Abstract
The cellular delivery of short interfering RNA (siRNA) is a main hurdle in therapeutic drug development. Here, we describe that phosphorothioate (PTO)-derived oligonucleotides stimulate the physical cellular uptake of siRNA in trans in human cells. This is reflected by an apparent dose-dependent siRNA-mediated suppression of lamin A/C in primary human umbilical vein endothelial cells. The PTO-stimulated cellular uptake in trans is concentration dependent, length dependent, related to the phosphorothioate chemistry but not sequence specific. We provide experimental evidence to support a caveolin-mediated uptake mechanism. In sum, this work strongly suggests the exploration of PTOs as facilitators in the delivery of biologically active siRNA to mammalian cells.
Keywords: caveolin; cellular uptake; nonviral vector; siRNA, delivery; siRNA, transport
Introduction
Small interfering RNAs (siRNAs) mediate RNA interference in mammalian cells (Elbashir et al, 2001a, 2001b). The use of siRNAs as tools and drugs to specifically suppress gene expression has moved from basic biological research to become the focus of applied molecular biology and molecular medicine (Hannon & Rossi, 2004). The increasing importance of siRNA as a potential therapeutic is reflected by the fact that clinical phase I and phase II studies in the use of synthetic siRNA have already started (Dorsett & Tuschl, 2004). For siRNAs as well as for other oligomeric nucleic acids used in target validation in vivo and therapeutic drug development including antisense oligonucleotides (ONs; Kalota et al, 2004), the growing group of small biologically active RNAs (Bachellerie et al, 2002), ribozymes (Scanlon, 1998), CpG-immunostimulatory ONs (Krieg & Davis, 2001) and aptamers (Toulmé et al, 2004), one of the main hurdles still remains: efficient delivery to target cells and tissues. In cell culture and in the use of primary tissues ex vivo, siRNA can be delivered by a variety of carrier systems including viral and nonviral vector systems (Wadhwa et al, 2004). For application in vivo, there are several efficient drug-delivery systems (Allen & Cullis, 2004). However, most delivery methods cannot be simply adapted to ONs and the use of those delivery systems seems to be complicated in humans until now, although very recently, the expression of apolipoprotein B in mice was suppressed by cholesterol-conjugated siRNA, which was delivered systemically at relatively high doses of 50 mg/kg (Soutschek et al, 2004).
Evidence suggests that, in vivo, some cell and tissue types take up nucleic acids without carriers (Laktionov et al, 1999; de Diesbach et al, 2000). From these observations, one might suggest that successful delivery of biologically active siRNA is possible in principle. This view is further supported by the recent finding of the spontaneous carrier-free cellular uptake of siRNA by mammalian cells, although at a very low efficiency (Lingor et al, 2004; Overhoff et al, 2004). For example, at a 200 nM extracelluar concentration of siRNA, between 101 and 103 endogenous copies can be detected per cell (Overhoff et al, 2004). Furthermore, the spontaneous cellular uptake of long-chain DNA was shown to occur at measurable amounts and, interestingly, this transport process could be increased, that is, stimulated in trans in the presence of specific physical forms of nucleic acids, including supercoiled DNA and single-stranded ON (Lehmann & Sczakiel, 2005). We therefore investigated whether the delivery of siRNA to mammalian cells can be stimulated by coincubation with specific nucleic acids in trans. Here, we provide conclusive evidence for the stimulation of the physical cellular uptake and increased biological effectiveness of siRNA by phosphorothioate (PTO)-modified ON in the use of two cellular systems, the cell line ECV304 and primary human umbilical vein endothelial cells (HUVECs).
Results and Discussion
When we tested the influence of a variety of nucleic acids on the cellular uptake of siRNA by human cells, a surprising effect was observed: the presence of ON carrying PTO-modified internucleotide linkages at either end in a serum-free culture of ECV304 cells led to an increased physical uptake of the coincubated siRNA. This was measured as described in detail elsewhere (Overhoff et al, 2004). It should be noted that this protocol measures uptake of internalized siRNA rather than cell-surface-bound siRNA. Furthermore, trypsin treatment of adherently growing cells before the preparation of endogenous RNA, which may be expected to substantially reduce the yields of RNA (Laktionov et al, 2004), and the direct scraping of cells before RNAs were prepared resulted in the same yields of detectable siRNA (see the supplementary information online).
