Abstract
The daily recurrence of activity and rest are so common as to seem trivial. However, they reflect a ubiquitous temporal programme called the circadian clock. In the absence of either anatomical clock structures or clock genes, the timing of sleep and wakefulness is disrupted. The complex nature of circadian behaviour is evident in the fact that phasing of the cycle during the day varies widely for individuals, resulting in extremes colloquially called 'larks' and 'owls'. These behavioural oscillations are mirrored in the levels of physiology and gene expression. Deciphering the underlying mechanisms will provide important insights into how the circadian clock affects health and disease.
Keywords: circadian rhythm; entrainment, phase; oscillator; clock
Introduction
'Birds do it, bees do it, even educated fleas do it.' Cole Porter was speaking of love, but, save the refrain, he could just as well have been referring to the circadian clock. Although he discussed only animals, organisms of all phyla show circadian rhythms. Rhythmic behaviour persists even in constant conditions, although with a period slightly shorter or longer than the day (hence, circa diem), demonstrating that the oscillation represents an endogenous temporal programme.
In animals, daily rhythms are typically measured as activity versus rest, but hundreds of other parameters from the level of behaviour to gene expression are also oscillating with a circadian period. Plants show numerous circadian rhythmic phenomena, including leaf movement, growth rate and stomatal opening, as well as the expression of much of the photosynthetic machinery. The fungus Neurospora crassa makes asexual spores every 22 h in constant darkness, and the prokaryotic genetic model organism—the cyanobacterium Synechococcus—separates the incompatible processes of photosynthesis and nitrogen fixation to opposite times of the day, thereby solving the problem using time rather than space as do other cyanobacteria. In short, daily biological rhythmicity is widespread in nature, with environmental cues (called zeitgebers), such as light and temperature, synchronizing internal time to the earth's 24-h rotation.
Characteristics of a circadian clock
Several properties unify circadian systems in all organisms so far studied. First is the self-sustained rhythm with its long period (Fig 1A). The circadian system has been routinely probed by plunging an organism into constant conditions, resulting in the (initially) surprising finding that has become fundamental to the field of clocks research: that all organisms have a remarkably precise, sustained, circadian rhythm. Persistent rhythms have been observed in unicells (Lakin-Thomas & Brody, 2004; Mergenhagen, 1980), as well as dissociated cells from the brain (Welsh et al, 1995). Even Rat-1 fibroblasts (an old cell-culture line) can be triggered to display circadian rhythms in gene expression (Balsalobre et al, 1998). Thus, the circadian period is generated at the level of the cell.
Figure 1.
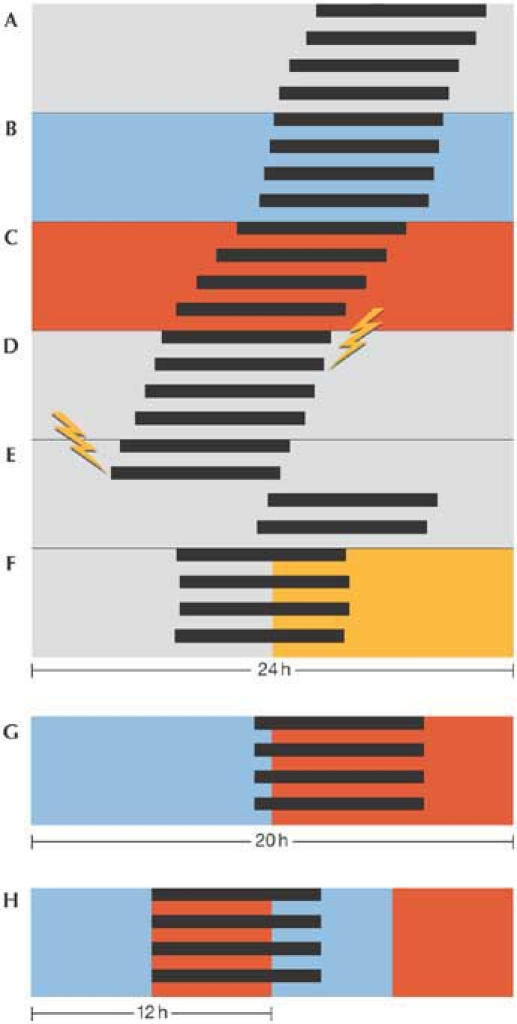
