Summary
Symposium on Ubiquitin and Signaling
Keywords: ubiquitin, signalling, covalent modification, protein targeting, protein degradation
Introduction
The 2004 Nobel Prize in Chemistry was awarded to I. Rose, A. Ciechanover and A. Hershko for their groundbreaking discovery of ubiquitin-dependent proteolysis in the late 1970s (Wilkinson, 2004). In the subsequent 25 years, we have become aware that the ubiquitin (Ub) domain acts as a targeting signal for numerous events in the cell. The addition of a single Ub to target proteins is involved in histone structure and transcriptional control, as well as receptor internalization and endosomal sorting. The addition of multiple Ub molecules, usually as a polyubiquitin chain, is involved in degradation by the proteasome, DNA repair, ribosomal function, signal transduction, and other less well understood processes.

The Ubiquitin and Signaling Meeting was a Keystone Symposium organized by T. Hunter, C. Joazeiro and M. Hochstrasser and was held in Taos, New Mexico, between 22 and 27 February 2005.
Besides Ub-mediated protein modification, the cell uses similar enzymatic machinery to conjugate several Ub-like proteins (Ubl) to target proteins (Schwartz & Hochstrasser, 2003). Like monoubiquitylation, most conjugation of Ubl to substrates seems to be associated with non-proteolytic functions. Examples include: Nedd8 (neuronally expressed during development 8), a Ubl that is conjugated to the cullin component of Ub ligases and regulates assembly of the active ligase complex; SUMO (small Ub-related modifier), a distant relative of Ub involved in modulating protein activity and nuclear localization; and ISG15 (interferon stimulated gene product 15), a 15 kDa protein containing two Ub domains that is conjugated to cellular proteins in response to interferon, lipopolysaccharide (LPS) and other inflammatory signals. Figure 1 summarizes the functions that have been ascribed to Ub-mediated protein modification.
Figure 1.

Assembly and fates of various forms of the ubiquitin domain. The E1 activating enzyme adenylates the carboxyl terminus of ubiquitin (Ub) and then forms a thiolester with the E2 conjugating enzymes that act as mobile carriers of activated Ub. Ub ligases are responsible for the specificity of attachment of Ub to the target protein through the recruitment of both an E2 thiolester and a specific substrate. Modification by a single Ub domain regulates localization and/or activity of conjugated proteins. PolyUb chains can be formed with different linkages and these direct proteins to different fates, presumably requiring chain-specific receptors. Note that more than one E2 can work with a given E3 and that several E3s can use a single E2. BRCA1, breast cancer 1; UEV, ubiquitin-conjugating enzyme E2 variant protein.
This meeting spanned the breadth of Ub metabolism: the structure of the signal; the enzymes involved and their genetics; diseases caused by a malfunction in the Ub pathway and the corresponding drug intervention. A unifying theme of the meeting was the complexity of protein–protein interactions that are required for Ub signal transduction. Every step of Ub metabolism (except the initial step of Ub activation) is cumulative, involving large families of proteins that are combined to form scaffolds, adaptors and receptors. This is both the beauty and the challenge of the system. Understanding and manipulating these interactions to treat disease is our mission for the next 25 years.
The Keynote address
C. Pickart (Baltimore, MD, USA) set the stage for the meeting and described quantitative studies on the recognition of different polyUb chains by the Ub-associated (UBA) domain. PolyUb chains are created following conjugation of Ub to any of the seven lysines of another Ub, and different chains are thought to specify different fates for proteins. PolyUb chains should have distinct structures depending on their linkage and Pickart's group has shown this by comparing K48- and K63-linked dimers. This implies that chain-specific binding proteins are involved in sorting these chains to their respective fates. Binding data showed that UBA domains can be classified into several categories depending on their chain-binding specificity. Furthermore, replacing a UBA domain in the Ub-specific protease USP5 (a deubiquitylating enzyme that exhibits little chain-binding specificity) with the K48-selective UBA domain from RAD23 results in a chimeric protein that binds to K48-linked chains. This suggests that genetic variation and combination of such modules might generate proteins that are capable of specifically recognizing the diverse signals of different polyUb chains.
