be more complex because the plasma membrane levels of Notch are similar in pIIa and pIIb. More recently, Numb was shown to form a complex with the four-pass transmembrane protein Sanpodo in vivo (O'Connor-Giles & Skeath, 2003). Sanpodo is expressed in central nervous system precursors undergoing asymmetric cell division. The protein is located at the cell membrane but is found in intracellular dots in daughter cells that inherit the Numb protein. As Sanpodo is required for Notch signalling during asymmetric cell division, Numb and α-Adaptin might in fact prevent Notch activation by inducing the endocytosis of Sanpodo in one of the two daughter cells. To test this model for the SOP lineage, we characterized Sanpodo function in developing ES organs. We found that Sanpodo is localized at the plasma membrane in pIIa but colocalizes with Notch and Delta on early and late endosomes in pIIb. In numb or α-adaptin mutants, Sanpodo remains at the plasma membrane throughout asymmetric cell division. Sanpodo binds to the phosphotyrosine-binding (PTB) domain of Numb, which has been shown to be essential for Numb function. We propose that endocytosis of Sanpodo might be the primary function of Numb during ES organ development.
Results and Discussion
Loss of Sanpodo leads to cell fate transformations
We used the ey-Flp/FRT system (Newsome et al, 2000) to screen for mutants affecting ES organ development (Berdnik & Knoblich, 2002), and isolated the mutation 3R6, which causes the loss of microchaetae on the head capsule and of eye interommatidial bristles (Fig 1A). To test whether this morphological defect is because of cell fate transformations during ES organ development, the four cell types were analysed by the expression of their different marker genes. In 3R6 mutant ey-Flp clones, 22% of all head ES organs (n=460) were composed of four neurons (Fig 1B), whereas 6% had two sheath cells and two neurons (Fig 1B). Thus, 3R6 mutant SOP cells frequently divide symmetrically into two pIIb cells, a cell fate transformation that is characteristic for genes involved in Notch activation (Guo et al, 1996).
Figure 1.
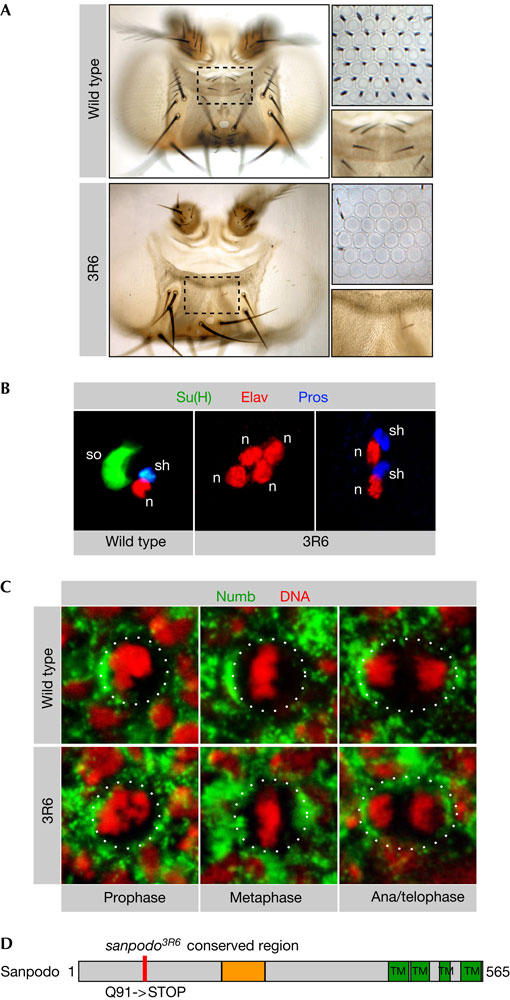
Sanpodo is required for asymmetric cell division. (A) 3R6 mutant external sensory (ES) organs on the head show a pronounced loss of microchaetae and interommatidial bristles compared with wild type (high-magnification images). (B) Wild-type ES organs contain one socket (so) cell (Su(H) positive), one neuron (n) cell (Elav positive) and one sheath (sh) cell (Pros positive). In 3R6 mutant ES organs, outer cells are transformed into inner cells (four neurons or two sheath cells and two neurons). (C) In wild-type eye sensory organ precursor (SOP) cells, Numb localizes to the anterior cell cortex throughout mitosis, whereas Numb is mislocalized in a subset of 3R6 mutant mitotic eye SOP cells. (D) The 3R6 mutation changes a glutamine at amino-acid position 91 of the protein Sanpodo into a stop codon. Sanpodo is a 64 kDa protein with four transmembrane domains in the carboxyl terminus and a region in the middle of the protein that is conserved between Drosophila and Anopheles.