Cartoon of a circadian rhythm in free-running and entrained conditions. The width of the panels is 24 h, except for (G), which represents 20 h. Each bar exemplifies a bout of activity (in this case, the 'activity' of spore formation in Neurospora crassa). (A) In constant darkness and temperature (25 °C), spores develop once per 22 h. The period changes only slightly with ambient temperature, lengthening when it is colder (20 °C; B), and shortening when warmer (30 °C; C). The phase of the rhythm is not changed significantly when a light pulse (flash) is delivered in the subjective mid-day (D), whereas large phase shifts are induced when light is given in the subjective night (E). The rhythm is entrained to a period of exactly 24 h in appropriate zeitgeber cycles of light and darkness (F). Entrainment to non-24-h T cycles is also possible, as shown in (G), in which the phase settles later in the short, 20-h cycle of warm and cold than it would in a 24-h cycle. If an entraining cycle is about one-half of the free-running period, a frequency demultiplication occurs, whereby one bout occurs per two cycles (H, showing 12-h temperature cycles).
Another typical feature of circadian clocks is compensation with respect to spurious changes in the environment, some of which can act as zeitgebers when changing on a regular (daily) basis. Although the clock mechanism is essentially a collection of biochemical reactions, it runs with the same period over a wide range of temperatures (temperature compensation; Fig 1B,C). However, temperature cycles in the 24-h range can synchronize circadian clocks (Brown et al, 2002; Merrow et al, 1999). A flash of lightning in the night will not reset the rhythm in many organisms, and even a substantial amount of light, if supplied at certain times during the cycle (that is, during the internal, subjective day when light is 'expected') might not affect the free-running rhythm in constant conditions (Fig 1D compared with Fig 1E).
The period of the free-running rhythm and its temperature compensation have been determined in constant conditions but, in nature, the clock is predominantly exposed to cycles of light, temperature, food availability, predator presence, and so on. Many of these can act as zeitgebers for circadian systems, synchronizing them through an active mechanism called entrainment (Fig 1F,G). All zeitgebers mirror the cycle length of the earth's rotation and they therefore shaped circadian systems as they evolved. To probe the mechanisms of entrainment (that is, how the internal, biological oscillator interacts with the external, physical one), circadian researchers often use non-24-h cycles (T cycles). As the period of the cycle is shortened, the phase of entrainment occurs later (Fig 1G). In longer cycles, the phase of entrainment advances. This was first shown using lizards in temperature cycles (Hoffmann, 1963). If the cycle is too short or too long, the endogenous circadian rhythm runs 'through', systematically reacting to the cycle (Holst, 1939) but without getting stably caught by it. The 'range of entrainment' within which the clock entrains is both species- and zeitgeberspecific. When the period of an imposed cycle is about one-half (one-third, one-quarter, etc.) of the endogenous period, circadian rhythms typically 'frequency demultiply', occurring once per two (three, four, etc.) external cycles (Fig 1H). All of these entrainment properties show that circadian clocks behave as robust oscillators rather than as hourglass timers, which are passively reset. In the latter case, with instant resetting properties, we would not be troubled by jet lag!
