Abstract
Innate immunity in vertebrates and invertebrates is of central importance as a biological programme for host defence against pathogenic challenges. To find novel components of the Drosophila immune deficiency (IMD) pathway in cultured haemocyte-like cells, we screened an RNA interference library for modifiers of a pathway-specific reporter. Selected modifiers were further characterized using an independent reporter assay and placed into the pathway in relation to known pathway components. Interestingly, the screen identified the Inhibitor of Apoptosis Protein 2 (IAP2) as being required for IMD signalling. Whereas loss of DIAP1, the other member of the IAP protein family in Drosophila, leads to apoptosis, we show that IAP2 is dispensable for cell viability in haemocyte-like cells. Cell-based epistasis experiments show that IAP2 acts at the level of Tak1 (transforming growth factor-β-activated kinase 1). Our results indicate that IAP gene family members may have acquired other functions, such as the regulation of the tumour necrosis factor-like IMD pathway during innate immune responses.
Keywords: innate immune responses, signalling, Drosophila, RNAi, functional genomics, apoptosis
Introduction
Innate immunity is essential as a first-line defence mechanism against pathogenic challenges in most metazoans. During recent years, it has become clear that the molecular mechanisms that control and execute innate immune responses in humans have well-conserved counterparts in genetically tractable organisms (reviewed by Hoffmann & Reichhart, 2002). In particular, the analysis of signalling pathways in model organisms such as Drosophila has opened new avenues with which to understand and genetically dissect cellular processes that initiate innate immune responses.
Drosophila immunity, which is devoid of an adaptive response, relies mainly on two nuclear factor kappa-B (NF-κB) signalling pathways, commonly referred to as Toll and immune deficiency (IMD) pathways. Following microbial challenge, these pathways regulate the production of antimicrobial peptides by the fat body (an equivalent of the mammalian liver) and blood cells. The induced peptides are subsequently secreted in the haemolymph (blood; Hoffmann, 2003). Additionally, as in mammals, microbial challenges activate Jun amino-terminal kinase (JNK) and Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathways, but their role in insect immune response is less understood (Boutros et al, 2002; Agaisse et al, 2003). The JAK–STAT pathway has also recently been implicated in antiviral responses (Dostert et al, 2005).
The IMD pathway in Drosophila is related to the mammalian tumour necrosis factor (TNF) signalling pathways, as it uses structurally and functionally similar components (Hoffmann & Reichhart, 2002). IMD signalling is mainly activated by Gram-negative bacteria, and loss-of-function mutants are susceptible to Gram-negative bacterial infection (for reviews, see Tzou et al, 2002; Hoffmann, 2003; Brennan & Anderson, 2004, and references therein). Bacterial patterns are recognized by the transmembrane receptor, peptidoglycan recognition protein-LC (PGRP-LC). Subsequently, activation of PGRP-LC initiates a signalling cascade that leads to the processing and nuclear translocation of the NF-κB protein Relish. Three proteins that contain the death domain (DD) are involved in Relish activation: the Drosophila homologue of Fas-associated death protein (FADD), the homologue of RIP adaptor proteins IMD and the caspase 8 homologue DREDD. Downstream of IMD, the Drosophila homologue of transforming growth factor-β-activated kinase 1 (Tak1) activates the signalosome equivalent consisting of IRD5 (IKKβ; immune response-deficient 5) and Kenny (IKKγ). Relish is phosphorylated by the active IKK complex and proteolytically cleaved. Activated Relish translocates to the nucleus and promotes transcription of a distinct set of antibacterial peptides genes, including Diptericin (Dipt), Cecropin (CecA1 and A2), Attacin (AttA, AttB and AttD) and Metchnikowin (Mtk). Another effector arm of IMD signalling, branching off the IMD pathway at the level of Tak1, links the detection of Gram-negative bacteria to JNK signalling and leads to a rapid upregulation of cytoskeletal genes (Boutros et al, 2002).