To test whether the 3R6 mutant phenotype is the result of a defect in Numb localization, we stained 3R6 mutant clones for Numb protein. Numb localization was normal in mutant SOP cells on the head or notum (data not shown). In a fraction of metaphase SOP cells within the mutant eye (27% of n=59), however, Numb is no longer asymmetrically localized (Fig 1C). Although this defect might contribute to the phenotype, it cannot account for the cell fate transformations because it is eye specific and the frequency of interommatidial bristle loss in 3R6 mutant eyes is higher than 27% (Fig 1A). Furthermore, mutants defective in Numb localization, such as aurora-A, cause cell fate transformations opposite to those observed in 3R6 mutant clones (Berdnik & Knoblich, 2002). We therefore conclude that 3R6 acts downstream of Numb in cell fate specification although it is also required for Numb localization in a small subset of SOP cells.
The 3R6 mutation was narrowed down to the cytological region 99–100 by P-element and deficiency mapping. This region includes the gene sanpodo, and complementation analysis confirmed that 3R6 is a new allele of sanpodo. Although sanpodo has been described to encode the Drosophila homologue of Tropomodulin (Dye et al, 1998), 3R6 mutants do not have any mutation in the tropomodulin gene. More recently, Sanpodo has been reported to be encoded by the gene CG31020 (O'Connor-Giles & Skeath, 2003). Sequence analysis indicated a C-to-T transition in 3R6 mutants that introduces a stop codon after amino acid (aa) 90 of the CG31020 open reading frame (Fig 1D). No Sanpodo protein could be detected in spdo3R6 mutant clones (data not shown), indicating that spdo3R6 is a strong hypomorph or a null allele.
Sanpodo localizes to endocytic vesicles
To analyse the subcellular distribution of Sanpodo, we generated a specific antibody. In interphase SOP cells, Sanpodo localizes in intracellular dots (Fig 2B) that might be cytoplasmic vesicles. During mitosis, Sanpodo is mostly cytoplasmic, probably as a result of the fragmentation of intracellular membrane compartments (Fig 2B). After cytokinesis, the vesicular Sanpodo staining re-appears in the pIIb cell, whereas the protein accumulates at the plasma membrane in pIIa (Fig 2B). To confirm this membrane localization, we analysed the colocalization of Sanpodo with the membrane-bound protein Gβ13F (Schaefer et al, 2001). In the pIIa cell, Sanpodo colocalized with Gβ13F, and high-magnification images also showed Sanpodo vesicles in close proximity with the plasma membrane, which are probably docking to the membrane (Fig 2A, arrowheads). In the pIIb cell, we detected Sanpodo on intracellular vesicles and it was absent from the plasma membrane (Fig 2A, arrowheads). A similar localization pattern for Sanpodo has been described for the MP2 neuroblast (O'Connor-Giles & Skeath, 2003). Thus, the subcellular localization of Sanpodo correlates with the presence or absence of Numb.
Figure 2.
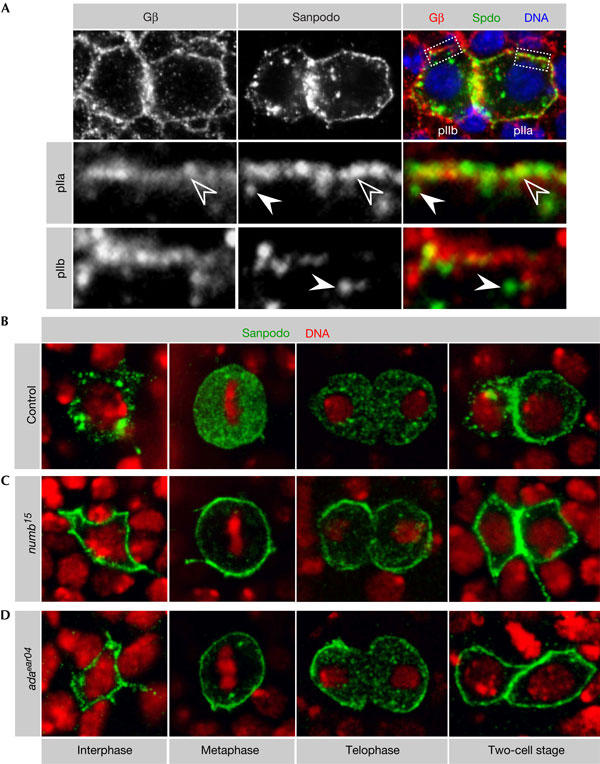
Numb and α-Adaptin inhibit Sanpodo localization to the plasma membrane. (A) After cell division, Sanpodo localizes to the membrane in the pIIa cell, where it colocalizes with the membrane marker Gβ13F (open arrowheads), and vesicles in close proximity to the plasma membrane, which can be distinguished from membrane-localized Sanpodo (arrowheads). In the pIIb cell, no colocalization with Gβ13F is observed and Sanpodo localizes to intracellular vesicles instead (arrowheads). (B) In control sensory organ precursor (SOP) cells, Sanpodo localizes to intracellular vesicles in interphase and to the cytoplasm during mitosis. After cell division, Sanpodo is found in endocytic vesicles in the pIIb cell and at the plasma membrane in the pIIa cell. In (C) numb15 and (D) adaear04 mutant SOP cells, Sanpodo localizes predominantly to the plasma membrane throughout the cell cycle.