Although circadian systems are all composed of cellular clocks, there are large differences in their assembly. For example, in plants, circadian rhythms in neighbouring cells are apparently running at independent phases (Thain et al, 2000), whereas in animals, they form a highly interdependent, hierarchical network (Fig 2). A dedicated 'pacemaker' resides in the nervous system that receives the environmental and temporal information from zeitgebers (for example, dawn and dusk), processes them and transmits this information through endogenous signals to the cellular clocks in the periphery, supporting coordinated timing (for example, liver, heart or kidney; Akhtar et al, 2001; Panda et al, 2002; Storch et al, 2002; Yoo et al, 2004). The mammalian pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus. Thus, cellular clocks throughout the body are synchronized with respect to the zeitgeber and each other, although not necessarily with the same phase; molecular oscillations in the SCN of mice held in constant darkness, for example, are ahead of those in the liver by approximately 4–6 h (Balsalobre et al, 2000). Furthermore, molecular oscillations of clock genes in the liver can be uncoupled from the SCN by scheduled feeding (Damiola et al, 2000).
Figure 2.
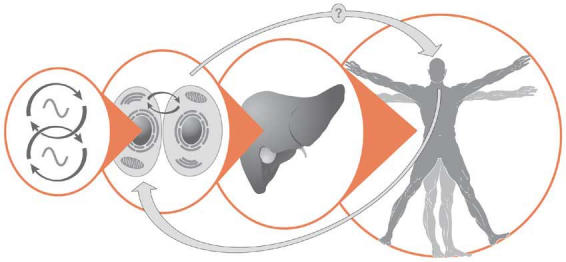
The circadian system in animals is organized hierarchically. Molecular oscillations are generated at the cellular level, in which clock components include transcription–translation feedback loops and possibly metabolic regulatory pathways (left). Organ or peripheral clocks develop a coordinated rhythm that is synchronized relative to a pacemaker in the brain. The most obvious manifestation of this timing system is the sleep–wake cycle, but hundreds of parameters—from cognitive functions to circulating hormone levels—are also changing over the 24-h day. Coupling between the brain and oscillators in the periphery has been shown, and it can be disrupted (Damiola et al, 2000), but feedback from peripheral clocks to the brain has yet to be formally demonstrated.
What is it good for?
Circadian clocks allow organisms to anticipate and 'prepare for' environmental changes, thereby increasing their fitness. For example, a photosynthetic machinery that is already geared up when the first sunrays appear will harvest more energy. A consequence of this logic is that the clock's benefits are most obvious under entrained conditions. By mixing wild-type cyanobacteria and mutant strains with either long or short free-running periods, systematic studies have shown that strains with an endogenous period close to that of the entraining cycle are most successful, whereas the strains did not out-compete each other in constant conditions (Yan et al, 1998). Ablation of the SCN pacemaker in mammals results in the loss of consolidated activity and rest. Accordingly, SCN-lesioned chipmunks are more prone to predators in the wild (DeCoursey & Krulas, 1998).
The seasonal changes of photoperiod become greater with increasing distance from the equator, and many examples of reproduction and physiology show photoperiodic regulation (Elliott & Goldman, 1981). Flowering and seed production in many plants is triggered by day length. Entire animal reproductive systems (from gene expression to anatomical structures) wax and wane with the seasons. Clever experimental light-cycle protocols have shown that the circadian system is at least part of the mechanism that regulates photoperiodism (Saunders, 1990), thereby increasing reproductive fitness (Beaver et al, 2002).
As the genetic era matures, so does the availability of and knowledge about clock mutants. Recent reports indicate that clock mutant animals can suffer from pathologies in addition to their aberrant circadian behaviour. Increased incidence of radiation-induced cancers occur in Period 2 (Per2) mutant mice (Fu et al, 2002), and Clock mutants tend to develop obesity (Turek et al, 2005). Whether and how these pathologies are related to defective clock functions is not known, but lack of coordinated metabolic regulation might be part of the cause. When constructing a network of feedback loops in computer simulation, it is quite difficult to establish conditions that prevent chaotic behaviour (Roenneberg & Merrow, 2002)—these special conditions seem to have evolved in the form of the circadian clock. Among them is not only endogenous rhythmicity but also conditional responsiveness to environmental stimuli, which is necessary to prevent chaotic behaviour.