Although most of the known components were identified by genetic screens or candidate gene approaches, significant gaps remain in the understanding of innate immune signalling pathways. A key advance in recent years has been the discovery and use of RNA interference (RNAi), which allows the silencing of genes through introduction of short, double-stranded RNAs (dsRNAs) homologous to endogenous messenger RNAs (Fire et al, 1998). RNAi has been successfully used to study gene function in invertebrate and mammalian cell culture (reviewed by Hannon & Rossi, 2004).
To identify new components of the IMD pathway, we performed an RNAi screen in cultured Drosophila haemocyte-like cells. We identified putative new regulators that were required to induce Rel-dependent reporter genes after Escherichia coli stimulation and mapped them in relation to known pathway components by cell-based epistasis analysis. We further characterized the function of the positive pathway regulator, Inhibitor of Apoptosis Protein 2 (IAP2). Surprisingly, our results show that IAP2, in contrast to other inhibitors of apoptosis protein family members, is not involved in apoptosis, but is required for the expression of NF-κB and JNK pathway-dependent target genes during innate immune responses.
Results and Discussion
Identification of new IMD pathway components
The exposure of Drosophila SL2 cells to Gram-negative bacteria leads to an upregulation of immune-responsive transcripts, including antimicrobial peptides (Samakovlis et al, 1992; Boutros et al, 2002). To monitor IMD pathway activity, we fused approximately 350 bp promoter sequence of Metchnikowin, which contains Relishspecific binding sites (Senger et al, 2004), to a firefly luciferase gene (supplementary Fig 1A online). On transfection of the mtk luc reporter into SL2 cells and treatment with heat-inactivated E. coli, we observed an approximately 22-fold induction of firefly reporter gene activity (supplementary Fig 1B online), whereas a Renilla co-reporter gene constitutively expressed under the control of the viral IZ promoter (Invitrogen) was not significantly induced. While monitoring mtk luc induction, we screened an RNAi library (Hild et al, 2003; Boutros et al, 2004) and identified all previously known IMD pathway modifiers, as well as putative new regulators of IMD–Rel pathway activity. To confirm the requirement of selected candidate genes, we resynthesized dsRNAs and re-tested them using a second reporter system derived from the IMD signalling-responsive attacinA enhancer (Tauszig et al, 2000; Fig 1A,B). Confirmed candidates included the GTPase-activating protein GAP1, which reduces the IMD pathway reporter activity to 35%, a level similar to that observed with dsRNAs targeting Key or Tak1. GAP1 has been previously implicated in early embryonic development as a regulator of small GTPase pathways (Gaul et al, 1992). Another signalling factor identified is CG7417/ORF1, a gene homologous to a family of mammalian Tak1-binding proteins (TAB), which are required for Tak1-mediated NF-κB activation (Shibuya et al, 1996; Takaesu et al, 2000). CG7417 could function as an activator or an adaptor for Tak1 in the IMD pathway; however its role still remains to be characterized in vivo (Fig 1; supplementary Table 1 online).
Figure 1.
