Abstract
Dicer is a key enzyme involved in RNA interference (RNAi) and microRNA (miRNA) pathways. It is required for biogenesis of miRNAs and small interfering RNAs (siRNAs), and also has a role in the effector steps of RNA silencing. Apart from Argonautes, no proteins are known to associate with Dicer in mammalian cells. In this work, we describe the identification of TRBP (human immunodeficiency virus (HIV-1) transactivating response (TAR) RNA-binding protein) as a protein partner of human Dicer. We show that TRBP is required for optimal RNA silencing mediated by siRNAs and endogenous miRNAs, and that it facilitates cleavage of pre-miRNA in vitro. TRBP had previously been assigned several functions, including inhibition of the interferon-induced double-stranded RNA-regulated protein kinase PKR and modulation of HIV-1 gene expression by association with TAR. The TRBP–Dicer interaction shown raises interesting questions about the potential interplay between RNAi and interferon–PKR pathways.
Keywords: Dicer, TRBP, PKR, RNA interference, microRNA
Introduction
RNA interference (RNAi)-mediated and microRNA (miRNA)-mediated reactions have emerged recently as important pathways regulating eukaryotic gene expression at various levels. Specificity of these processes is dependent on 20- to 25-nt small interfering RNAs (siRNAs) and miRNAs, acting as guides that recognize sequences of target nucleic acids. To perform their effector function, siRNAs and miRNAs are incorporated into ribonucleoprotein (RNP) complexes, referred to as si- or mi-RISCs (acting post-transcriptionally) or RITS (acting at the chromatin level). Dozens of different proteins have been identified as either essential or regulatory factors for RNAi and miRNA reactions (Tomari & Zamore, 2005). Moreover, it has become increasingly apparent that both pathways intersect with several other cellular processes, such as chromosome segregation (Matzke & Birchler, 2005), RNA editing (Scadden, 2005) and nonsense-mediated degradation (Domeier et al, 2000; Kim et al, 2005).
MicroRNAs are generated from the genome-encoded precursor hairpins by the sequential action of two ribonuclease (RNase) III-type nucleases, Drosha and Dicer. Dicer is also responsible for the excision of siRNAs from double-stranded (ds)RNA (Kim, 2005; Tomari & Zamore, 2005). However, Dicer is not confined to miRNA and siRNA biogenesis. Each of the two Drosophila Dicers, Dcr1 and Dcr2, also seems to be essential for the effector step of RNAi (Tomari & Zamore, 2005). Dcr2, which functions primarily in RNAi, heterodimerizes with a dsRNA-binding domain (dsRBD) protein R2D2 (Liu et al, 2003). The resulting complex senses the differential stability of the ends of the siRNA duplex and determines which siRNA strand will enter the RISC (Tomari et al, 2004). Recent studies have shown that Drosha and Dcr1, the Drosophila Dicer specializing in miRNA biogenesis, function in complexes with dsRBD protein partners DGCR8/Pasha (reviewed by Tomari & Zamore, 2005) and Loquacious (Loqs; Förstemann et al, 2005; Saito et al, 2005), respectively.
In contrast to Drosophila, mammals and Caenorhabditis elegans express only a single Dicer protein. Like most other Dicers, mammalian enzymes are large, ∼200 kDa, proteins containing ATPase/RNA helicase, DUF283, PAZ domains, two catalytic RNase III domains and a carboxy-terminal dsRBD. Although the biological importance and biochemistry of mammalian Dicer have been intensively studied, little is known about its protein partners. The only proteins known to interact directly with Dicer in mammals are members of the Argonaute (Ago) family, which represent siRNA/miRNA-associating core components of RISCs (reviewed by Tomari & Zamore, 2005). The interaction, involving the RNase III domain of Dicer and a PIWI domain of Argonautes (Tahbaz et al, 2004), is probably central to the handover of the products of Dicer catalysis (siRNAs and miRNAs) to Argonautes during RISC assembly.