The facilitated cellular uptake of siRNA is PTO-specific
First, we addressed the role of the ON chemistry in increased uptake of siRNA and compared the completely thio-derived ON TM6-4 with non-modified ribo, deoxyribo, 2′-OMe or thio-ribo analogues of TM6-4 with the same nucleotide sequence. This comparison indicates that in fact the internucleotide PTO group in a deoxyribo backbone is necessary to stimulate cellular uptake of siRNA, although to a minor extent the all-thio ribo derivative also shows functional effects (Fig 1A).
Figure 1.
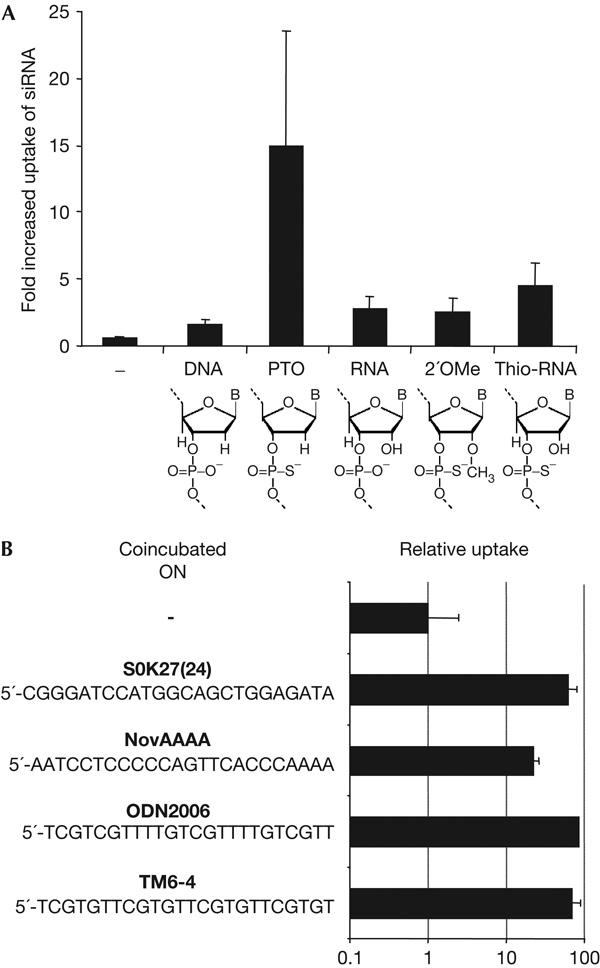
Influence of nucleic acid chemistry and nucleotide sequence on the stimulated uptake of short interfering RNA (si2B) by ECV304 cells at 200 nM extracellular concentration. (A) The backbone chemistry of the oligonucleotide (ON) TM6-4 ([5′-TCGTGT-3′]4) at 600 nM is indicated below each bar, which indicates the factor of increased uptake of si2B. This factor is set as 1 in the absence of ON. The coincubation time was 16 h and uptake of short interfering RNA (siRNA) was quantified by liquid hybridization analysis as described. (B) ECV304 cells were incubated with si-scr (200 nM) and phosphorothioates (PTOs; 600 nM) with the indicated sequences for 16 h. The relative uptake was standardized to the value obtained with si-scr alone.
The uptake mechanism is not sequence dependent
The influence of the nucleotide sequence of ON on the ON-stimulated cellular uptake of siRNA was studied using a set of all-thioate-derived ONs sharing the length of 24 nucleotide (nt) but differing substantially in sequence (Fig 1B). Some of these ONs have biological activities. For example, the motif TM6 was originally identified by a combinatorial approach in which cellular uptake in cis was studied (data not shown) and ‘ODN2006' exerted immunostimulatory effects on B cells (Hartmann & Krieg, 2000). However, no significant difference was observed for all the 24-mers shown in Fig 1B. Furthermore, we did not find any influence of the nucleotide sequence of siRNA on its PTO-stimulated cellular uptake (data not shown).
Characteristics of stimulatory PTOs
To further characterize and possibly improve the PTO-stimulated physical cellular uptake of siRNA, we investigated the length and concentration dependence in ECV304 and HUVEC cells (Fig 2). In both cell types, there is an increasing stimulation of the uptake of siRNA with increasing length of the coincubated PTO. Interestingly, it seems that this effect reaches a plateau in the laboratory cell line ECV304 at a length of approximately 30 nt (Fig 2A, right panel), whereas stimulation of siRNA uptake increases to an ON length of 72 nt in primary human HUVEC cells without any indication of a plateau at this length (Fig 2A, left panel). For the concentration dependence of the PTO-stimulated uptake of siRNA by ECV304 and HUVEC cells, there is an apparent value for half-maximal effects in the range of 100–200 nM of the coincubated PTO at an siRNA concentration of 200 nM (Fig 2B).