The genes in the clock
Practically all of the success stories in deciphering a biological mechanism include genetic approaches. Furthermore, numbers are the geneticist's best friend, and the circadian system offers highly quantifiable traits: period length in constant conditions and phase in entrainment. A genetic basis for the free-running circadian rhythm was suggested as early as 1932, by breeding bean plants and selecting for subpopulations with short or long periods (Bünning, 1932). The clock's complex genetic nature could already have been predicted by the bellshaped distribution of the progeny, obtained from crossing long- and short-period individuals. Numerous examples of cellular circadian systems argued that the clock is not simply an emergent property of a complex (multi-organ) system but is based within individual cells. The complexity of the clock was viewed as an obstacle to its genetic analysis. Eventually, pioneering mutagenesis screens revealed the first clock gene (Period) in Drosophila melanogaster (Konopka & Benzer, 1971), followed by the frequency gene in N. crassa (Feldman & Hoyle, 1973). Many clock genes have since been identified in these and other model systems—for example, mice, Xenopus, Arabidopsis and Synechococcus.
Clock proteins in animals, plants, fungi and bacteria are largely unrelated by sequence. However, similar molecular mechanisms are apparent: clock genes and their proteins form a transcription–translation regulatory loop with positive and negative feedback controls (Young & Kay, 2001). As a result, many clock genes and clock-controlled genes—pure outputs of the clock so that their mutations do not affect rhythm generation—cycle with a coordinated period throughout the organism, both in vivo (Akhtar et al, 2001; Storch et al, 2002), as well as in culture (Balsalobre et al, 1998).
Other clock components, such as kinases or transcription factors that drive the feedback loop, are constitutively expressed. Post-transcriptional and post-translational modifications (predominantly phosphorylation so far) are also key regulators in the clock mechanism. Phosphorylation is generally thought to regulate the clock by modulating clock-protein half-life (Görl et al, 2001; Liu et al, 2000; Young & Kay, 2001), but it could also regulate subcellular compartmental shuttling or the activation/inactivation of transcription factor activity.
Putative clock genes (identified through mutagenesis, interaction and homology) are typically probed with reverse genetic approaches, in which candidates are knocked out and the resultant mutants assayed for clock properties. Co-regulatory effects on the expression of other clock genes are used to establish how a gene functions in the clockwork. For example, expression levels of Cryptochromes 1 (Cry1) and 2 (Cry2) in Clock mutant mice are non-rhythmic and low, indicating that these components of the transcription–translation feedback loop are downstream of the activator complex formed by the Clock and Brain and Muscle Arnt-like protein 1 (BMAL1; Kume et al, 1999). Molecular clock models have accordingly become an extensive network of interlocking loops, without revision of the basic feedback loop hypothesis.
A recent report may help to promote a re-assessment of this view: circadian oscillations have been reconstituted in a test tube using just three Synechococcus proteins together with ATP (Nakajima et al, 2005). The test tube oscillation is temperature-compensated with little apparent damping and even adopts appropriate periods when mutant proteins are added. The measured output was the phosphorylation of the cyanobacterial clock protein KaiC in the presence of its modifiers KaiA and KaiB. The authors concluded that this metabolic phosphorylation oscillator—not the transcriptional–translational feedback loop formed by KaiA, KaiB and KaiC and their respective protein products—generates the circadian rhythms, but that this could be a peculiarity of a prokaryotic clock. However, similar to the readout in the Synechococcus experiment, every molecular clock network described so far has at least one component that is rhythmically phosphorylated in a predictable way. Indeed, kinase mutants represent some of the most severe clock phenotypes (Lowrey et al, 2000; Price et al, 1998).