The molecular mechanisms that recognize different versions of the Ub signal are poorly understood and form the central question in this field. Binding of the monomeric Ub or Ubl domain frequently occurs through interactions with a hydrophobic face near its carboxy terminus (Fig 2). However, it seems likely that other faces of Ub can be recognized as they are so highly conserved. Numerous other presentations discussed below dealt with the issue of specific recognition of the Ub domain in different contexts and the identification of the relevant binding proteins.
Figure 2.
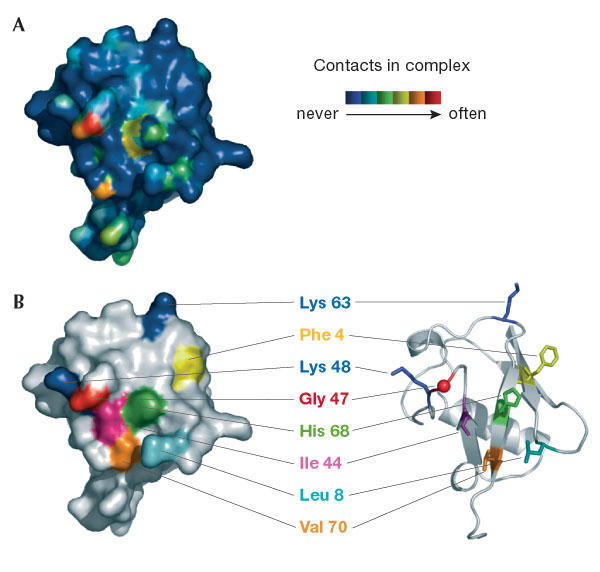
Summary of interactions between ubiquitin and its binding proteins. (A) The surface of ubiquitin is coloured according to the frequency of contacts with binding proteins and deubiquitylating enzymes (see key). (B) Surface and ribbon representation of ubiquitin in the same orientation and with key residues labelled for orientation. Figure adapted with permission from H. Schubert, University of Utah.
Ubiquitin and Ub-like ligases
Three classes of enzyme are involved in the ubiquitylation reaction: Ub-activating enzymes (E1), Ub-conjugating enzymes (E2) and Ub-protein ligases (E3). The same chemistry is also used in the conjugation of other Ubl proteins (Fig 3 and below). E1 adenylate the C terminus of Ub and then form thiolesters with E2, which act as mobile carriers of activated Ub. E3 are responsible for the specificity of Ub attachment to the target protein through the recruitment of both an E2 thiolester and a specific substrate (Robinson & Ardley, 2004). All known Ub ligases use one of two catalytic domains; the HECT (homologous to E6-AP C terminus) or the RING (really interesting new gene) domains. A large fraction of such proteins that have been examined so far have Ub-ligase activity. C. Joazeiro (San Diego, CA, USA) has annotated the loci that encode proteins with a HECT or RING domain in the human genome (∼600) and found that they outnumber the 518 protein kinases. Indeed, the central role of Ub ligases in the cell was emphasized by two plenary sessions devoted to detailing their components, structures, activity, modulation and specificity.
Figure 3.

Ubiquitin-proteasome-dependent proteolysis. There are four basic reactions: activation, conjugation, deconjugation and proteolysis.
Whereas substrate recognition by Ub ligases seems to involve extended conformational epitopes, SUMOylation can be targeted by specific consensus sequences. F. Melchior (Göttingen, Germany) reported that the Ub-conjugating enzyme E2-25K itself might be regulated by SUMO modification (a collaborative study with T. Sixma). The crystal structure of an E2-25K fragment with SUMO suggested that SUMOylation of the protein prevents Ub thioester formation, and this has been confirmed in vitro. The SUMO acceptor site on E2-25K did not conform to the consensus site found in most SUMO targets (ΨKXE, in which Ψ is a large hydrophobic amino acid and X is any residue) and required a helical context. Conversely, classical consensus sites in the amino-terminal helix of E2-25K were not modified unless presented as an unstructured peptide. These findings highlight the importance of the secondary structure context of putative sequences that are the target of SUMOylation.