Figure 2.
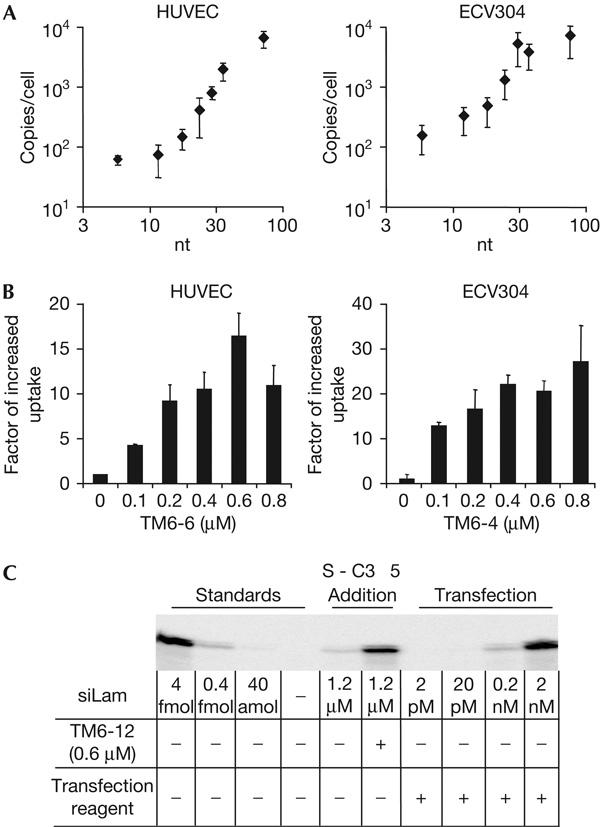
Length and concentration dependency of the phosphorothioate-stimulated physical cellular uptake of short interfering RNA as quantified by the liquid hybridization protocol described in the Methods section. (A) Human umbilical vein endothelial cells (HUVEC) or ECV304 cells were incubated with 200 nM si-scr or si2B, respectively, and 600 nM of phosphorothioates (PTO) that were TM6-derived oligonucleotides of the homologous series of TM6-n/[5′-TCGTGT-3′]n (n=1,2,3,4,5,6,12) for 16 h. (B) Concentration dependence of the uptake of si-scr. HUVEC cells were incubated with 200 nM of si-scr and the indicated concentrations of the PTO TM6-6 (36 nt) for 16 h. ECV304 cells were incubated with 200 nM si2B and the indicated concentrations of TM6-4 (24 nt) for 16 h. In both cases, the increased uptake was standardized to the value obtained by incubation of cells with short interfering RNA alone. (C) Comparison of the physical uptake of siLam mediated by PTOs or by the transfectant lipofectamine at the cell culture concentrations indicated below the gel.
We then compared the efficiency of the PTO-stimulated delivery of siRNA with lipofectamine-mediated delivery in the renal carcinoma cell line SK-RC35. This experiment indicates a similar efficiency of uptake at 2 nM of siRNA after transfection and 1.2 μM of siRNA at PTO stimulation (Fig 2C), which corresponds to approximately 5 × 105 copies of siRNA per cell.
Evidence for a caveolin-mediated uptake mechanism
To understand the mechanism of the PTO-stimulated delivery of siRNA, we investigated specific inhibitors of different uptake pathways and quantified its physical uptake (Fig 3A). These data are consistent with an energy-driven and caveolin-mediated endocytotic uptake pathway of siRNA. The hypothesis supports the increased uptake in the presence of okadaic acid, an activator of the caveolin pathway (Fig 3A). The use of chloroquine does not provide evidence for the crucial involvement of acidic compartments in the PTO-stimulated uptake of siRNA (data not shown).
Figure 3.