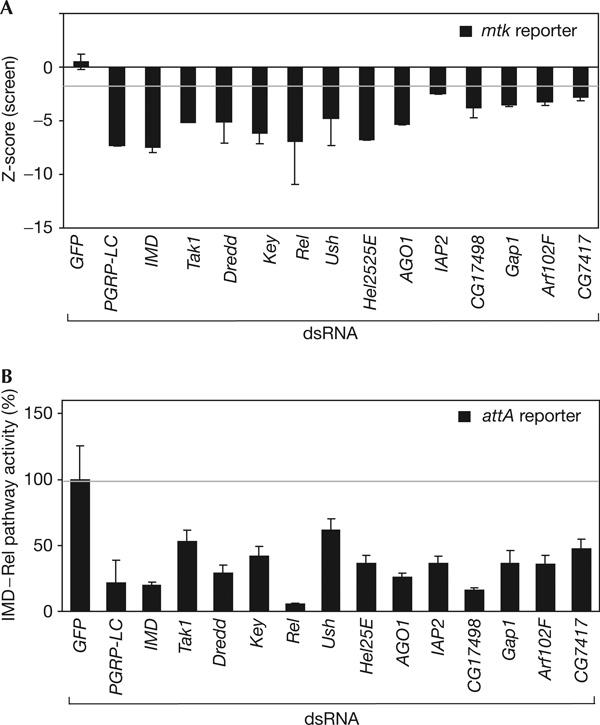
Identification of new immune deficiency pathway components. (A) Effect of depletion of candidate transcript on mtk reporter induction. Depletion of known and new immune deficiency (IMD) pathway components by RNA interference (RNAi) leads to a significant reduction of IMD–Rel pathway activity (black bars). Zscores were calculated as the number of median-adjusted standard deviations that a particular RNAi experiment differed from the median of all RNAi experiments in a 96-well plate. (B) Depletion of known and newly identified components leads to a reduction of the IMD pathway-dependent attacineA (attA) reporter. The percentage reduction of pathway activity after Escherichia coli stimulation as compared with a control double-stranded RNA (dsRNA; green fluorescent protein (GFP)) is shown. The pathway-specific reporter is normalized against a constitutive co-reporter. Error bars represent the standard deviation of three experiments.
We further analysed candidates by mapping their position within the IMD signalling cascade. We performed a cell-based epistasis analysis, activating the pathway ectopically by overexpression of IMD (Georgel et al, 2001) or constitutively active Rel (RelΔPEST; Stoven et al, 2003), whereas IMD pathway components and candidate genes were depleted by RNAi. Signalling activity was monitored with the mtk luc reporter. This approach correctly predicted the position of already known pathway components (Fig 2A,B). We found that the homologue of the mammalian Friend of GATA (FOG) protein Ushaped (Ush) acts downstream of IMD and at the same level as Relish (Fig 2A,B). Ush has been implicated in haemocyte differentiation (Fossett et al, 2001). Its depletion could lead to a misdifferentiation of the cells, which would then be unable to mount an immune response. Furthermore, Argonaute1 (AGO1), a component of the eukaryotic translation initiation factor 2 complex implicated in microRNA processing (Okamura et al, 2004), was identified as a modifier of the IMD signalling pathway, reducing its activity to 25% as compared with dsRNA against GFP (Fig 1). Its requirement in cultured haemocyte cells seems to be IMD pathway specific, as it was not identified as a factor for cell viability or other signalling pathways from screens (our unpublished data). Epistasis experiments place its action between IMD and Rel (Fig 2). Other factors that were identified are shown in Fig 2 and supplementary Table 1 online.
Figure 2.
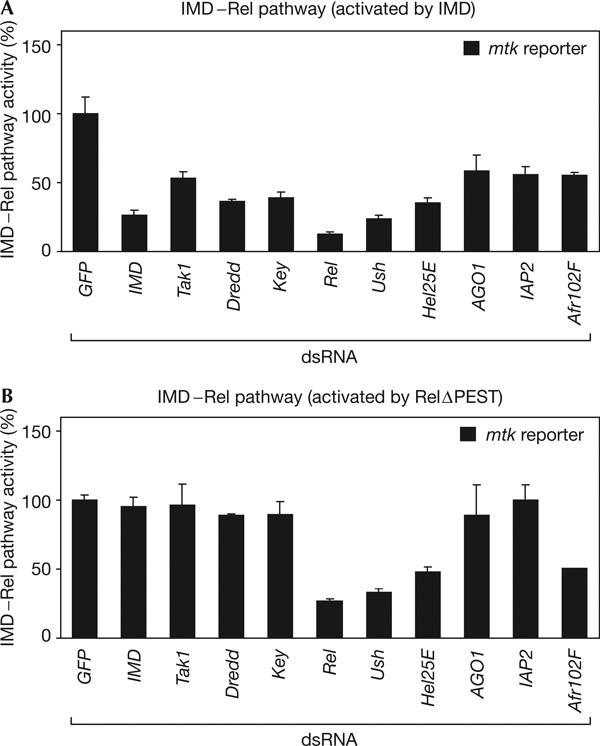
Epistasis analysis mapping the position of candidates in the immune deficiency pathway. Immune deficiency (IMD)/Rel signalling is activated by expression of IMD (A) or a dominant active Relish protein (RelΔPEST) (B). Depletion of IMD–Rel cascade members downstream of IMD and upstream of Relish can block reporter induction by IMD but not by RelΔPEST (IMD, Tak1, Dredd, Key; mtk reporter). Doublestranded RNAs (dsRNAs) against Ush, Hel25E and Afr102F block the activation of the mtk luc reporter induced by IMD and Relish, indicating that they act at the level of Relish. IAP2 and Argonaute1 (AGO1) act between IMD and Relish, as their knockdown results in the loss of reporter induction by IMD. Error bars represent the standard deviation of four experiments.