The catalytic mechanism of ubiquitylation has been controversial. Some HECT-domain Ub ligases form an intramolecular thiolester with Ub as an intermediate stage in the ubiquitylation of the target protein, but no such covalent catalytic intermediate is formed by RING-domain ligases. On the basis of a crystal structure of the SUMO-RanGAP1–Ubc9–Nup358–RanBP2 complex, D. Reverter (New York, NY, USA) presented evidence that the SUMO E3 ligase activity of Nup358–RanBP2 is largely due to the optimization of SUMO orientation within the E2-thiolester complex. Thus, RING ligases might function as adaptors that orientate the thiolester bond and align the lysine of a target protein to facilitate nucleophilic attack. C. Patterson (Chapel Hill, NC, USA) discussed the CHIP (C-terminal of Hsp70-interacting protein) ligase. The interaction of CHIP and Hsp70 results in a decrease in Hsp70 ATPase and chaperone activity and is believed to divert the Hsp70–substrate complex from a folding to a degradation pathway when the substrate protein cannot be refolded. Indeed, Chip-knockout mice are more sensitive to protein damage and have an ageing phenotype. P. Brzovic (Seattle, WA, USA) speculated on the unusual means by which the breast-cancer-associated Ub ligase (BRCA1) is regulated. UbcH5c, an E2 for BRCA1, was found to have a non-covalent Ub site (in addition to the site of Ub thiolester attachment) that allows activated UbcH5c–Ub to self-assemble into larger complexes. Mutations that selectively disrupt the non-covalent interaction between UbcH5c and Ub result in the loss of polyubiquitylation activity directed by BRCA1, but not monoubiquitylation. Thus, self-assembly of UbcH5c–Ub might be required for BRCA1-directed K6-linked polyubiquitylation. Alternatively, the non-covalent binding site may position activated UbcH5c–Ub near the distal end of a growing polyubiquitylated protein to facilitate polyUb chain elongation. Although controversial, this mechanism is reminiscent of the heterodimerization of Mms2 and Ubc13 that is required to synthesize K63-linked polyUb. Oligomerization might therefore be a more general mechanism for assembling polyUb chains.
Modification of the substrate can also target it to a waiting ligase. It is well established that phosphorylation of some substrates can trigger their ubiquitylation. A. Varshavsky (Pasadena, CA, USA) described the recent work of his group on the arginylation branch of the N-end rule pathway. They now have evidence that oxidation of an N-terminal cysteine triggers arginylation and degradation of the protein. A second example of regulation by oxidation status was provided by W. Kaelin (Boston, MA, USA). In mammalian cells, oxygen-dependent hydroxylation of the transcription factor HIF-1α allows ubiquitylation of its oxygen sensor degradation domain (ODD) by the von Hipple Lindau Ub ligase, resulting in HIF-1α degradation. During hypoxia, HIF-1α is stable and activates the transcription of many genes that promote survival under low oxygen conditions. Fusing the ODD of HIF-1α to luciferase, Kaelin was able to show that it functions as an oxygen-sensitive targeting signal in mice. Using whole-body imaging techniques this sensor can now be used to indicate areas of hypoxia in live animals. Drugs that prevent HIF-1α hydroxylation could therefore be useful in cases of heart attack and stroke when cells are recovering from hypoxia.
Deubiquitylating enzymes
Deubiquitylating enzymes (DUBs) reverse the conjugation of Ub and regulate many processes such as cellular signalling, DNA repair and vesicular transport (Amerik & Hochstrasser, 2004). It has been estimated that there are more than 80 mammalian DUBs that act on Ub or Ubl conjugates. In an exciting new approach for investigating DUB cellular function, S. Nijman (Amsterdam, the Netherlands) generated an RNA interference library and used it to knock down selected DUBs. He showed that knocking down the DUB CYLD (the mutation of which causes the benign tumour syndrome cylindromatosis) caused an upregulation of NFκB activity through TRAF2, which is a substrate for CYLD. Knockdown of the DUB USP1 caused an increase in the monoubiquitylation of FANCD2 (a protein defective in some types of Fanconi's anaemia) and proliferating cell nuclear antigen (PCNA). In its monoubiquitylated form, PCNA stimulates DNA repair by translesion synthesis. Such approaches promise to reveal many new DUB functions.