To test whether Numb is required for Sanpodo localization, we analysed mitotic clones mutant for the null allele numb15. The vesicular staining of Sanpodo is lost from numb15 mutant interphase SOP cells (Fig 2C). During mitosis, Sanpodo remains at the plasma membrane and, after mitosis, it is not found in vesicles in pIIa (Fig 2C). To test whether Numb exerts its function on Sanpodo through α-Adaptin, we analysed adaear04 mutant SOP cells. This mutation deletes the Numb-binding domain of α-Adaptin. It causes phenotypes in SOP cells that are similar to numb, but leaves the cell-essential functions of α-Adaptin intact (Berdnik et al, 2002). As in numb15 mutants, Sanpodo is cortical throughout the cell cycle in adaear04 mutant SOP cells (Fig 2D). This is surprising and indicates that Numb and α-Adaptin functions are not restricted to the two-cell stage and that Numb-dependent endocytosis of Sanpodo already occurs before mitosis.
As in embryos (O'Connor-Giles & Skeath, 2003), Sanpodo colocalizes with Notch and Delta on intracellular vesicles (Fig 3A,B). These vesicles might be endocytic, as their formation depends on α-Adaptin. We therefore determined the identity of Sanpodo vesicles by colocalization with markers for endocytic compartments. Sanpodo showed strong colocalization with Rab5 (Fig 3C) and Rab7 (Fig 3D), whereas Rab11 hardly ever overlapped Sanpodo (Fig 3E). This suggests that Sanpodo is present on early and late endosomes but not on recycling endosomes. Together with the Notch and Delta colocalization, these data suggest that Sanpodo might act in regulating the transport of Notch and Delta to lysosomes through the degradative pathway.
Figure 3.
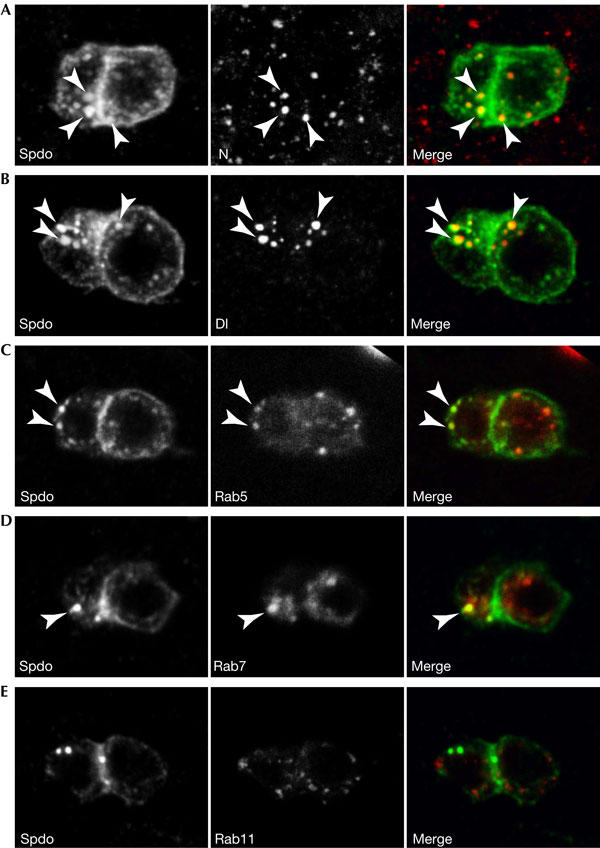
Sanpodo colocalizes with Notch and Delta in endocytic vesicles in the pIIb cell. (A–E) Sanpodo (Spdo) localizes to the membrane in the pIIa cell but is found in intracellular dots in the anterior pIIb cell. Sanpodo colocalizes strongly with (A) Notch (N) and (B) Delta (Dl) primarily in the pIIb cell. Sanpodo colocalizes with (C) Rab5 (marker for early endosomes) and (D) Rab7 (late endosomes) in the pIIb cell but barely with (E) Rab11 (recycling endosomes). Arrowheads indicate colocalization. Note that these images are projections of several optical slices to better illustrate the difference in the number of vesicles, which are generally found more apical. Therefore, the membrane localization of Sanpodo in the pIIa cell is not as clearly visible as in the single optical slices shown in Fig 2A.