In this work, we describe the identification of TRBP (human immunodeficiency virus (HIV-1) transactivating response (TAR) RNA-binding protein; Gatignol et al, 1991) as a dsRBD protein partner of human Dicer, and show that TRBP is required for optimal RNA silencing mediated by siRNAs and endogenous miRNAs. TRBP has previously been assigned several functions, including inhibition of the interferon (IFN)-induced dsRNA-regulated protein kinase PKR (Daher et al, 2001), modulation of HIV-1 gene expression through its association with TAR (Dorin et al, 2003) and control of cell growth (Benkirane et al, 1997; Lee et al, 2004). A mouse TRBP homologue, Prbp, was shown to function as a translational regulator during spermatogenesis, and mice that have this deletion, in addition to being male sterile, usually died at the time of weaning (Zhong et al, 1999). The TRBP–Dicer interaction established in this work raises the possibility of crosstalk between RNAi and IFN–PKR pathways in normal and virus-infected cells.
Results and Discussion
Dicer and TRBP interact in vivo and in vitro
We raised monoclonal antibodies (mAbs) against human Dicer (supplementary Fig S1 online). The mAbs 33, 73 and 83, which effectively immunoprecipitate Dicer from extracts of different cultured cells (data not shown), were used to identify proteins associated with Dicer in human embryonic kidney (HEK)293 cells. Proteins retained with either mAbs 33/73/83 or anti-Myc mAb, used as a control, were separated using two-dimensional gel electrophoresis, and spots enriched in Dicer immunoprecipitates were processed for mass spectrometry analysis. One protein reproducibly co-purified with Dicer was identified as TRBP, a protein containing three dsRBDs (supplementary Fig S1 online). Members of the Argonautes family were also among the selected proteins, as were some others, but the reproducibility and significance of their interactions with Dicer were not further investigated (data not shown).
To validate the Dicer–TRBP interaction, we performed co-immunoprecipitation experiments using either extracts from human HEK293 cells or purified proteins. Two anti-Dicer antibodies (Abs), mAb 73 and polyclonal Ab 347, but not the control mAb isotypic with mAb 73, immunoprecipitated endogenous TRBP that was present in HEK293 cells, as shown by western blotting with anti-TRBP Abs (Fig 1A; several forms of TRBP, with apparent molecular masses of 43–46 kDa, are expressed in human cells (see below)). Treatment with RNases digesting both double- and singlestranded RNAs did not eliminate the association (Fig 1B), indicating that the interaction is not mediated by RNA. As immunoprecipitating anti-TRBP Abs were not available, we generated a stable HEK293 cell line, HEK293/HA–TRPB2, expressing the haemagglutinin (HA)-tagged version of the beststudied isoform of TRBP, TRBP2. Co-immunoprecipitation experiments performed with the HEK293/HA–TRPB2 extract and either anti-HA or anti-Dicer Abs showed that each Ab was able to pull down the partner protein (Fig 1C). Further indication that TRBP and Dicer form part of the same complex was provided by gradient sedimentation experiments. Analysis of cytoplasmic extracts prepared from either human HEK293 or mouse teratoma P19 cells showed that Dicer and TRBP, or their mouse counterparts, cosediment in a region corresponding to a molecular mass of ∼250 kDa (Fig 2A,B). Notably, miR-17, an abundant miRNA in HEK293 cells, was also enriched in this region, as was the activity of processing a 30-bp dsRNA to siRNA (Fig 2B,C). Taken together, the data indicate that Dicer and TRBP interact with each other in mammalian cells.
Figure 1.