G. Odorizzi (Colorado Springs, CO, USA) discussed the role of Doa4, a DUB that is involved in the deubiquitylation of endosomal cargo proteins during their sorting into multi-vesicular bodies (MVBs). By using electron tomography, he showed that there are two populations of MVBs in yeast cells: large (∼40 nm diameter) and small (∼25 nm diameter), and that Doa4 was required for formation of the former. These results suggest a different genesis and/or fate of the two vesicle types.
Several DUBs are associated with the proteasome and are involved in the release and editing of polyUb chains during degradation. If these chains are inadequately disassembled, they are degraded by the proteasome. R. Cohen (Iowa City, IA, USA) discussed the DUB USP14, which is bound to the S2 subunit in the base of the 19S proteasome regulator, but the activity of which requires two further ATPase subunits, S4 and S7 in an S2–S4–S7 subcomplex. X-ray crystallography of the USP14 catalytic domain (reported by Y. Shi, Princeton, NJ, USA) suggests that association with this complex might promote a conformational change in a loop structure that occludes the active site in free USP14. D. Finley (Boston, MA, USA) extended this analysis by suggesting that the yeast orthologue of USP14 (Ubp6) might have a specialized role in trimming or remodelling chains assembled by a loosely bound Ub ligase, Hul5.
The proteases involved in deconjugating Ubl proteins were also discussed. K. Wilkinson (Atlanta, GA, USA) reported the synthesis of substrates, inhibitors and ligands specific for DUBs that act on Ubl proteins, and their use to identify other DUBs that act on the same proteins. He also detailed studies using non-hydrolysable polyUb analogues to characterize the chain-binding specificity of DUBs and other binding proteins. These tools allow detailed molecular characterization of the binding and catalytic specificity of DUBs. K. Orth (Dallas, TX, USA) described the structural characterization of XopD, which is a SUMO-specific DUB from the plant pathogen Xanthomonas. The enzyme has no activity on mammalian SUMO conjugates, but it works efficiently on tomato SUMO. These results suggest that interference with SUMO metabolism is a possible mechanism of pathogenicity of the plague organism Yersinia pestis, as its YopD protein is similar in structure to XopD. D. Zhang (La Jolla, CA, USA) discussed the cellular role of the ISG15-specific isopeptidase Ub-specific processing protease 43 (UBP43) with the aid of a Ubp43-knockout mouse model. ISG15 is induced and conjugated to a variety of proteins in response to interferon, LPS and other inflammatory signals. This demonstration showed that not only did UBP43−/− cells have increased protein conjugation by ISG15, but also that Ubp43−/− mice are hypersensitive to treatment with LPS, show enhanced phagocytosis, and are resistant to fatal encephalitis caused by lymphocytic choriomeningitis virus and to Salmonella infection. LPS treatment of Ubp43−/− mice increased the phosphorylation of STAT1 (a substrate for ISG15 modification) followed by an increase in interferon signalling.
Non-proteasomal functions of ubiquitin
It has recently become clear that the covalent attachment of Ub to a target protein serves as a localization signal, rather than a signal for destruction, by acting as a protein–protein interaction domain in endocytosis, membrane fusion and transcriptional control (Schnell & Hicke, 2003). For instance, monoubiquitylation causes cellsurface receptors to undergo endocytosis, and progression through the endosomal sorting complex required for transport (ESCRT) system depends on this signal. The Ub is removed by Doa4 on formation of MVBs, and thus acts as a vectoral signal assuring the internalization and degradation of these receptors. Receptors that are not monoubiquitylated are recycled back to the cell surface.
To define the specific Ub-binding proteins that are involved in non-proteasomal function, L. Hicke (Evanston, IL, USA) and I. Dikic (Frankfurt, Germany) have independently developed screens to identify proteins, and subsequently domains, that bind to Ub. Hicke screened known endocytic pathway components for their Ub-binding ability and found that the Src homology 3 (SH3) domain of Sla1 is a Ub-binding domain. Hicke showed that this variant of the domain binds to Ub but fails to bind to several proline-rich SH3 ligands. As with all previously identified Ub-binding domains, this atypical SH3 domain interacts with Ub through a hydrophobic patch centred around isoleucine 44.