Sanpodo binds to the PTB domain of Numb
Sanpodo and Numb interact in vivo (O'Connor-Giles & Skeath, 2003), which suggests that Numb links Notch by means of Sanpodo to α-Adaptin to promote the endocytosis of the trimeric complex. Numb contains a PTB domain in the middle of the protein (Fig 4A), which has been reported to associate with the intracellular domain of Notch in vitro (Guo et al, 1996). Moreover, this domain has been shown to be required for Notch inhibition and Numb function in vivo (Frise et al, 1996). To determine whether Sanpodo binds to a region essential for Numb function, we performed co-immunoprecipitation experiments with Drosophila S2 cells. Cells were transfected with different Numb deletion constructs (Fig 4A; Frise et al, 1996) and a truncated form of Sanpodo (SanpodoΔTM) that lacks the transmembrane domains. Full-length Numb and amino-terminal (NumbΔN) and carboxy-terminal (NumbΔ5) deletions are able to co-precipitate Sanpodo (Fig 4B). A Numb deletion lacking the PTB domain (NumbΔPTB), in contrast, is unable to interact with Sanpodo (Fig 4B). In addition, β-galactosidase (β-Gal) fusions of the Numb N terminus, which include the PTB domain, can co-precipitate full-length Sanpodo from embryonic lysates of transgenic flies (Fig 4C). We conclude that the PTB domain is required for the interaction with Sanpodo in vivo. Although we cannot exclude that the Numb PTB domain can bind directly to Notch as well, our results are consistent with a model in which Numb and Sanpodo act as a scaffolding complex linking α-Adaptin to the Notch receptor to promote its endocytosis.
Figure 4.
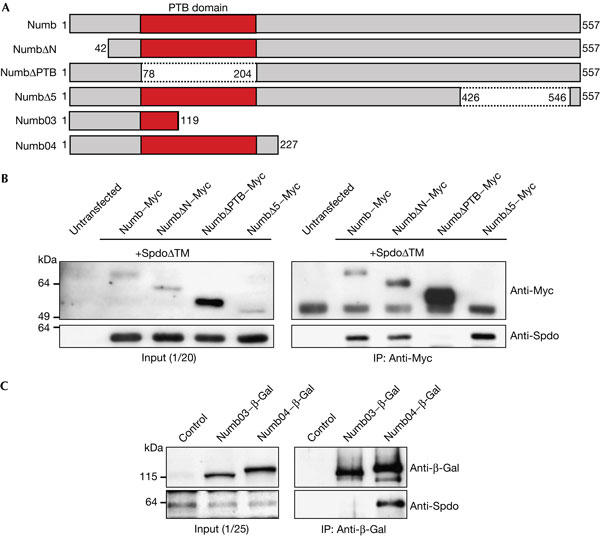
Sanpodo binds to the phosphotyrosine-binding domain of Numb. (A) Numb contains a phosphotyrosine-binding (PTB) domain. Different Numb deletion constructs were used for binding assays. (B) NumbΔPTB alone cannot co-precipitate SanpodoΔTM from S2 cells. IP: immunoprecipitation. (C) Numb04–β-Gal, but not Numb03–β-Gal, can co-precipitate Sanpodo from embryonic lysates. Spdo, Sanpodo.