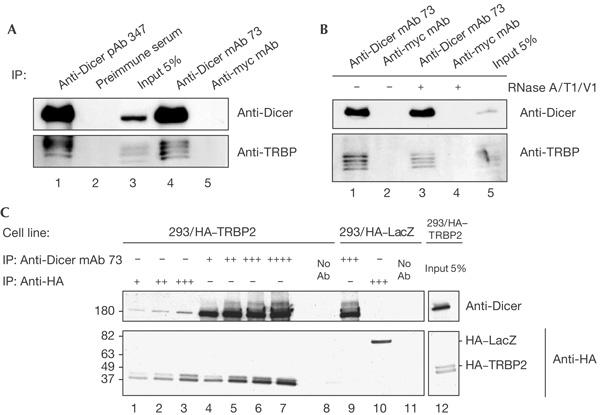
TRBP and Dicer co-immunoprecipitation. (A,B) Anti-Dicer antibodies (Abs) pull down endogenous TRBP (human immunodeficiency virus (HIV-1) transactivating response (TAR) RNA-binding protein) in extracts of human embryonic kidney (HEK)293 cells, and co-immunoprecipitation of Dicer and TRBP is not sensitive to ribonuclease (RNase) treatment (B). mAb, monoclonal antibody; pAb, polyclonal antibody. (C) Dicer–TRBP interaction studied with a HEK293 cell line expressing haemagglutinin (HA)-tagged TRBP2. A HEK293/HA–LacZ cell line was used as a control. The identity of the Abs, some at increasing concentrations (+ through ++++), used for immunoprecipitation (IP) is indicated at the top of the panels. Abs used for western blots are indicated on the right.
Figure 2.
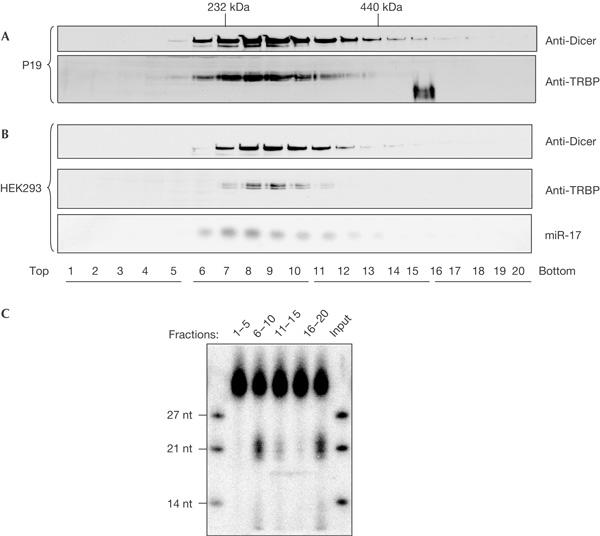
TRBP and Dicer co-sedimentation on glycerol gradients. (A,B) Sedimentation of cytoplasmic extracts from P19 cells (A) and human embryonic kidney (HEK)293 cells (B). Gradient fractions were analysed for Dicer and TRBP (human immunodeficiency virus (HIV-1) transactivating response (TAR) RNA-binding protein) by western blotting and for miR-17 by northern blotting. (C) Activity of fractions analysed in (B), pooled as indicated (top of the panel), in processing 30-bp double-stranded RNA into small interfering RNA.
To find out whether the Dicer–TRBP interaction was direct, we purified both proteins, as recombinant fusions with His6 and a maltose-binding protein (MBP) from insect cells and Escherichia coli, respectively (Fig 3A). The proteins were incubated together and applied either to Protein G–Sepharose beads coated with different Abs or to amylose beads. Dicer mAb 73, but not control anti-HA mAb, effectively pulled down TRBP2. Likewise, TRBP2 retained on amylose beads pulled down Dicer (Fig 3B). The low efficiency of the latter pull-down could be the result of a sterical hindrance caused by the MBP tag or owing to the propensity of TRBP to form homodimers (see below). To eliminate the possibility that proteins co-purifying with either Dicer or MBP–TRBP2 preparations are involved in binding, we studied the Dicer–TRBP interaction in the yeast two-hybrid (2H) assay (Fig 3C). As the budding yeast does not encode TRBP or Dicer homologues, any interaction detected in this system would probably result from direct binding. Plasmids encoding full-length TRBP2, or different regions thereof, fused to the Gal4 DNA-binding domain, and a plasmid encoding Dicer appended to the Gal4 activation domain, as well as several control plasmids, were transformed into yeast. We detected interactions between Dicer and TRBP2 and all its mutants encompassing amino acids 228–366. This region of TRBP includes the dsRBD C domain, suggesting that this domain mediates the interaction (see Conclusions). We also detected TRBP2 interacting with itself, which was consistent with its ability to homodimerize (Daher et al, 2001). Taken together, our results indicate that Dicer and TRBP interact directly with each other.