Ubiquitylated cargo requires sequential binding to a series of Ub-binding proteins for sorting into the MVB. This suggests that Ub may be moved from one receptor to the next by binding proteins that bind to a region other than the hydrophobic patch. Indeed, the entire Ub molecule is conserved, which is consistent with such a model. To identify Ub-binding proteins that do not interact with the hydrophobic patch of Ub, Dikic used a two-hybrid screen with I44A–Ub as the bait. This led to the identification of cellular proteins involved in post-replicative DNA repair that have two such binding domains, dubbed UBM domains. In addition, 12 other proteins were cloned that bind to wild-type Ub, and by using a bioinformatics approach, Dikic classified these into four distinct families of Ub-binding domains. Coupled with other studies on Ub-binding domains (Pickart, Wilkinson), it is apparent that Ub binding has independently evolved many times and that combinatorial assembly of different Ub-binding domains is an efficient way to generate the necessary diversity to use Ub as a targeting signal in many pathways.
The targeting of ubiquitylated proteins is also important for an earlier step of endocytosis: the internalization of cell-surface receptors. Internalization can occur by clathrin-dependent or -independent mechanisms. S. Polo (Milan, Italy) showed that, in the case of epidermal growth factor receptor (EGFR), the choice of these pathways is determined by the intensity of EGFR stimulation and its ubiquitylation status. Low levels of receptor stimulation lead to clathrin-dependent internalization, whereas high levels lead to increased EGFR ubiquitylation and clathrin-independent internalization (that is, through lipid raft/caveolae structures). The caveolar pathway requires involvement of the Ub-interaction motif proteins, epsin, eps15 and eps15R. It is tempting to speculate that these two internalization pathways lead to alternate fates: either sorting into signalling vesicles and/or receptor recycling, or sorting into MVBs and subsequent lysosomal degradation.
During the past few years, much interest has been generated by the observation that Ub binds to complexes formed by p97/VCP/Cdc48, which is an AAA-ATPase that is implicated in membrane fusion events such as those required for endosomal sorting. H. Meyer (Zurich, Switzerland) summarized his work regarding p97/VCP/Cdc48, its Ub-binding cofactor p47, and VCIP135, a DUB that contains the ovarian tumour (OTU) domain. He showed that p47 and active VCIP135 are necessary for optimal reassembly of Golgi cisternae after mitosis. I44A–Ub inhibits this process, which suggests that p47 mediates this process by binding to the hydrophobic patch of Ub. The identification of substrates of the VCIP135 DUB will lead to a better understanding of this process. In addition, H. Teo (Cambridge, UK), J. Hurley (Bethesda, MD, USA) and W. Sundquist (Salt Lake City, UT, USA) discussed Ub binding by the ESCRT complexes and pointed out that the same machinery is required for endosomal sorting and viral budding.
Monoubiquitylation is also important in transcriptional control and chromatin-structure regulation. It is known that ubiquitylation and deubiquitylation of histone H2B by Rad6/Bre1 and Ubp8, respectively, have essential roles in H2B methylation and transcription (Muratani & Tansey, 2003). However, the molecular details and the timing of these events have remained elusive. M. Osley (Albuquerque, NM, USA) and colleagues used chromatin immunoprecipitation to elucidate the order of events necessary for transcription of the reporter gene GAL1. Their data suggest that a continuous cycle of ubiquitylation (by Rad6 and Bre1) and deubiquitylation (by Ubp8 or another DUB) of histone H2B moves down the DNA with the polymerase complex, and that this action facilitates chromatin remodelling and transcriptional elongation. P. Kaiser (Irvine, CA, USA) presented data showing a non-proteasomal function of K48-linked Ub. Under normal culture conditions, the yeast transcription factor Met4 is modified by K48-linked polyUb on K163 by the Met30/Skp1 Ub ligase complex. K48-linked Ub is the canonical polyUb signal for proteasomal degradation of the target protein. However, K48-linked polyubiquitylation does not lead to Met4 degradation, but instead to its inactivation. During heavy-metal stress, the F-box protein Met30 remains associated with Met4, but the binding of Met30 to Skp1 is prevented. This blocks Met4 ubiquitylation, which allows for Met4 phosphorylation and attainment of its active state.