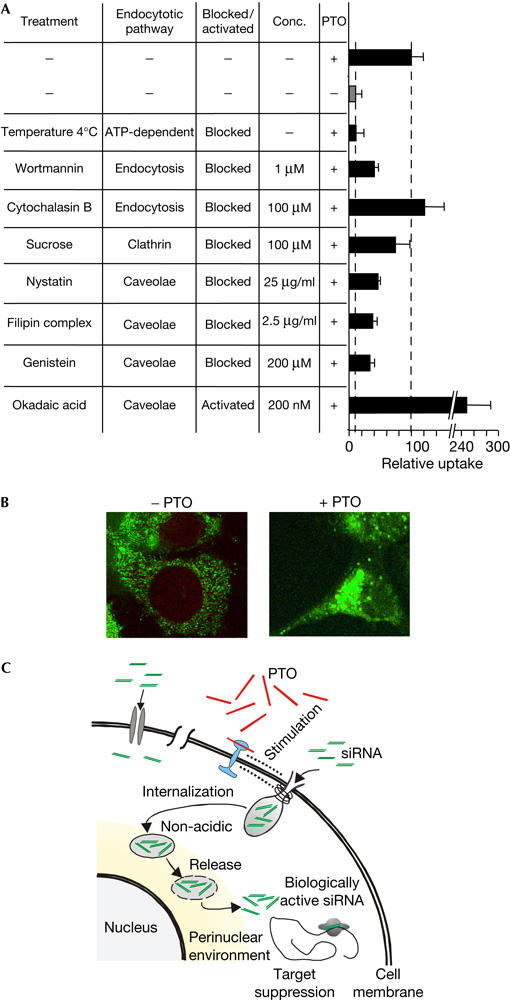
Characteristics and a model of the phosphorothioate-stimulated uptake of short interfering RNA. (A) Effects of temperature and specific inhibitors and activators of uptake pathways of siLam (200 nM) and the phosphorothioate (PTO) TM6-6 (600 nM). (B) Confocal laser scanning microscopy of ECV304 cells that were incubated with Rh110-labelled short interfering RNA (siRNA; siLam, 1.2 μM) in the absence (left panel) or presence of PTOs (TM6-6, 1.0 μM). (C) Schematic depiction of the presumed PTO-stimulated cellular uptake of siRNA (upper right). siRNA (green) is internalized in caveosomal vesicles, transported to the perinuclear environment and released to become active as a suppressor of gene expression. This uptake pathway is stimulated by PTOs (red), hypothetically by interacting with PTO-specific cellular components (blue) that do not give rise to increased endogenous levels of PTO. A presumed low-efficient uptake pathway of siRNA that is not influenced by PTOs is depicted on the upper left of this schema.
Further support for a caveolin-mediated mechanism comes from studies with lymphoid cells that are considered not to support a caveolin-mediated pathway (Fra et al, 1995). Neither the T-lymphoid cell line Jurkat (Schneider et al, 1977) nor the B-lymphoid cell line BJA-B (Menezes et al, 1975) supports the PTO-stimulated uptake of siRNA at the conditions studied in this work (data not shown).
With regard to the coincubated PTOs, which are assumed to enter cells by an endocytotic receptor-mediated pathway at the concentrations used here, we find that their intracellular abundance is not altered at varying conditions of incubation and is close to the detection limits (data not shown).
In addition, we looked at the influence of the physical state of the stimulating nucleic acid. This includes the long- and short-chain forms of single- and double-stranded DNA and RNA, which is of relevance, as various forms of nucleic acid have been recently shown to influence the spontaneous uptake of naked double-stranded DNA (Lehmann & Sczakiel, 2005). It turns out that none of the long-chain forms of DNA or the single-stranded RNA has a stimulatory effect on the cellular uptake of siRNA as the PTOs have (Table 1). In particular, it is noteworthy that the PTO-promoted uptake of siRNA is apparently different from the promotion of cellular uptake of long-chain DNA by specific DNA segments as described recently (Beltinger et al, 1995).
Table 1.
Effect of different physical forms of nucleic acids on the phosphorothioate-stimulated uptake of siLam at 200 nM
| Coincubated nucleic acid | Length | Concentration (nM) | Copies/cell of siRNA |
|---|---|---|---|
| – | – | – | ⩽250 |
| PTO: TM6-12 | 36 nt | ∼420 | ∼3,000 |
| PTO: TM6-12 | 72 nt | ∼210 | ∼7,000 |
| Unmodified: ssRNA | 3 kb | ∼5 | ⩽250 |
| Unmodified: ds DNA | 250 bp | ∼30 | ⩽250 |
| Circular plasmid DNA | ∼4 kb | ∼2 | ⩽250 |
| Linear plasmid DNA | ∼4 kb | ∼2 | ⩽250 |
The concentration of all coincubated nucleic acids was 2.0 μg/ml, which corresponds to the molarity indicated in the third column. The detection limit here was 250 copies of siRNA per cell. dsRNA, double-stranded RNA; PTO, phosphorothioate; siRNA, short interfering RNA; ssRNA, single-stranded RNA.