Figure 3.
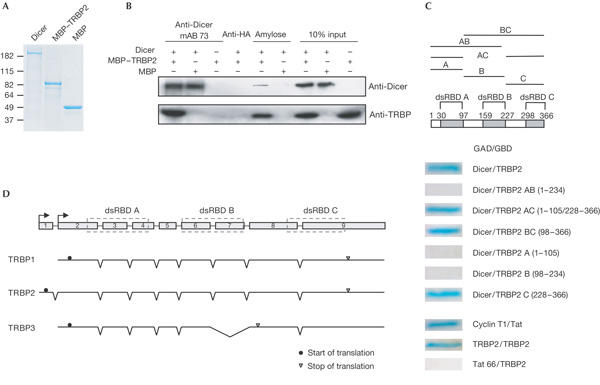
Interaction of TRBP with Dicer studied with purified proteins (A,B) and in the yeast two-hybrid system (C), and schematic representation of different TRBP transcripts expressed in human cells (D). (A) SDS–polyacrylamide gel electrophoresis (SDS–PAGE) analysis of indicated purified recombinant proteins. MBP, maltose-binding protein. (B) Purified TRBP (human immunodeficiency virus (HIV-1) transactivating response (TAR) RNA-binding protein) and Dicer interact with each other. Added proteins, antibodies (Abs) and amylose beads are indicated at the top. Abs used for western blotting are shown on the right. HA, haemagglutinin; mAb, monoclonal antibody. (C) Dicer and TRBP interact in a yeast two-hybrid (2H) assay through the carboxy-terminal domain of TRBP. Top: scheme of TRBP2 and its fragments included in 2H constructs. Double-stranded RNA-binding domains (dsRBDs) are shown as grey boxes. Bottom: β-galactosidase staining diagnostic of the interaction between proteins expressed as fusions with either Gal4 DNA activation domain in pGADGH (GAD) or Gal4 DNA binding domain in pGBT9 (GBD). Cyclin T1/Tat and TRBP2/TRBP2 are positive controls. Tat/TRBP2 is a negative control. (D) Scheme of the TRBP gene and its encoded transcripts. Intron positions and exon regions encoding three dsRBDs are indicated. Two alternative transcription starts, and translation initiation and stop codons, are indicated by arrows, circles and triangles, respectively. Skipping of exon 7 in TRBP3 causes translation to terminate in exon 8.
The 45 kDa TRBP2 consists of three dsRBDs. Another previously described TRBP isoform, TRBP1, differs from TRBP2 at the amino terminus (Bannwarth et al, 2001; see Fig 3D). By complementary DNA cloning and by inspecting EST databases, we identified another TRBP splice variant, potentially encoding a TRBP3 isoform, which would miss the C-terminal dsRBD (Fig 3D). Interestingly, one of the three isoforms of Loqs, the probable Drosophila homologue of TRBP, is also devoid of the C-terminal dsRBD (Förstemann et al, 2005). The biological function of individual TRBP variants remains unknown. The alignment of TRBP2 with dsRBD Dicer protein partners from other species and with a TRBP-related mammalian protein PACT is shown in supplementary Fig S3A online. High sequence conservation of the C-terminal dsRBD in Loqs, TRBP2 and PACT (supplementary Fig S3B online) suggests that this domain has a function distinct from two other dsRBDs (see below).