How might a metabolic oscillator be consistent with the negative feedback mechanism involving transcription and translation? In a single organism, two oscillators can run free almost independently of each other. This has been dramatically shown in temporal isolation experiments in humans: the intervals between activity, rest, meals, defaecation and even subjective estimations of when an hour has passed can be stretched out to a circa 48-h (or longer) day in these constant conditions. However, the period of the core body temperature rhythm remains around 25 h (Aschoff et al, 1967). More recently, using jet-lag simulations in mice, the rhythm of clock-gene expression was dissociated from that of neuronal firing rate in the SCN pacemaker (Vansteensel et al, 2003).
Physiological experiments show that there are multiple oscillators even in single-cell and syncitial organisms: in the fungus N. crassa or the dinoflagellate Gonyaulax polyedra, two oscillators can run independently under constant conditions (Correa et al, 2003; Roenneberg & Morse, 1993). Indications of mechanisms beyond the transcription–translation feedback loop are not only observed when circadian systems run free in constant conditions. In Neurospora, it is now broadly acknowledged that frequency (frq)-less Neurospora strains are systematically entrained by temperature cycles (Merrow et al, 1999), and selfsustained rhythms persist under some conditions in clock mutant strains (Lakin-Thomas & Brody, 2000). In a wild-type strain, entrained phase in Neurospora correlates with clock protein levels, but not necessarily with the respective RNA expression (Tan et al, 2004), which suggests uncoupling of the feedback loop (although not all of its components) from clock function. A coupling 'agent' might be suggested by work showing that clock transcription factor activity can be regulated by the redox state of the cell (Rutter et al, 2001).
A synthesis of these findings indicates a conceptual framework that incorporates at least two main oscillatory systems (Fig 2, left), one stringently based on rhythmic transcription, the other apparently not. As coupling between oscillatory systems is altered (due to photoperiod, nutrition, temperature, and so on), different components in the system might become dominant over others. The necessity to accommodate changing conditions or seasons might explain the complexity of the circadian clock.
The clock in our genes
Clock genes in humans were revealed through their homology to those in mice. However, at this time, there are no (published) circadian methods using human cells in tissue culture that would facilitate functional clock studies. In humans, we rely on entrainment to allow genetic studies. Entrained phase is described by 'chronotype', a term that reflects the preferred timing of activity and rest during the day. Chronotype can be assessed with a simple questionnaire asking about sleep and wake times on work and on free days (Roenneberg et al, 2004). The chronotype distribution in the population is almost normal, with the mean midsleep time on free days at about 04:30. Few individuals lie at the extreme ends (pronounced morning and evening types, larks and owls, respectively): 0.2% of people are more than 3 h earlier than the mean (that is, go to bed around 21:30 or earlier) and 4.5% prefer to sleep 3 h later (that is, go to bed around 03:30 or later). The bell-shaped distribution contrasts with discrete Mendelian inheritance patterns, which suggests the involvement of many genes (or at least many polymorphisms in several genes), making the search for human clock genes more difficult.
But what does chronotype tell us about the circadian clock? The relationship between free-running period and chronotype (entrained phase) was established long ago, showing that organisms with short free-running periods entrain earlier in identical zeitgeber cycles compared with those that have long free-running periods (Pittendrigh & Daan, 1976). Similarly, chronotype (or diurnal preference) and free-running periods show a highly significant correlation in humans (Duffy et al, 2001). At least in one human subject, a highly penetrant per2 allele confers early chronotype as well as short period (Toh et al, 2001). In addition to a genetic disposition, chronotype changes with age, becoming progressively later during adolescence and then gradually shifting earlier again (Roenneberg et al, 2004).