New trends
One of the most striking recent advances in this field is the profusion of 'omics' approaches that are beginning to bear fruit. For instance, several proteins that contain Ub-binding sites are known to act as receptors that direct ubiquitylated proteins to the proteasome. However, which receptors target specific ubiquitylated substrates is less clear. R. Deshaies (Pasadena, CA, USA) has developed a method to determine the specificity of such receptors. Using a proteomics approach, his group has identified about 50 yeast proteins, including the substrates Gcn4 and Sic1, which accumulate as ubiquitylated species in Rpn10 proteasomal mutants but not in wild-type yeast. In addition, Wilkinson used a proteomics approach to identify yeast proteins that bind to non-hydrolysable polyUb analogues. Two of these also bind to the cell division control 48 (CDC48) protein and are candidate Ub-binding adaptors that may assist in CDC48 functions.
Identifying substrates for Ubl modification is one of the most informative approaches for elucidating their non-proteolytic roles. C. Zhao (Austin, TX, USA) discussed targets for ISG15 modification. Purification of ISG15 conjugates and mass-spectrometry analysis led to the identification of 12 interferon-induced proteins, including the anti-viral proteins MxA and RIG-I; Ub and SUMO ligases Ubc13 and PC2, respectively; a SUMO protease SENP1; proteins that are involved in the oxidative stress response, RNA processing, chromatin remodelling and RNA polymerase II transcription; and ∼150 constitutively expressed proteins. C. Stefan (San Diego, CA, USA) described a more classical genetic screen for genes that affect the internalization of plasma-membrane proteins and their transport through the Golgi–lysosomal pathway, whereas S. Gygi (Boston, MA, USA) described an innovative proteomics approach used to identify ubiquitylated proteins in cell lysates. He used catch-and-release reagents and an absolute quantification (AQUA) strategy to determine quantitatively the types of polyUb chains present in cell extracts and to identify new ubiquitylation substrates. Finally, another new tool is the development of methods for monitoring ubiquitylation in living cells. T. Kerppola (Ann Arbor, MI, USA) used a Ub-mediated fluorescence complementation strategy to tag Ub and putative target proteins. Fluorescence is only achieved when the Ub-attached half-green fluorescent proteins are brought into juxtaposition with the target protein fused to the other half of the green fluorescent protein by ubiquitylation, and in this way, the sites of ubiquitylated substrate accumulation can be uniquely identified.
Summary
The field of Ub-dependent signalling has come of age. We are now focused on understanding its involvement in specific diseases and in basic pathways of regulation and control. Genomics and proteomics are being applied in numerous ways and a variety of tools are being developed to monitor ubiquitylation in cells, tissues and whole animals. Pharmacological agents are widely sought and promise to move the field forward even further and faster. The genetic diversity of many of the gene families suggests that selective inhibitors might be involved, as in the cases of kinases and phosphatases. Alternatively, we are getting glimpses of the mechanisms of recognition of the Ub signal, and there is a deep appreciation that these pathways are regulated by numerous multi-protein complexes with the involvement of adaptors, scaffolds and substrate-recognition modules. The opportunities to perturb specific subsets of these pathways may well involve the search for drugs that interfere with these multi-protein complexes.

Acknowledgments
We acknowledge the support of National Institutes of Health (NIH) grants GM30308 and GM66355 (K.D.W.) and GM000680 (K.L.F.), and a Fellowship from the American Heart Association (J.E.M.).
References
- Amerik AY, Hochstrasser M (2004) Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695: 189–207 [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP (2003) How the ubiquitin–proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Ubiquitin enters the new millennium. Mol Cell 8: 499–504 [DOI] [PubMed] [Google Scholar]
- Robinson PA, Ardley HC (2004) Ubiquitin-protein ligases. J Cell Sci 117: 5191–5194 [DOI] [PubMed] [Google Scholar]
- Schnell JD, Hicke L (2003) Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem 278: 35857–35860 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Hochstrasser M (2003) A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci 28: 321–328 [DOI] [PubMed] [Google Scholar]
- Wilkinson KD (2004) Ubiquitin: a Nobel protein. Cell 119: 741–745 [DOI] [PubMed] [Google Scholar]