IAP2 is specifically required for IMD signalling
We further evaluated IAP2 because of its domain composition, which is suggestive of a role as an anti-apoptotic factor (Hay et al, 1995). IAP2 and DIAP1 are members of a two-gene family in Drosophila, with highly conserved homologues in insects, mice and humans (Fig 3A; reviewed by Vaux & Silke, 2005). DIAP1 is essential for cell survival and was shown to bind to, and thereby inhibit, effector caspases (Meier et al, 2000; Muro et al, 2002).
Figure 3.
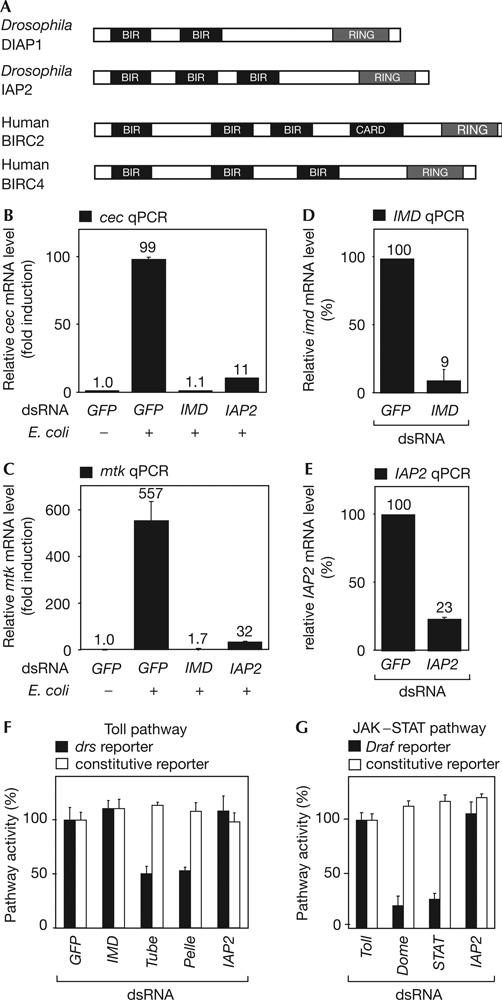
Inhibitor of Apoptosis Protein 2 is specifically required for immune deficiency signalling. (A) Domain structure of Inhibitor of Apoptosis Protein 2 (IAP2). Drosophila proteins DIAP1 and IAP2 and two human IAPs, BIRC2 (BIR-containing protein 2) and BIRC4 are depicted. Two or three BIR motifs located close to the amino terminus are characteristic features of all IAPs. (B,C) Quantitative real-time reverse transcription–PCR (qPCR) experiments monitoring the effect of IAP2 RNA interference (RNAi) on endogenous immune deficiency (IMD)–Rel target gene induction after an immune stimulus. The messenger RNA levels of the IMD–Rel target genes cecropinA2 (cec) and Metchnikowin (Mtk) are induced 99- and 557-fold in response to bacterial induction, respectively (GFP, +). Depletion of IMD by RNAi inhibits induction of cec and mtk expression (IMD, +). Similarly, IAP2 RNAi significantly reduces expression of both peptides in response to a bacterial challenge (IAP2, +). (D,E) IMD and IAP2 levels are strongly reduced by the corresponding doublestranded RNA (dsRNA). (F) Test for requirement of IAP2 in the Toll pathway in SL2 cells. Toll signalling is reduced when components of the Toll pathway are knocked down by RNAi (Tube, Pelle). Depletion of unrelated factors (GFP), components of the IMD pathway (IMD) or IAP2 does not affect Toll signalling activity (black bars, Drosomycin reporter). (G) Test of IAP2 activity in the Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathway in S2R+ cells. JAK–STAT signalling is disturbed when components of the JAK–STAT cascade are depleted by RNAi (Dome, STAT92E; black bars, 2x6xDraf luc reporter). Signalling is not blocked when unrelated factors (Toll) or IAP2 are knocked down by RNAi (IAP2). Error bars represent standard deviation of three independent replicates.