We can start to understand and probe the strategy of using chronotype to find human clock genes by studying the mouse model system (despite the fact that mice are nocturnal). Mutant mice have been generated for many putative clock genes, and their activity rhythms have been recorded both in constant and entrained conditions. One study has specifically and systematically addressed entrained phase in clock mutant mice (Spoelstra et al, 2004). The differences in entrained phase (using traditional 12 h light:12 h dark cycles) are minimal in Cry1 and Cry2 mice compared with background-matched wild-type controls (less than 0.2 h), despite large differences in free-running periods. The lack of correspondence between phase and period in these mutant animals still has to be scrutinized experimentally (for example, by varying the zeitgeber strengths and lengths, which should both alter entrained phase). Other mouse mutants do 'obey' the phase-period law. The free-running period of the Per1 Cry2 double mutant is 30 min longer and becomes active about 4 h later in light:dark cycles than that of the wild type (Oster et al, 2003).
Ironically, the same double mutant illustrates the inherent complications of approaching human clock genes by using chronotype as a clock phenotype. Mouse Per1 Cry2 mutants result from a cross between a short period (Per1, 1 h shorter than wild type) and a long period (Cry2, 0.8 h longer) mutant. The entrained phase of Per1 complies with the phase-period rule (0.4 h earlier), whereas the chronotype of Cry2 is like that of wild type. These results show that neither phase nor period is inherited in a simple and predictable manner. Investigations into how this works will help to genetically characterize the bell-shaped chronotype distribution.
As our society moves towards a worldwide '24/7' culture, with shift work and jet lag almost the norm, circadian clock research is becoming highly relevant to human health, behaviour and quality of life. Chronotype is our only handle on human clock genetics in real life. Understanding the complex genetics (and possibly epigenetics) in laboratory-reared mice, Arabidopsis, Neurospora and Synechococcus will also help in the understanding of the rich variation in circadian clocks in humans, who—besides inheriting certain clock properties—are exposed to seasons, cultures, social pressures and age-related chronotype changes.



Acknowledgments
We thank S. Daan for comments. Our work is supported by the Deutsche Forschungsgemeinschaft, the Nederlandse Organisatie voor Wetenschaappelijk Onderzoek, the European Commission and the Gottlieb Daimler und Karl Benz-Stiftung.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smoth AG, Gant TW, Hastings MH, Kyriacou CP (2001) Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–550 [DOI] [PubMed] [Google Scholar]
- Aschoff J, Gerecke U, Wever R (1967) Desynchronization of human circadian rhythms. Japan J Physiol 17: 450–457 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U (1998) A serum shock induces gene expression in mammalian tissue culture cells. Cell 93: 929–937 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U (2000) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–2347 [DOI] [PubMed] [Google Scholar]
- Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM (2002) Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc Natl Acad Sci USA 99: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U (2002) Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12: 1574–1583 [DOI] [PubMed] [Google Scholar]
- Bünning E (1932) Über die Erblichkeit der Tagesperiodizität bei den Phaseolus Blättern. Jb wiss Bot 81: 411–418 [Google Scholar]
- Correa A, Lewis ZA, Greene AV, March IJ, Gomer RH, Bell-Pedersen D (2003) Multiple oscillators regulate circadian gene expression in Neurospora. Proc Natl Acad Sci USA 100: 13597–13602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Minh NL, Preitner N, Kornmann B, Fleury-Olela F, Schibler U (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey PJ, Krulas JR (1998) Behavior of SCN-lesioned chipmunks in natural habitat: a pilot study. J Biol Rhythms 13: 229–244 [DOI] [PubMed] [Google Scholar]
- Duffy JF, Rimmer DW, Czeisler CA (2001) Association of intrinsic circadian period with morningness–eveningness, usual wake time, and circadian phase. Behav Neurosci 115: 895–899 [DOI] [PubMed] [Google Scholar]
- Elliott JA, Goldman BD (eds) (1981) Seasonal Reproduction: Photoperiodism and Biological Clocks. New York, USA: Plenum [Google Scholar]