Speculation
Our analysis shows that Sanpodo regulates Notch signalling during Drosophila ES organ development. In the pIIa cell, Sanpodo is localized at the plasma membrane and is required for Notch activation. In the pIIb cell, Sanpodo is removed from the plasma membrane by Numb- and α-Adaptin-dependent endocytosis. This correlates with the inability of this daughter cell to activate Notch signalling, suggesting that it is the plasma-membrane-localized Sanpodo protein that activates the Notch receptor. Previous epistasis experiments have suggested that Sanpodo acts during the intramembranous (S3) cleavage of the Notch receptor (O'Connor-Giles & Skeath, 2003). Assuming that this cleavage occurs at the plasma membrane, it is possible that Notch needs to bind to Sanpodo to become a substrate for the protease Presenilin, which carries out the S3 cleavage.
Although this model is attractive, it does not explain why Sanpodo colocalizes with Notch in endocytic vesicles and why these vesicles are found in both pIIa and pIIb cells. Furthermore, we found that ectopic expression of Sanpodo during neurogenesis (where Numb is expressed but not asymmetric) causes a neurogenic phenotype (data not shown). Thus, Sanpodo can both activate and inhibit Notch signalling depending on the absence or presence of Numb. These observations are more consistent with an alternative model in which Sanpodo regulates the endocytosis of Notch. It was recently shown that ubiquitination and subsequent endocytosis can downregulate Notch (Sakata et al, 2004). Conversely, endocytosis can also positively influence Notch signalling and was shown to be required for Notch activation in vertebrates (Gupta-Rossi et al, 2004). We speculate that Sanpodo might have a general role in Notch endocytosis. In the absence of Numb, endocytosis could be required for Notch signalling, whereas in its presence, the inhibitory endocytic pathway could prevail. Although this model is speculative, it would also explain why expression of Numb in tissues that do not express Sanpodo has little or no influence on Notch signalling.
Methods
Identification of spdo3R6. Sanpodo was identified in a genetic screen (Berdnik & Knoblich, 2002). The mutation was mapped between 99F and 100B on the basis of lethality over deficiency Df(3R)tll-g and recombination mapping (Berdnik et al, 2002). spdo3R6 is lethal over the known sanpodo allele spdoC55. For sequence analysis, DNA was isolated from homozygous embryos identified by the absence of a yellow+ marker on the balancer chromosome, and the sanpodo coding region was analysed by PCR sequencing.
Flies. spdo3R6, numb15 and adaear04 clones were generated using the ey-Flp/FRT/cell-lethal system (Newsome et al, 2000) or Ubx-Flp. Ubx-Flp was generated by inserting two copies of the Ubx enhancer fragment PBX-41 (gift from M. Bienz) into pCaSpeR-hsFlp, which carries the Flp recombinase under the control of a complete hsp70 promoter. Ubx-Flp induces recombination in all imaginal discs. Neuralized-Gal4 (Bellaiche et al, 2001) was used to overexpress UAS–Rab5–GFP (green fluorescent protein; Wucherpfennig et al, 2003), UAS–Rab11–GFP (gift from M. Gonzalez-Gaitan) and UAS–Rab7–GFP (Entchev et al, 2000). Maternal–Gal4–V32 (gift from D. St Johnston) was used to overexpress UAS–Numb03–β-Gal and UAS–Numb04–β-Gal (Knoblich et al, 1997).
Immunofluorescence and antibodies. Pupae were aged for 24–26 h (for lineage) or 15–18 h (for Sanpodo and Numb localization) after puparium formation, dissected in 8% paraformaldehyde, fixed for a further 20 min on ice and stained, essentially as described (Rhyu et al, 1994). Antibodies used were mouse anti-Elav (1:30, mAB9F8A9; Developmental Studies Hybridoma Bank (DSHB, Iowa City, IA, USA)), rabbit anti-Prospero (1:1,000; Vaessin et al, 1991), rat antisu(H) (1:2,000; gift from F. Schweisguth), rabbit anti-Gβ13F (1:150; Schaefer et al, 2001), rabbit anti-Numb (1:100; Schober et al, 1999), rat antisanpodo (1:100; O'Connor-Giles & Skeath, 2003), rabbit anti-GFP (affinity purified, 1:500; Abcam, Cambridge, UK), mouse anti-Notch (1:200, C594.9B; DSHB), mouse anti-Delta (1:200, C458.2H; DSHB), mouse anti-Myc (1:500, 9E10) and mouse anti-β-Gal (1:100; Promega, Madison, WI, USA). Guinea-pig antisanpodo was generated against an N-terminal MBP (maltose binding protein) fusion of aa 11–232 and used at a dilution of 1:1,000. Hoechst 33258 was used to visualize DNA. Images were recorded on a Zeiss LSM510 confocal microscope and processed with Adobe Photoshop.