To confirm that IAP2 is a positive regulator of the IMD pathway, we tested the effect of IAP2 depletion on the induction of endogenous target genes by quantitative real-time reverse transcription–PCR (qPCR). SL2 cells were treated with dsRNA against GFP as a negative control, IMD as a positive control or IAP2. We then monitored expression levels of the IMD–Rel target genes cec and mtk in immune-stimulated and unstimulated cells. As shown in Fig 3B,C, RNAi against IAP2 significantly reduced the levels of both target genes similarly to RNAi against IMD (Fig 3D,E).
In addition to the IMD pathway, Drosophila immunity relies on the Toll and the JAK–STAT signalling pathways. We tested for a putative implication of IAP2 in Toll or JAK–STAT signalling using luciferase reporter assays (Tauszig et al, 2000; Muller et al, 2005; see the supplementary information online). Knockdown of known Toll or JAK–STAT pathway components showed a significant reduction of reporter induction, whereas dsRNA directed against IAP2 or GFP, as a control, did not influence Toll or JAK–STAT pathway activity (Fig 3F,G). These results indicate that IAP2 is specifically involved in the IMD signalling pathway but no other known immune-responsive pathways.
As IAPs have been widely implicated as regulators of cell death (Deveraux & Reed, 1999), we investigated whether IAP2 functions as a regulator of cell viability. Therefore, we quantified cell proliferation after 5 days of RNAi against an unrelated factor (GFP), DIAP1 and IAP2. As shown in Fig 4A, RNAi targeting DIAP1 led to a decrease of cell numbers from 0.5 million seeded cells to less than 20,000 live cells after 5 days. This is consistent with previous reports, which showed the essential role of DIAP1 in cell viability (Muro et al, 2002). In contrast, IAP2-depleted samples, similarly to GFP RNAi, showed an increase in cell numbers from 0.5 to 2.2 million. This indicates that IAP2, in contrast to DIAP1, is dispensable for cell viability. These results were confirmed by propidium iodide (PI) staining using fluorescence-activated cell sorting (FACS) analysis (Fig 4B) and viability assays monitoring intracellular ATP levels (supplementary Fig 2 online). Taken together, these results show that IAP2 is a regulator of IMD signalling and is not required for cell growth or survival.
Figure 4.