- Feldman JF, Hoyle MN (1973) Isolation of circadian clock mutants of Neurospora crassa. Genetics 75: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee CC (2002) The circadian gene Period2 plays an important role in tumor suppression and DNA damage responses in vivo. Cell 111: 41–50 [DOI] [PubMed] [Google Scholar]
- Görl M, Merrow M, Huttner B, Johnson J, Roenneberg T, Brunner M (2001) A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J 20: 7074–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K (1963) Zur Beziehung zwischen Phasenlage und Spontanfrequenz bei der endogenen Tagesperiodik. Z Naturforschg 18b: 154–157 [Google Scholar]
- Holst E (1939) Die relative Koordination als Phänomen und als Methode zentralnervöser Funktionsanalyse. Ergebn Physiol 42: 228–306 [Google Scholar]
- Konopka R, Benzer S (1971) Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 68: 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas PL, Brody S (2000) Circadian rhythms in Neurospora crassa: lipid deficiencies restore robust rhythmicity to null frequency and white-collar mutants. Proc Natl Acad Sci USA 97: 256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin-Thomas PL, Brody S (2004) Circadian rhythms in microorganisms: new complexities. Annu Rev Microbiol 58: 489–519 [DOI] [PubMed] [Google Scholar]
- Liu Y, Loros J, Dunlap JC (2000) Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci USA 97: 234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergenhagen D (1980) Circadian rhythms in unicellular organisms. Curr Topics Microbiol Immunol 90: 123–147 [DOI] [PubMed] [Google Scholar]
- Merrow M, Brunner M, Roenneberg T (1999) Assignment of circadian function for the Neurospora clock gene frequency. Nature 399: 584–586 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415 [DOI] [PubMed] [Google Scholar]
- Oster H, Baeriswyl S, Horst GTJvd, Albrecht U (2003) Loss of circadian rhythmicity in aging mPer1−/− mCry2−/− mutant mice. Genes Dev 17: 1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Millar BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S (1976) A functional analysis of circadian pacemakers in nocturnal rodents. IV. Entrainment: pacemaker as clock. J Comp Physiol A 106: 291–331 [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95 [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M (2002) Life before the clock—modeling circadian evolution. J Biol Rhythms 17: 495–505 [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Morse D (1993) Two circadian oscillators in one cell. Nature 362: 362–364 [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Kuehnle T, Pramstaller PP, Ricken J, Havel M, Guth A, Merrow M (2004) A marker for the end of adolescence. Curr Biol 14: R1038–R1039 [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, Wu LC, McKnight SL (2001) Regulation of Clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293: 510–514 [DOI] [PubMed] [Google Scholar]
- Saunders DS (1990) The circadian basis of ovarian diapause regulation in Drosophila melanogaster: is the period gene causally involved in photoperiodic time measurement? J Biol Rhythms 5: 315–332 [DOI] [PubMed] [Google Scholar]
- Spoelstra K, Albrecht U, van der Horst GTJ, Brauer V, Daan S (2004) Phase responses to light pulses in mice lacking functional per or cry genes. J Biol Rhythms (in press) [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83 [DOI] [PubMed] [Google Scholar]
- Tan Y, Dragovic Z, Roenneberg T, Merrow M (2004) Entrainment of the circadian clock: translational and post-translational control as key elements. Curr Biol 14: 433–438 [DOI] [PubMed] [Google Scholar]
- Thain SC, Hall A, Millar AJ (2000) Functional independence of circadian clocks that regulate plant gene expression. Curr Biol 10: 951–956 [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH (2001) An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Turek FW et al. (2005) Obesity and metabolic syndrome in circadian clock mutant mice. Science 308: 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansteensel MJ, Yamazaki S, Albus H, Deboer T, Block GD, Meijer JH (2003) Dissociation between circadian Per1 and neuronal and behavioral rhythms following a shifted environmental cycle. Curr Biol 13: 1538–1542 [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM (1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697–706 [DOI] [PubMed] [Google Scholar]
- Yan OY, Andersson CR, Kondo T, Golden SS, Johnson CH, Ishiura M (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA 95: 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH et al. (2004) PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 101: 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]