The subcellular localization of siRNA after PTO-promoted cellular uptake is different when compared with the spontaneous uptake in the absence of PTOs. Confocal laser scanning microscopy of ECV304 cells indicates a perinuclear and cytoplasmic spot-wise localization of siRNA, whereas the fluorescence signal is diffuse and homogeneously distributed over the cytoplasm without PTOs (Fig 3B).
In sum, the data described here (Fig 3A,B; Table 1) are compatible with a hypothetical mode of uptake that is schematically depicted in Fig 3C. Essentially, we assume a caveolin-related pathway including non-acidic compartments that may guide siRNA to intracellular structures accumulating around the nuclear membrane. Such presumed structures might include the Golgi apparatus or the endoplasmic reticulum, or other non-acidic compartments such as caveosomes (Parton & Richards, 2003; Pelkmans et al, 2004). In the absence of PTOs, we also observed the uptake of naked siRNA, although at very low levels and showing a diffuse intracellular distribution, which clearly differs from the subcellular localization of PTO-promoted uptake of siRNA, suggesting different modes of uptake in the presence and absence of PTOs.
Physical uptake of siRNA is related to biological activity
The amount of siRNA in HUVEC cells in the absence of PTOs is in the order of the experimental detection limit, which is approximately 50 molecules per cell (Overhoff et al, 2004). In the case of PTO stimulation, this is increased by a factor ranging between 30 and 50, which corresponds to approximately 2,000 molecules per cell at 600 nM of PTO. When considering that a copy number per cell in the order of 900–9,000 of the intercellular adhesion molecule-1 (ICAM-1)-directed siRNA si2B is related to half-maximal target suppression (Overhoff et al, 2004), it is conceivable that PTO-enhanced cellular delivery of siRNA could be sufficient to observe target suppression.
To investigate this possibility, we chose the well-established siRNA-suppressed lamin A/C system (Elbashir et al, 2002) as a model and coincubated HUVEC cells from independent sources with the lamin A/C-directed siRNA ‘siLam' and PTOs (Fig 4A). For both batches of HUVEC cells shown in Fig 4A, a dose-dependent decrease of apparent expression of lamin A/C was observed at increasing concentrations of extracellular siLam. It should be noted that, in the presence of unrelated siRNA and PTO at the same conditions, an increase of lamin expression was observed, as is the case at low doses of siLam in the presence of PTO for donor 2 (Fig 4A). Furthermore, we observed a donor-related variability of the siLam-dependent target suppression (Fig 4A), which indicates that further unidentified factors might also influence the stimulation of the PTO-mediated cellular uptake mechanism of siRNA.
Figure 4.
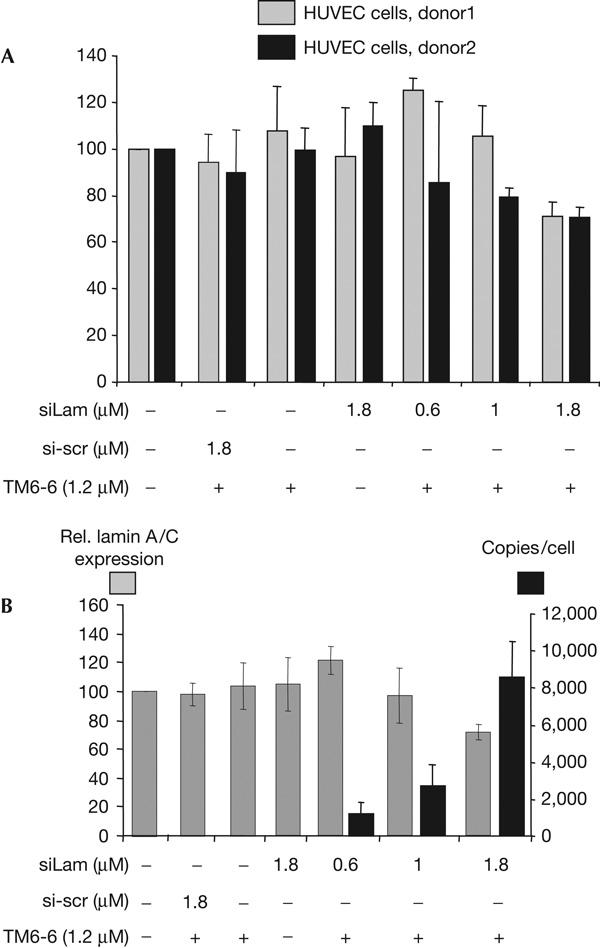
Donor-dependent suppression of lamin A/C expression by phosphorothioate-delivered siLam (A, mean and s.d. of six experiments for each donor) and relationship between physical uptake and biological activity in human umbilical vein endothelial cells (HUVEC) (B, mean and s.d. of six experiments for each of two donors). HUVEC cells were incubated with the indicated concentrations of siLam and 1.2 μM of the phosphorothioate TM6-6 (36 nt) for 24 h, the supernatant was substituted by medium 199 supplemented with 20% FCS and 25 μg/ml of endothelial cell growth supplement and the cells were incubated for another 24 h before analysis. The lamin A/C target messenger RNA was quantified by reverse transcription–PCR and the physical cellular uptake of siLam was measured by liquid hybridization analysis as described. The detection limit for siLam was 40 copies/cell.