TRBP is required for miRNA and siRNA silencing
To assess the potential role of TRBP in RNAi and miRNA pathways, we generated stable HEK293T-REx cell lines, in which the expression of TRBP is knocked down by RNAi. In cell lines 293/TRBPkd1 and 293/TRBPkd2, the expression of stably integrated short hairpins targeting different regions in TRBP messenger RNA is controlled by a pol III promoter with tetracycline (Tet) operator sites. Real-time PCR and western analyses indicated that TRBP expression was about fivefold lower at both mRNA and protein levels. In either cell line, partial decrease of the protein had already occurred in the absence of Tet induction, indicating some leakiness of the system (Fig 4A; supplementary Fig S2 online). TRBP depletion had no appreciable effect on cell growth (data not shown).
Figure 4.
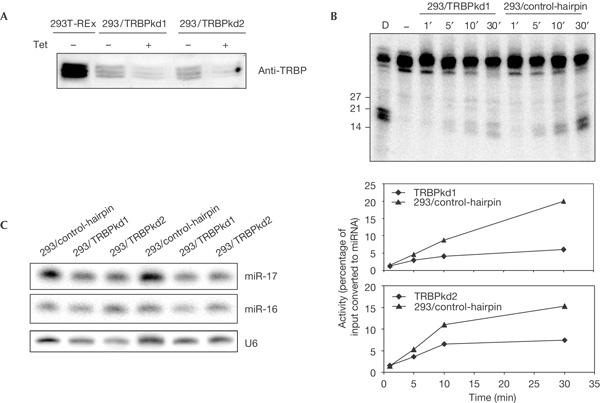
Depletion of TRBP affects pre-microRNA processing in vitro but has no effect on accumulation of mature microRNAs in vivo. (A) Levels of TRBP (human immunodeficiency virus (HIV-1) transactivating response (TAR) RNA-binding protein) in 293/TRBPkd1/2 and human embryonic kidney (HEK)293T-REx control cell lines after treatment with tetracycline (Tet) for 72 h as indicated. Anti-TRBP antibody was used for western blotting. (B,C) Effect of TRBP knockdown on processing of pre-let-7 RNA in vitro (B) and on accumulation of mature microRNAs (miRNAs) in vivo (C). (B) Processing of pre-let-7 RNA was assayed in extracts from 293/TRBPkd1, 293/TRBPkd2 and 293/control-hairpin cell lines. Phosphorimage of 293/TRBPkd1 (upper panel) and quantification of data for both TRBPkd cell lines (bottom panels) are shown. (C) Total RNA isolated from TRBPkd cell lines, from two independent cultures and control cell lines was analysed by northern blot using probes specific for indicated RNAs. Quantification of data from four independent northern blots showed no significant differences in miR-16 and miR-17 levels between TRBPkd and control cultures. A representative experiment is shown. Similar results were obtained for miR-19B and let-7 miRNAs (data not shown).
We compared the miRNA-precursor-processing activity of extracts prepared from different cell lines (Fig 4B). Extracts from either TRBPkd cell line processed pre-let-7 RNA less efficiently than extracts from control cells. The decrease in activity was not due to destabilization of Dicer, as its level was similar in control and TRBPkd cells (Fig 5B). Despite extracts from TRBPkd cells being deficient in pre-miRNA processing, steady levels of several miRNAs in these cells were not significantly different from control HEK293 cells, and there was no apparent accumulation of pre-miRNAs (Fig 4C; data not shown). Notably, as in the case of TRBP, depletion of Loqs in Drosophila S2 cells had no principal effect on mature miRNA levels although extracts of Loqs knockdown cells were deficient in pre-miRNA processing. However, in contrast to TRBP, depletion of Loqs resulted in accumulation of pre-miRNAs in S2 cells (Förstemann et al, 2005; Saito et al, 2005).
Figure 5.