Constructs, cell culture and biochemistry. Drosophila S2 cells were propagated in Schneider's medium (Gibco, Carlsbad, CA, USA) containing 10% FCS, 50 U/ml penicillin and 50 g/ml streptomycin. UAS constructs were expressed by co-transfection with actin–Gal4 (gift from T. Volk) using Cellfectin (Invitrogen, Carlsbad, CA, USA). UAS constructs used were Numb–Myc, NumbΔN–Myc, NumbΔPTB–Myc and NumbΔ5–Myc (Frise et al, 1996). UAS–SanpodoΔTM was generated by introduction of a stop codon after aa 424 by PCR. Immunoprecipitations were carried out essentially as described (Betschinger et al, 2003).
Acknowledgments
We thank J. Betschinger and F. Wirtz-Peitz for comments on the manuscript, E. Kleiner for technical assistance, D. Berdnik for generation of Ubx-Flp and M. Bienz, M. Gonzalez-Gaitan, D. St Johnston, F. Schweisguth, T. Volk, the Bloomington Drosophila Stockcenter and the DSHB for reagents. Work in J.A.K.'s lab was supported by the Austrian Academy of Sciences and the Austrian Research Fund (FWF).
References
- Bailey AM, Posakony JW (1995) Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev 9: 2609–2622 [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Le Borgne R, Schweisguth F (2004) Asymmetric localization and function of cell-fate determinants: a fly's view. Curr Opin Neurobiol 14: 6–14 [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F (2001) Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol 3:50–57 [DOI] [PubMed] [Google Scholar]
- Berdnik D, Knoblich JA (2002) Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr Biol 12: 640–647 [DOI] [PubMed] [Google Scholar]
- Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA (2002) The endocytic protein α-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev Cell 3: 221–231 [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA (2003) The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422: 326–330 [DOI] [PubMed] [Google Scholar]
- Dye CA, Lee JK, Atkinson RC, Brewster R, Han PL, Bellen HJ (1998) The Drosophila sanpodo gene controls sibling cell fate and encodes a tropomodulin homolog, an actin/tropomyosin-associated protein. Development 125: 1845–1856 [DOI] [PubMed] [Google Scholar]
- Entchev EV, Schwabedissen A, Gonzalez-Gaitan M (2000) Gradient formation of the TGF-β homolog Dpp. Cell 103: 981–991 [DOI] [PubMed] [Google Scholar]
- Frise E, Knoblich JA, Youngershepherd S, Jan LY, Jan YN (1996) The Drosophila Numb protein inhibits signaling of the Notch receptor during cell–cell interaction in sensory organ lineage. Proc Natl Acad Sci USA 93: 11925–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M, Bellaiche Y, Schweisguth F (1999) Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell. Development 126: 3573–3584 [DOI] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN (1996) Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17: 27–41 [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Six E, LeBail O, Logeat F, Chastagner P, Olry A, Israel A, Brou C (2004) Monoubiquitination and endocytosis direct γsecretase cleavage of activated Notch receptor. J Cell Biol 166: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY (2001) Asymmetric cell division in the Drosophila nervous system. Nat Rev Neurosci 2: 772–779 [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN (1997) The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc Natl Acad Sci USA 94: 13005–13010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F (1998) Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr Biol 8: 771–774 [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ (2000) Analysis of Drosophila photoreceptor axon guidance in eyespecific mosaics. Development 127: 851–860 [DOI] [PubMed] [Google Scholar]
- O'Connor-Giles KM, Skeath JB (2003) Numb inhibits membrane localization of sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev Cell 5: 231–243 [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN (1994) Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76: 477–491 [DOI] [PubMed] [Google Scholar]
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S (2004) Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol 14: 2228–2236 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA (2001) Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107: 183–194 [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA (1999) Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402: 548–551 [DOI] [PubMed] [Google Scholar]
- Schweisguth F (2004) Notch signaling activity. Curr Biol 14: R129–R138 [PubMed] [Google Scholar]
- Spana EP, Doe CQ (1996) Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17: 21–26 [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A (1998) Nuclear access and action of notch in vivo. Cell 93: 649–660 [DOI] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN (1989) numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58: 349–360 [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN (1991) prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67: 941–953 [DOI] [PubMed] [Google Scholar]
- Wucherpfennig T, Wilsch-Brauninger M, Gonzalez-Gaitan M (2003) Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J Cell Biol 161: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]