In subsequent experiments, the physical cellular uptake of siLam was related to apparent expression of lamin A/C in HUVEC cells. This experiment shows a concentration-dependent accumulation of intracellular siLam, which is related to decreased gene expression of lamin A/C, reaching expression levels in the order of 50% when compared with low doses of siLam (0.6 μM; Fig 4B). This is clear from the ratios of relative expression of lamin A/C and intracellular copy numbers of siRNA, which differ by more than a factor of 10 between 0.6 μM of siLam per cell and 1.8 μM of siRNA. A similar observation has been made in the use of the ICAM-1-directed siRNA si2B (Kretschmer-Kazemi Far & Sczakiel, 2003) in HUVEC cells. In this case, however, we observed a strong intra-individual variability in the extent of PTO-stimulated uptake of siRNA, and in apparent target suppression by siRNA (data not shown). In sum, these experiments indicate a relationship between the biological activity of lamin A/C- and ICAM-1-directed siRNA in HUVEC cells and their increased physical cellular uptake facilitated by PTOs.
We would like to mention that at low levels of uptake of siRNA and, hence, moderate levels of siRNA-mediated target suppression, we find a close relationship between physical uptake and biological activity. At high levels of PTO-stimulated uptake of siRNA, however, target suppression seems to be limited and is less pronounced, as extrapolated from low doses. This indicates that crucial steps that are different from, and subsequent to, the physical uptake seem to be limiting for the suppression of the target, which might include the release from vesicular compartments in which siRNA could be captured.
This work describes a PTO-stimulated delivery of siRNA to human target cells at concentrations that give rise to measurable target suppression. The delivery of naked siRNA by coincubated PTOs does not include carriers, transfectants or other kinds of mediators. It is reasonable to assume that this minimal treatment scenario minimizes undesired side effects in cell culture and, presumably also in vivo where effective doses might be reduced from the order of 50 mg/kg (Soutschek et al, 2004) to the 1 mg/kg range or even below. It is speculative, but possible, that the PTO-stimulated delivery of siRNA in vivo is related to some kind of cell type or tissue specificity.
On a more basal level, this phenomenon indicates that different physical and chemical forms of nucleic acids may positively interfere with the uptake of the same or other classes of nucleic acids, which opens a new possibility for substantially improving the delivery of nucleic acid drugs and tools to target cells and tissues. It is important to consider that PTO-modified ONs do not seem to exist in nature. Thus, they can be regarded as typical drugs or carriers usually applied in pharmacology rather than compounds that are closely related to naturally occurring nucleic acids. This implies that interference with biologically relevant and important mechanisms in vivo is not very likely to occur, with the exception of the well-known and characterized thio-specific side effects of PTOs. However, it seems to be conceivable that there are basically similar naturally occurring pathways that may be successfully explored to improve the system described in this work and to apply other forms of nucleic acids to mammalian cells.
Methods
Oligonucleotides. PTO-modified internucleotide linkages are depicted by ‘s'. The siRNAs used here are described in detail elsewhere (si2B and si-scr, Kretschmer-Kazemi Far & Sczakiel, 2003; siLam, Elbashir et al, 2002). It is mentioned in the text or figure legends whether homologues of the ON TM6-n are chemically modified.