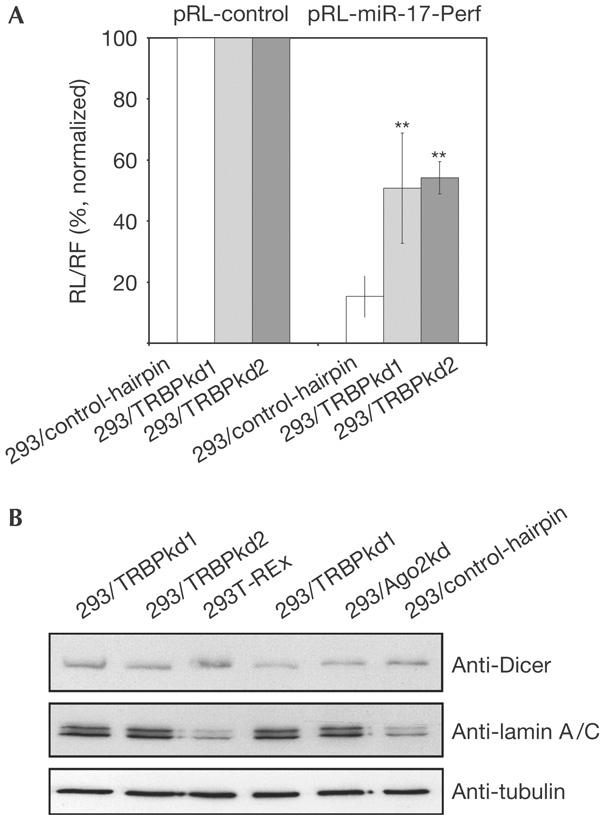
Effect of TRBP depletion on RNA silencing. Depletion of TRBP (human immunodeficiency virus (HIV-1) transactivating response (TAR) RNA-binding protein) decreases the efficiency of RNA interference mediated by the endogenous microRNA miR-17 (A) and by transfected anti-lamin A/C small interfering RNA (B). Cell lines used for analysis are indicated. In (A), activities of Renilla luciferase (RL)-miR-17-Perf reporter in every cell line are expressed in relation to activities of RL-control reporter (set as 100%). Values are means±s.d. of four transfections (**P<0.01). Similar results were obtained in several independent experiments. Antibodies used for western blotting in (B) are indicated on the right.
We used the miRNP-mediated mRNA-reporter-cleavage assay to find out whether depletion of TRBP had an effect on the activity of endogenous miRNPs in HEK293 cells. TRBPkd and control cells were transiently transfected with constructs expressing either control Renilla luciferase (RL) reporter mRNA or a reporter harbouring the site perfectly complementary to miR-17 in its 3′ untranslated region. In control cells, insertion of the miR-17 site repressed RL expression by ∼80%. However, in TRBPkd cells, the miRNA-mediated inhibition was about threefold less pronounced (Fig 5A), indicating that TRBP is required for either the assembly or functioning of miRNPs.
To investigate whether TRBP has a role in the RNAi reaction mediated by exogenous siRNA, we determined the efficiency of the lamin A/C RNAi in TRBPkd and various control cells. The siRNA treatment had a strong effect on lamin A/C level in parental HEK293T-REx cells or cells stably expressing a control hairpin. However, lamin A/C depletion was largely abolished in TRBPkd cells. Similar suppression of the lamin A/C knockdown was observed in a HEK293 cell line in which Ago2 was depleted by expression of a specific hairpin (Fig 5B).
Taken together, our data indicate that TRBP is primarily required for the assembly and/or functioning of si- or mi-RISCs in mammalian cells, but it may also facilitate the cleavage of pre-miRNAs by Dicer. The apparent discrepancy between the effect of TRBP knockdown on pre-miRNA processing in cells and cell extracts is readily explained by incomplete depletion of the protein, allowing for the manifestation of processing deficiency in vitro but not in vivo. It is worth noting that, as in the case of Drosophila Loqs (Förstemann et al, 2005; Saito et al, 2005) but in contrast to R2D2 (Liu et al, 2003), depletion of TRBP did not lead to appreciable destabilization of Dicer (Fig 5B).