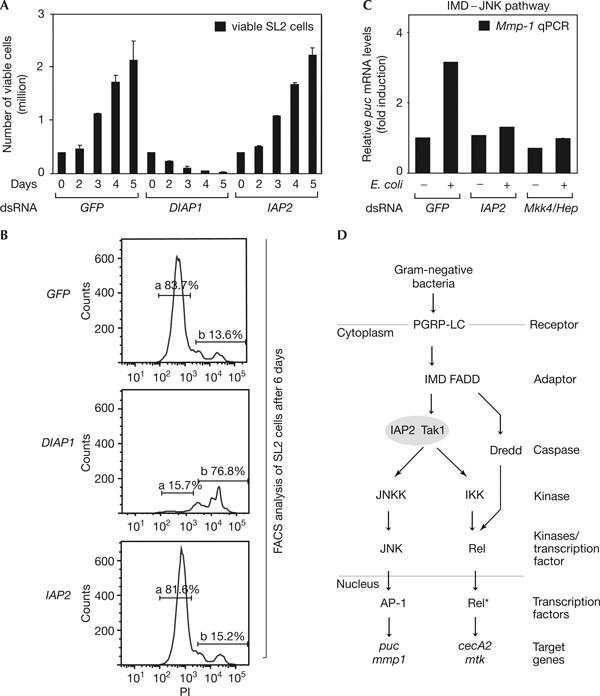
Inhibitor of Apoptosis Protein 2 is dispensable for cell viability and required for immune deficiency/Jun amino-terminal kinase branch. (A) Haemocytometer count monitoring the growth of SL2 cells treated with GFP, DIAP1 and IAP2 doublestranded RNA (dsRNA) from 0 to 5 days. RNA interference (RNAi) targeting a factor not involved in maintenance of viability (GFP) does not affect cell growth, whereas depletion of DIAP1 by RNAi causes severe growth defects. Knockdown of IAP2 by RNAi does not influence cell growth. (B) Fluorescence-activated cell sorting (FACS) assays comparing the effects of GFP, DIAP1 and IAP2 RNAi on SL2 cell viability. At 6 days after RNAi treatment, cells were stained with propidium iodide (PI) and analysed by FACS. About 84% of cells were viable when treated with an unrelated control dsRNA (GFP; gate a, top panel). Only 16% of cells treated with dsRNA depleting DIAP1 were viable on day 6 (gate a, middle panel), in contrast to 82% living cells in IAP2 dsRNA-treated samples (gate a, bottom panel). Gate a: PI-negative events, viable cells; gate b: PI-positive events, dead cells and debris. (C) Quantitative real-time reverse transcription–PCR (qPCR) analysis of Inhibitor of Apoptosis Protein 2 (IAP2) function in JNK signalling in SL2 cells. Messenger RNA levels of the JNK target gene Matrix metalloproteinase 1 (Mmp-1) increase after stimulation with heat-inactivated Escherichia coli for 1 h (GFP, +). Knockdown of JNK components (Mkk4/hep) blocks induction of the IMD–JNK cascade (Mkk4/hep, +). Similarly to Mkk4/hep RNAi, IAP2 knockdown suppresses induction of IMD–JNK signalling (IAP2, +). Error bars represent standard deviation of two replicates. If not indicated otherwise, error bars represent standard deviation of four replicates. (D) Model placing IAP2 function at the level of Tak1 (transforming growth factor-β-activated kinase 1) in the IMD signalling pathway.
IAP2 acts upstream or at the same level as Tak1
Our results indicated that IAP2 is required for the IMD–Rel branch and cell-based epistasis mapped it downstream of IMD and upstream of Relish (Figs 1, 2). As the IMD pathway branches at the same level or downstream of Tak1 into Rel- and JNK-dependent signalling, we examined whether IAP2 is also required for activation of the JNK branch. Thus, we monitored the expression of the IMD/JNKspecific target genes Puckered and Matrix metalloproteinase 1 (Mmp-1; Boutros et al, 2002) by qPCR. As shown in Fig 4C and supplementary Fig 3 online, depletion of IAP2 by RNAi disrupts Mmp-1 and puc induction after innate immune stimuli to a level similar to that of knockdown of known factors specific for the IMD–JNK branch (Mkk4/hep). These experiments, together with the epistasis experiments, support a model whereby IAP2 acts, similarly to Tak1, downstream of IMD and upstream or at the level of the branching point of the IMD–Rel and IMD–JNK signalling arms (Fig 4D).