- s2B
5′-GCCUCAGCACGUACCUCUAtt-3′
5′-UAGAGGUACGUGCUGAGGCtt-3′
5′-CGAACUCACUGGUCUGACCtt-3′
5′-GGUCAGACCAGUGAGUUCGtt-3′
5′-CUGGACUUCCAGAAGAACAtt-3′
5′-UGUUCUUCUGGAAGUCCAGtt-3′
[5′-TCGTGT-3′]n
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- as2B
5′-UAGAGGUACGUGCUGAGGCtt-3′
5′-CGAACUCACUGGUCUGACCtt-3′
5′-GGUCAGACCAGUGAGUUCGtt-3′
5′-CUGGACUUCCAGAAGAACAtt-3′
5′-UGUUCUUCUGGAAGUCCAGtt-3′
[5′-TCGTGT-3′]n
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- s-scr
5′-CGAACUCACUGGUCUGACCtt-3′
5′-GGUCAGACCAGUGAGUUCGtt-3′
5′-CUGGACUUCCAGAAGAACAtt-3′
5′-UGUUCUUCUGGAAGUCCAGtt-3′
[5′-TCGTGT-3′]n
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- as-scr
5′-GGUCAGACCAGUGAGUUCGtt-3′
5′-CUGGACUUCCAGAAGAACAtt-3′
5′-UGUUCUUCUGGAAGUCCAGtt-3′
[5′-TCGTGT-3′]n
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- sLam
5′-CUGGACUUCCAGAAGAACAtt-3′
5′-UGUUCUUCUGGAAGUCCAGtt-3′
[5′-TCGTGT-3′]n
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- asLam
5′-UGUUCUUCUGGAAGUCCAGtt-3′
[5′-TCGTGT-3′]n
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- TM6-n
[5′-TCGTGT-3′]n
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- S0K27
5′-CsGsGsGsAsTsCsCsAsTsGsGsCsAsGsCs TsGsGsAsGsAsTsA-3′
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- NovAAAA
5′-AsAsTsCsCsTsCsCsCsCsCsAsGsTsTsCs AsCsCsCssAsAsA-3′
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
- ODN2006
5′-TsCsGsTsCsGsTsTsTsTsGsTsCsGsTsTs TsTsGsTsCsGsTsT-3′
Exposure of cells to short interfering RNA. Cells were seeded in six-well culture plates at a density of 2 × 105 cells/well 16 h before the uptake experiment. Then, cells were washed twice with pre-warmed PBS. To deliver siRNA, cells were incubated with 200 nM siRNA±PTOs in a final volume of 1 ml OptiMEM medium lacking fetal calf serum (FCS).
Detection of short interfering RNA. The detection of siRNA in cellular extracts is described by Overhoff et al (2004). Briefly, after a 16 h incubation, cells were washed five times with 1.5 ml of pre-warmed PBS and trypsin-treated with 250 μl of a trypsin/EDTA (Linaris, Bettingen aM, Germany) solution for 3 min at 37°C. The trypsin treatment was stopped by addition of 750 μl of medium 199 (Sigma-Aldrich, Taufkirchen, Germany) supplemented with 10% FCS. Subsequently, cells were centrifuged for 3 min at 800g. The cell pellet was resuspended in 200 μl PBS containing 1% NP-40 (LKB, Bromma, Sweden) and incubated for 10 min on ice followed by phenol–chloroform extraction. Samples were extracted carefully once with 200 μl phenol (pH 4.5–5.0; Roth, Karlsruhe, Germany) and twice with 200 μl chloroform/i-amylalcohol (24:1, vol/vol) followed by precipitation with 2.5 × volume of ethanol (100%), 0.1 × volume of 3 M sodium acetate (pH 5.2) and 20 μg of glycogen. RNA pellets were re-suspended in 30 μl of hybridization buffer (100 mM NaCl and 20 mM Tris–HCl (pH 7.4)).
Expression of lamin A/C in HUVEC cells. For quantification of siRNA-suppressed expression of the lamin A/C target messenger RNA, we carried out reverse transcription of total RNA prepared from treated cells, followed by quantitative PCR. The synthesis of complementary DNA was primed by a random pool of hexamer primers and Superscript II RNase H− reverse transcriptase according to the manufacturer's specifications (Invitrogen, Karlsruhe, Germany). Quantitative PCR was accomplished with SYBR green PCR core reagents (Eurogentec, Köln, Germany). For the detection of lamin A/C cDNA, the following primers were used: forward primer, 5′-AATGATCGCTTGGCGGTCTA-3′; reverse primer, 5′-GCCCTGCGTTCTCCGTTT-3′. To standardize the samples, a quantitative PCR for glyceraldehyde 3-phosphate dehydrogenase was performed (forward primer, 5′-AACAGCGACACCCACTCCTC-3′; reverse primer, 5′-GGAGGGGAGATTCAGTGTGGT-3′. The expression level of lamin A/C in cells cultured in OptiMEM medium was set to 100%.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank W. Wünsche for excellent technical assistance, F. Warwsinek for initial experiments and F. Eckstein, A. Gewirtz and C. Stein for stimulating and helpful suggestions.