Conclusions
Our findings that mammalian Dicer forms a complex with a dsRBD protein TRBP add support to the idea that large RNase III-type Drosha and Dicer nucleases generally require dsRBD protein partners for their function. Drosophila R2D2 and Loqs are two Drosophila dsRBD proteins that work in conjunction with Dcr2 and Dcr1, acting in RNAi and miRNA pathways, respectively (Liu et al, 2003; Förstemann et al, 2005; Saito et al, 2005). The R2D2–Dcr2 association is required for asymmetric loading of siRNAs into RISC (Tomari et al, 2004), whereas Loqs and Dcr1 are essential for efficient pre-miRNA processing, and also participate in gene silencing that is triggered by artificial dsRNA hairpins and endogenous Supressor of Stellate repeats (Förstemann et al, 2005; Saito et al, 2005). The observation that TRBP is required for efficient cleavage of pre-miRNA in vitro and for the function of RISC programmed with either endogenous miRNA or transfected siRNA in cells indicates that TRBP combines at least some functions that are performed separately by R2D2 and Loqs in Drosophila. However, it should be noted that TRBP is structurally more related to Loqs than to R2D2 (supplementary Fig S3 online; Förstemann et al, 2005). We investigated, using both cell extracts and recombinant proteins, whether Dicer and TRBP are involved in sensing the thermodynamic stability of the 5′ ends of the siRNA strands in the same way as Dcr2 and R2D2. These experiments, using 5-iodo-U-modified siRNAs, have not produced conclusive results (L.J., unpublished results).
After the submission of this work, another work describing the TRBP–Dicer partnership has been reported (Chendrimada et al, 2005). Two observations described in the Chendrimada et al paper are in disagreement with our results. First, they found that depletion of TRBP resulted in a decrease of steadystate levels of miRNAs, whereas in our analysis the miRNA content was not significantly changed. Second, in contrast to our findings, Chendrimada et al report that knockdown of TRBP causes destabilization of Dicer, and vice versa. However, we note that the analysis of TRBP and Dicer levels was carried out not with total extracts from cells depleted in either protein but with Argonaute (Ago2) immunoprecipitates. Hence, it is possible that depletion of Dicer or TRBP affects the ability of the partner protein to form a complex with Ago2 rather than destabilizing the protein. This interpretation would be consistent with a relatively mild phenotype of mice with a knockout of Prbp, the mouse homologue of TRBP (Zhong et al, 1999), contrasting with the lethal phenotype of the Dicer knockout (Bernstein et al, 2003). It would also support our observations that the efficient depletion of Dicer in HEK293 cell lines has no principal effect on the level of TRBP (A.H., K. Tang & W.F., unpublished results).
Another three-dsRBD protein, PACT, 42% identical to TRBP, is expressed in mammals. In contrast to TRBP, which inhibits PKR, PACT (or its mouse homologue Rax) has a stimulatory effect on PKR (Gupta et al, 2003; Bennett et al, 2004, and references therein). The effects of TRBP and PACT on PKR activity are mediated by the C-terminal dsRBDs, which are devoid of detectable dsRNA-binding properties (Gupta et al, 2003, and references therein). In addition to effects on PKR, the C-terminal domains of PACT and TRBP can mediate heterodimerization of both proteins and also homodimerization of PACT (Hitti et al, 2004; G. Laraki & A.G., unpublished results). The apparent involvement of the C-terminal TRBP region in association with Dicer (Fig 3C) raises the possibility that RNAi/miRNA and PKR pathways are subject to reciprocal regulation by a rather complex network of protein–protein interactions. As both RNAi and IFN–PKR pathways have a role in antiviral responses, communication between them would not be surprising. In the future, it would be interesting to find out whether Dicer also associates with PACT, and how these protein interactions affect RNA silencing and other defence pathways in normal and virus-infected cells. The latter question is particularly interesting in the light of a recent report that the HIV-1 TAR-binding protein Tat functions as an RNAi suppressor, possibly compromising the activity of Dicer (Bennasser et al, 2005).