In conclusion, we identified several new components of the IMD innate immune pathway. Our experiments implicate several signalling factors in the control of IMD-dependent responses in haemocyte-like cells, including a GTPase-activating protein, a homologue of the mammalian Tak1-binding protein, and several proteins involved in RNA binding and processing. Their role in Drosophila immune response in vivo remains to be characterized. Strikingly, the screen identifies IAP2, a member of the Inhibitor of Apoptosis Protein family, as being required for Drosophila innate immune signalling. We show that IAP2 is specifically involved in the IMD signalling pathway, as it disrupts the induction of the IMD–Rel and IMD–JNK pathway target genes and is not required for other immune-induced pathways, such as Toll and JAK–STAT. Cell-based epistasis analysis and qPCR experiments monitoring the IMD–JNK branch suggest a function of IAP2 downstream of IMD and upstream or at the same level as Tak1. Although most previously characterized IAPs were shown to act as inhibitors of caspases (Deveraux & Reed, 1999), it is unlikely that the role of IAP2 is to inhibit DREDD, the caspase implicated in IMD signalling. If this were correct, depletion of IAP2 should lead to an enhancement of pathway activity after immune stimulus or to a constitutive expression of target genes without an immune stimulus, which is not the case. As human Tak1 has been shown to be activated by polyubiquitination (Wang et al, 2001), and it has recently been shown that ubiquitination is required for the activation of Tak1 and the IKK complex in Drosophila (Zhou et al, 2005), we might speculate that IAP2 may have a role in Tak1 ubiquitination through its RING domain. Whether mammalian IAPs have a role in innate immune responses remains to be established.
Methods
Cell culture. Drosophila SL2 and S2R+ cells were cultured in Schneider's Drosophila medium (Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal calf serum (PAA) and 1% penicillin–streptomycin (Invitrogen) at 25°C.
RNA synthesis, RNA interference and luciferase assay. dsRNA synthesis, RNAi treatment and luciferase experiments were performed, as described before (Muller et al, 2005). Complete primer and amplicon sequence information can be found at http://rnai.dkfz.de. Details on used reporter constructs, dsRNA concentration and cells can be found in the supplementary information online. SL2 cells were transiently transfected with specific reporter and expression constructs using Effectene or Cellfectin according to the manufacturer's instructions. When applicable, immune stimulation of cells was performed by adding heat-inactivated E. coli (DSM498) to a final concentration of 20 μg/ml. At 16 h after stimulation, luciferase activity was measured.
RNA extraction and quantitative real-time reverse transcription–PCR Total RNA from SL2 cells was isolated using Qiagen Shredder and RNeasy Mini columns or Trizol (Invitrogen) according to the manufacturer's description. A 5 μg portion of total RNA was treated with DNase I (Fermentas, St Leon-Rot, Germany) for 30 min before reverse transcription with Superscript II (Invitrogen) and oligo(dT)12–18 (Invitrogen). qPCR was performed using LightCycler 1.0 and LightCycler 480 instruments, TaqMan Master Kit, and the Drosophila Universal ProbeLibrary (Roche Applied Science, Mannheim, Germany). Rp49 levels were used to normalize the data. Protocols for RNAi treatment and induction, as well as primer sequences and probe information, are provided in the supplementary information online.
Fluorescence-activated cell sorting and cell viability analysis. Flow cytometry analysis was performed in SL2 cells after RNAi treatment in triplicate in 96-well tissue culture plates (Falcon, BD Biosciences, Heidelberg, Germany; as described in the supplementary information online). Cells were incubated for 6 days at 25°C. Cells were carefully resuspended in 100 μl complete medium and diluted 1:1 with staining solution (PBS, 4 μg/ml PI) to a final volume of 200 μl. Samples were analysed by flow cytometry (FACSArray, BD Biosciences), sampling 40 μl with a maximum of 20,000 ungated events per probe. To determine total PI-negative and PI-positive cell numbers, events were gated using FlowJo (TreeStar, Ashland, OR, USA) software.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Note in proof. Kleino et al (2005) Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J (in press; doi:10.1038/sj.emboj.7600807).