References
- Allen TM, Cullis PR (2004) Drug delivery systems: entering the mainstream. Science 303: 1818–1822 [DOI] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaille J, Hüttenhofer A (2002) The expanding snoRNA world. Biochimie 84: 775–790 [DOI] [PubMed] [Google Scholar]
- Beltinger C et al. (1995) Binding uptake, and intracellular trafficking of phosphorothioate-modified oligodeoxynucleotides. J Clin Invest 95: 1814–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Diesbach P et al. (2000) Identification, purification and partial characterisation of an oligonucleotide receptor in membranes of HepG2 cells. Nucleic Acids Res 28: 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Tuschl T (2004) siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov 3: 318–329 [DOI] [PubMed] [Google Scholar]
- Elbashir SM et al. (2001a) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T (2001b) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199–213 [DOI] [PubMed] [Google Scholar]
- Fra AM, Williamson E, Simons K, Parton RG (1995) De novo formation of caveolae in lymphocytes by expression of VIP21-caveolin. Proc Natl Acad Sci USA 92: 8655–8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ (2004) Unlocking the potential of the human genome with RNA interference. Nature 431: 371–378 [DOI] [PubMed] [Google Scholar]
- Hartmann G, Krieg AM (2000) Mechanism and function of a newly identified CpG DNA motif in human primary B cells. J Immunol 164: 944–952 [DOI] [PubMed] [Google Scholar]
- Kalota A, Shetzline SE, Gewirtz AM (2004) Progress in the development of nucleic acid therapeutics for cancer. Cancer Biol Ther 3: 4–12 [DOI] [PubMed] [Google Scholar]
- Kretschmer-Kazemi Far R, Sczakiel G (2003) The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res 31: 4417–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AM, Davis HL (2001) Enhancing vaccines with immune stimulatory CpG DNA. Curr Opin Mol Ther 3: 15–24 [PubMed] [Google Scholar]
- Laktionov PP et al. (1999) Characterisation of membrane oligonucleotide-binding proteins and oligonucleotide uptake in keratinocytes. Nucleic Acids Res 27: 2315–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laktionov PP et al. (2004) Cell-surface-bound nucleic acids: free and cell-surface-bound nucleic acids in blood of healthy donors and breast cancer patients. Ann NY Acad Sci 1022: 221–227 [DOI] [PubMed] [Google Scholar]
- Lehmann M, Sczakiel G (2005) Spontaneous uptake of biologically active recombinant DNA by mammalian cells via a selected double-stranded DNA segment. Gene Ther 12: 446–451 [DOI] [PubMed] [Google Scholar]
- Lingor P, Michel U, Schöll U, Bähr M, Kügler S (2004) Transfection of ‘naked' siRNA results in endosomal uptake and metabolic impairment in cultured neurons. Biochem Biophys Res Comm 315: 1126–1133 [DOI] [PubMed] [Google Scholar]
- Menezes J, Leibold W, Klein G, Clements G (1975) Establishment and characterization of an Epstein–Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-negative African Burkitt's lymphoma. Biomedicine 22: 276–284 [PubMed] [Google Scholar]
- Overhoff M, Wünsche W, Sczakiel G (2004) Quantitative detection of siRNA and single-stranded oligonucleotides: relationship between uptake and biological activity of siRNA. Nucleic Acids Res 32: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Richards AA (2003) Lipid rafts and caveolae as portals for endocytosis: new insight and common mechanisms. Traffic 4: 724–738 [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Burli T, Zerial M, Helenius A (2004) Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell 118: 767–780 [DOI] [PubMed] [Google Scholar]
- Scanlon KJ (ed). (1998) Therapeutic Applications of Ribozymes. Totowa, NJ, USA: Human Press Inc. [Google Scholar]
- Schneider U, Schwenk HU, Bornkamm G (1977) Characterization of EBV-genome negative ‘null' and ‘T' cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer 22: 204–213 [DOI] [PubMed] [Google Scholar]
- Soutschek J et al. (2004) Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432: 173–178 [DOI] [PubMed] [Google Scholar]
- Toulmé JJ, Di Primo C, Boucard D (2004) Regulating eukaryotic gene expression with aptamers. FEBS Lett 567: 55–62 [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Kaul SC, Miyagishi M, Taira K (2004) Vectors for RNA interference. Curr Opin Mol Ther 6: 367–372 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