Methods
Co-immunoprecipitations. Anti-Dicer mAbs 33, 73 and 83 and control mAbs were crosslinked to Protein G–Sepharose 4 Fast Flow (Amersham Biosciences, Little Chalfont, UK) and used to purify Dicer complexes from HEK293 cytoplasmic extracts (S10). Co-immunoprecipitates were washed five times with lysis buffer (20 mM Tris–HCl, pH 7.5, 300 mM NaCl, 0.5% NP-40, 2.5 mM MgCl2) and analysed by liquid chromatography tandem mass spectrometry (LC-MSMS) and western blot.
293/TRBPkd1, 293/TRBPkd2 and 293/control-hairpin cell lines. Plasmids pTER-TRBPsh1, pTER-TRBPsh2 and pTER-control-hairpin were co-transfected with a puromycin resistance plasmid into HEK293T-REx cells (Invitrogen, Carlsbad, CA, USA) to generate stable cell lines.
Other procedures. Detailed methods can be found in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Material
Acknowledgments
We thank S. Schenk and T. Zoller for help with antibodies, K. Tang for help with knockdown cell lines, R. Portmann for assistance with mass spectrometry analysis and D. Schmitter for the Ago2kd cells. We also thank Antoine Peters and all members of the Filipowicz group for discussions.
References
- Bannwarth S, Talakoub L, Letourneur F, Duarte M, Purcell DF, Hiscott J, Gatignol A (2001) Organization of the human tarbp2 gene reveals two promoters that are repressed in an astrocytic cell line. J Biol Chem 276: 48803–48813 [DOI] [PubMed] [Google Scholar]
- Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A, Jeang KT (1997) Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with PKR. EMBO J 16: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT (2005) Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22: 607–619 [DOI] [PubMed] [Google Scholar]
- Bennett RL, Blalock WL, May WS (2004) Serine 18 phosphorylation of RAX, the PKR activator, is required for PKR activation and consequent translation inhibition. J Biol Chem 279: 42687–42693 [DOI] [PubMed] [Google Scholar]
- Bernstein E et al. (2003) Dicer is essential for mouse development. Nat Genet 35: 215–217 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher A et al. (2001) Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J Biol Chem 276: 33899–33905 [DOI] [PubMed] [Google Scholar]
- Domeier ME, Morese DP, Knight SW, Portereiko M, Bass BL, Mango SE (2000) A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289: 1928–1931 [DOI] [PubMed] [Google Scholar]
- Dorin D, Bonnet MC, Bannwarth S, Gatignol A, Meurs EF, Vaquero C (2003) The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit PKR. J Biol Chem 278: 4440–4448 [DOI] [PubMed] [Google Scholar]
- Förstemann K et al. (2005) Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a doublestranded RNA-binding domain protein. PLoS Biol 3: 1187–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A, Buckler-White A, Berkhout B, Jeang KT (1991) Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 251: 1597–1600 [DOI] [PubMed] [Google Scholar]
- Gupta V, Huang X, Patel RC (2003) The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology 315: 283–291 [DOI] [PubMed] [Google Scholar]
- Hitti EG, Sallacz NB, Schoft VK, Jantsch MF (2004) Oligomerization activity of a dsRNA-binding domain. FEBS Lett 574: 25–30 [DOI] [PubMed] [Google Scholar]
- Kim JK et al. (2005) Functional genomic analysis of RNA interference in Caenorhabditis elegans. Science 308: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Lee JY et al. (2004) Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J Biol Chem 279: 30265–30273 [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X (2003) R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC (2005) Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol 3: 1202–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD (2005) The RISC subunit TudorsN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 12: 489–496 [DOI] [PubMed] [Google Scholar]
- Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC (2004) Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep 5: 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD (2005) Perspective: machines for RNAi. Genes Dev 19: 517–529 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD (2004) A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Zhong J, Peters AH, Lee K, Braun RE (1999) A doublestranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet 22: 171–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