Supplementary Material
Supplementary Methods
Acknowledgments
We thank J.-M. Reichhart, J.-L. Imler, A. Goto, S. Wasserman, D. Hultmark and M. Zeidler for plasmids and K. Bartscherer for the Rp128-RL co-reporter construct. We are grateful to T. Horn and Z. Arziman for help with bioinformatics analysis, M. Stricker and B. Mosterman for technical support and all lab members for fruitful discussions. N.P. was supported by Alexander-von-Humboldt and Association pour la Recherche sur le Cancer postdoctoral fellowships. This work was supported by an Emmy-Noether grant from the Deutsche Forschungsgemeinschaft to M.B.
References
- Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N (2003) Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell 5: 441–450 [DOI] [PubMed] [Google Scholar]
- Boutros M, Agaisse H, Perrimon N (2002) Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell 3: 711–722 [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Consortium HF, Paro R, Perrimon N (2004) Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 [DOI] [PubMed] [Google Scholar]
- Brennan CA, Anderson KV (2004) Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol 22: 457–483 [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC (1999) IAP family proteins—suppressors of apoptosis. Genes Dev 13: 239–252 [DOI] [PubMed] [Google Scholar]
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler J-L (2005) The Jak–STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol 6: 946–953 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by doublestranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA (2001) The Friend of GATA proteins Ushaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc Natl Acad Sci USA 98: 7342–7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul U, Mardon G, Rubin GM (1992) A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell 68: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA (2001) Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell 1: 503–514 [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rossi J (2004) Unlocking the potential of the human genome with RNA interference. Nature 431: 371–378 [DOI] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM (1995) Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83: 1253–1262 [DOI] [PubMed] [Google Scholar]
- Hild M et al. (2003) An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol 5: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA (2003) The immune response of Drosophila. Nature 426: 33–38 [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM (2002) Drosophila innate immunity: an evolutionary perspective. Nat Immunol 3: 121–126 [DOI] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI (2000) The Drosophila caspase DRONC is regulated by DIAP1. EMBO J 19: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M (2005) Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 436: 871–875 [DOI] [PubMed] [Google Scholar]
- Muro I, Hay BA, Clem RJ (2002) The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 277: 49644–49650 [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samakovlis C, Asling B, Boman HG, Gateff E, Hultmark D (1992) In vitro induction of cecropin genes—an immune response in a Drosophila blood cell line. Biochem Biophys Res Commun 188: 1169–1175 [DOI] [PubMed] [Google Scholar]
- Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, Levine M (2004) Immunity regulatory DNAs share common organizational features in Drosophila. Mol Cell 13: 19–32.s [DOI] [PubMed] [Google Scholar]
- Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K (1996) TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science 272: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D (2003) Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc Natl Acad Sci USA 100: 5991–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, Ninomiya-Tsuji J, Matsumoto K (2000) TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell 5: 649–658 [DOI] [PubMed] [Google Scholar]
- Tauszig S, Jouanguy E, Hoffmann JA, Imler JL (2000) Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci USA 97: 10520–10525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou P, De Gregorio E, Lemaitre B (2002) How Drosophila combats microbial infection: a model to study innate immunity and host–pathogen interactions. Curr Opin Microbiol 5: 102–110 [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J (2005) IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol 6: 287–297 [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- Zhou R, Silverman N, Hong M, Liao DS, Chung Y, Chen ZJ, Maniatis T (2005) The role of ubiquitination in Drosophila innate immunity. J Biol Chem [epub ahead of print; doi:10.1074/jbc.M506655200] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods


